Abstract
As portable chest X-rays are an efficient means of triaging emergent cases, their use has raised the question as to whether imaging carries additional prognostic utility for survival among patients with COVID-19. This study assessed the importance of known risk factors on in-hospital mortality and investigated the predictive utility of radiomic texture features using various machine learning approaches. We detected incremental improvements in survival prognostication utilizing texture features derived from emergent chest X-rays, particularly among older patients or those with a higher comorbidity burden. Important features included age, oxygen saturation, blood pressure, and certain comorbid conditions, as well as image features related to the intensity and variability of pixel distribution. Thus, widely available chest X-rays, in conjunction with clinical information, may be predictive of survival outcomes of patients with COVID-19, especially older, sicker patients, and can aid in disease management by providing additional information.
Similar content being viewed by others
Introduction
COVID-19 has resulted in more than eighty-five million cases and over one million deaths in the United States1. With ongoing concerns of future resurgences2,3, and in an effort to improve the treatment and management of infected patients, principled methods for risk stratification and survival prognostication are critically important4,5. Early reports outlined diagnostic guidance for assessing chest X-ray abnormalities in emergency department settings, including patchy or diffuse reticulonodular ‘ground-glass’ opacities (GGOs) and consolidation with basal, peripheral, and bilateral predominance6,7. Recent studies advocated for the use of chest X-rays in grading patients with COVID-198,9 via scoring systems such as the COVID-specific Brixia score, which rates lung involvement on a scale from 0 to 18, or percentage of lung involvement10,11,12. While automated approaches for disease classification have attained a high (> 90%) diagnostic accuracy13, there is a dearth of research using radiomic features to predict clinical outcomes for patients admitted with COVID-19 due to their high-dimensional and heterogeneous nature, as well as data unavailability14. The added utility of such features for predicting in-hospital mortality, beyond clinical risk factors, is largely unknown14.
The University of Michigan Health System (or Michigan Medicine), as one of the primary regional centers managing the care of patients with COVID-19 during the pandemic, has collected a wealth of X-ray image data, in addition to demographic and clinical data, via the Electronic Health Record (EHR)15,16. Portable chest X-ray, with its availability and ease of use, has been routinely used for monitoring patients in need of urgent care at Michigan Medicine, even prior to the pandemic17. However, analysis of chest X-ray images is complicated by the data structure, particularly in the COVID-19 setting18. Leveraging machine learning techniques, we proposed a workflow for the extraction and selection of features from COVID-related X-ray images. By using survival information directly, our framework decomposes raw images into texture features and identifies those features that are most related to COVID-19 mortality. We used several machine learning techniques to assess the predictability of demographic and clinical factors and the radiomic texture features on in-hospital mortality, a primary endpoint for patients hospitalized with COVID-1919. Subgroup analyses revealed that chest X-ray images offered more prognostic utility for vulnerable (e.g., older or sicker) patients.
Results
Patient outcomes and characteristics
Of the 3313 hospitalized patients with X-rays, we analyzed a total of 3310 patients with anterior–posterior or posterior-anterior images, which provided clear views of the lungs; excluded were only three patients whose X-ray provided unclear views and could not be analyzed. During follow-up, we observed 590 (17.8%) in-hospital deaths and 20 (0.6%) discharges to hospice. Median age was 61 (interquartile range: 46–73) years, and the majority of patients were male (56%), with an over-representation of Black patients (21%) as compared to the surrounding population. Median respiratory rate was 18.8 (17.5–21.7) breaths per minute and median oxygen saturation was 95.5% (94.0–97.2%). There was a high proportion of patients with cardiac arrhythmias (70%), hypertension (70%), and fluid and electrolyte disorders (70%) at admission (Supplement E). Seven radiomic features and seven clinical features were included in the final model.
Prediction performance
We first compared the predictive performance of the following five algorithms using the clinical predictors only. The algorithms were the Cox proportional hazards model20,21, survival support vector machines22, random survival forests23, survival gradient boosting24, and ensemble averaging of the first four algorithms25. The average C-index across one hundred experiments ranged from 78.1 to 80.3%, with ensemble averaging performing the best. We then compared the algorithms using both the clinical and radiomic features and noted that ensemble averaging still outperformed the other methods, again achieving the highest average C-index of 81.0%. Moreover, incremental improvements were observed across all five algorithms, ranging from a 0.5% increase in C-index (random survival forests) to a 2.0% increase (survival gradient boosting; Table 1). This motivated us to conduct subgroup analyses to examine which subgroups would benefit more with the added image features; see the later section of “Subgroup analysis and risk stratification”.
Feature importance
Figure 1a gives the feature importance for the top clinical and imaging features under the five predictive approaches. The most important clinical features were age, indications of fluid and electrolyte disorders, respiratory rate, diastolic blood pressure, metastatic cancer, and solid tumor cancer without metastasis. Important imaging texture features included dependence non-uniformity, zone entropy, median pixel intensity, large area high gray level emphasis, maximal correlation coefficient, pixel intensity kurtosis, and robust mean absolute deviation. Patients with higher dependence non-uniformity, zone entropy, and maximal correlation coefficients had more heterogeneity or complexity in the texture patterns for their images. Those with higher median pixel intensity and large area high gray level emphasis had greater concentrations of high gray level values in their images, and those with higher pixel intensity kurtosis and robust mean absolute deviations had more outlying values in their pixel intensities26.
Results from predictive analysis of in-hospital mortality. (a) Average feature importance of clinical and imaging features based on one hundred testing datasets with standard errors, sorted by highest feature importance in ensemble averaging. (b) Kaplan–Meier curves for in-hospital mortality, stratified by patient age and risk group (defined by the median risk score; high risk = solid, low risk = dashed); risk scores defined either by clinical or clinical plus imaging features within each age group. (c) Kaplan–Meier curves for in-hospital mortality, stratified by comorbidity burden and risk group (defined by the median risk score; high risk = solid, low risk = dashed); risk scores defined either by clinical or clinical plus imaging features within each comorbidity burden group.
Adjusted associations between clinical/imaging features and survival
We fit a Cox regression model with the important features, presenting the hazard ratios (HR) and 95% confidence intervals (CI) in Table 2. Older age (HR: 2.33; 95% CI 2.07–2.63), higher respiratory rate (1.41; 1.28–1.55), and indications of fluid and electrolyte disorders (2.57; 1.98–3.34), metastatic cancer (1.41; 1.10–1.80), and solid tumor cancer without metastasis (1.32; 1.03–1.68) were significantly associated with higher in-hospital mortality. Conversely, higher diastolic blood pressure (0.81; 0.75–0.88), never smoking (0.46; 0.32–0.67) and former smoking (0.62; 0.43–0.90) were associated with lower mortality. Among the radiomic texture features, greater dependence non-uniformity (1.21; 1.08–1.36), large area high gray level emphasis (1.14; 1.04–1.25), and median pixel intensity (1.14; 1.05–1.25) were significantly associated with higher hazards for mortality, while lower maximal correlation coefficients (0.91; 0.83–0.99) were marginally associated with higher mortality hazards.
Subgroup analysis and risk stratification
We used ensemble averaging, which was the most predictive, to construct risk scores with and without the addition of the radiomic features. We compared how these scores could distinguish patients within certain subgroups, defined by age or comorbidity burden (Fig. 1b,c). Two findings are worth noting. First, the scores, based on clinical indicators only or both clinical and image features, could well distinguish patients across all the subgroups, highlighting the usefulness of clinical and image features in profiling the risk of patient mortality.
Second, patients were then classified as ‘high’ versus ‘low’ risk based on median risk scores defined by using both clinical and clinical + radiomic features. Within certain subgroups (e.g., patients older than 65 years or those with seven or more comorbid conditions), the separation between the survival curves of the high- and low-risk patients defined with the addition of the imaging features was more obvious than that between those of the high- and low-risk patients, defined using clinical features alone. This exemplifies the added prognostic utility of radiomic features in these subgroups. In contrast, the separation was not as apparent in the other subgroups, e.g., among those younger than 65 years or with fewer than seven comorbidity conditions.
To confirm our findings, we compared the increase in C-index with the addition of the radiomic features between these different subgroups. Table 3 shows a significantly higher increase in C-index among older patients than younger patients with the addition of radiomic features. There was a 2.3–3.1% increase in C-index among older patients across the different algorithms with the addition of the radiomic features. This increment is clinically meaningful27,28 and significantly larger (p < 0.001) than the 0.5–1.0% increase among younger patients. Similarly, a 1.6–2.5% increase in C-index was achieved among patients with a higher comorbidity burden, as compared to a 0.2–1.4% increase among patients with a lower comorbidity burden. This increment was clinically meaningful and statistically significant (p < 0.01).
Discussion
Many recent studies have discussed the potential of integrative models for discovery and prognostication in a wide range of clinical settings, including breast29 and lung30 cancers, coronary artery disease31, and pulmonary embolisms32. Each of these studies have demonstrated that multimodal prediction methods, which combine radiomic and clinical features, allow for improved predictive performance in a range of clinical settings. Further, recent works have suggested radiomic phenotypes from medical imaging are linked with molecular phenotypes, such as genomics and histopathology, and therefore may provide important and clinically relevant information33,34. In our setting, radiologic imaging plays an important role in grading and managing patients with COVID-19, as portable chest X-rays are an efficient and convenient means of triaging emergent cases. This work addresses the question as to whether imaging carries any additional prognostic utility in the management of patients with COVID-19. We observed a slight increase in prediction performance with the added X-ray features, which motivated us to further study which patient subgroups would benefit more from the additional image features.
Across all patients, we saw modest improvements in the predictive accuracy of the methods under comparison, but we found that older patients and those with higher comorbidity burden at admission saw significantly larger gains in C-index with the added radiomic features. Though the magnitudes of these improvements are relatively small, we recognize the potential clinical impact. Even a slight increase in prediction accuracy could benefit patient outcomes by helping clinicians identify high-risk patients and initiate timely interventions. Not every patient’s physiological derangements correlate closely with their images35,36; for instance, younger patients with bilateral peripheral pulmonary infiltrates can be oxygenating well and appear fine clinically, whereas older or sicker patients may be less able to compensate for the same degree of imaging abnormalities. Hence, while the metrics identified in our study may not be useful in every case, they may help pinpoint which patient subpopulations can more reliably benefit from imaging as a predictor of mortality.
There is a growing body of literature to support the use of imaging data for in-hospital mortality prognostication. Kim et al. found that X-ray grade was significantly associated with both length of stay in hospital and higher odds of intubation7. Garrafa et al. predicted in-hospital mortality using the COVID-specific Brixia score37, and the predictiveness for their testing data ranged from 0.52 (logistic regression) to 0.78 (random forests and gradient boosting), which was close to our results. Schalekamp et al. graded chest X-rays on a severity scale from zero to eight points38 and developed an image-based risk score to predict critical illness in patients with COVID-19. Soda et al. modeled patient survival with clinical and imaging features in an Italian cohort39 and obtained an accuracy of 0.68–0.76 across different methods with only clinical information and increasing to a range of 0.75–0.77 with both clinical and imaging features, an increment similar to our report. They found that age, oxygen saturation, respiratory rates, and active cancer were of the most importance, which was consistent with our findings.
Lung involvement and COVID-19 severity, assessed by visual examination of the raw X-ray images, were reported to be predictive of mortality10,11,12,40. However, visual approaches may be prone to subjectivity and inaccuracy. Recent works have shown that texture features associated with image heterogeneity are predictive of clinical outcomes when visual assessment of imaging data may not be correlated with these same endpoints41. In contrast, our method provides an objective means of extracting image features for aiding in survival prognostication. Our work also addresses the challenge of analyzing variable-size images, which cannot be processed by deep learning algorithms like AlexNet42 or ResNet43. Rather than directly feeding images into the models, we derived relevant texture features with maximal image differentiation for predicting COVID-19 survival based on a standard workflow13,14,44,45,46,47. These texture features may also be more interpretable than those derived from deep learning models48.
Further, our method enabled us to leverage patient survival information when selecting the image features, leading to some interesting discoveries. We found that median pixel intensity and large dependence high gray level emphasis, features corresponding to greater concentrations of high gray level values in the images, were important predictors of patient survival. Greater heterogeneity in the texture features, characterized by zone entropy and dependence non-uniformity was also predictive. These findings align with the current literature. For example, similar to our results, Varghese et al. showed the importance of certain first and second order texture features, namely, histogram and intensity, followed by the gray level size zone matrix and grey level co-occurrence matrix, for predicting intensive care unit utilization, intubation, and death49. Iori et al. identified important texture features, including entropy and dependence non-uniformity, for mortality prediction50.
We detected that predictions on certain subgroups of patients benefited more from the addition of these radiomic features. In particular, greater improvement in survival prediction was observed for older (> 65 years) patients and those with higher (> median 7/29 comorbidities) comorbidity burden. Our results agree with previous findings that the severity of disease in the images is associated with comorbidity burden and age51,52,53, hinting that radiomic features coming from older or sicker patients are likely to contain more information relevant to survival. In contrast, younger or healthier patients are at a lower risk of death, so the additional radiomic features do not add much to their prognostication54.
We note some limitations and areas of future work for the current study. First, only hospitalizations at Michigan Medicine were included in the analysis, potentially limiting the generalizability of the results. However, our workflow provides a general and useful framework for analyzing EHR data with chest X-ray images, and our results may generate hypotheses for larger-scale investigations, and potentially in other disease areas as well. As some improvement was observed among older patients and patients with a higher comorbidity burden, external validation is necessary to confirm these results and their clinical importance. Further investigations are also needed to assess the optimality of our feature extraction and screening techniques and the predictive accuracy of our approach as compared to standard clinical practice. We selected clinical predictors which were known to be predictive of worsened COVID-19 outcomes based on clinical practice and the current body of literature. However, risk factors which are viewed as important may differ across institutions with varying and complex patient populations. Further, to assess whether our proposed computer-based multi-modal approach can augment the practice of medicine, a next step would be to design a study in which predictions generated from our model are compared to those generated from clinicians based on a common a set of imaging and clinical features. This focus group approach would be an important next step in the continuation and validation of the current work. Lastly, comparisons to other, automated approaches such as deep learning may yield additional benchmarks for the accuracy of the proposed method.
Second, as with most EHR studies, there might be an inherent selection bias among those presenting to Michigan Medicine and subsequently admitted for COVID-19 related complications. Causal inference approaches may be explored to address observable and unobservable confounders. Third, comorbidities taken at admission were not differentiated from chronic conditions preceding infection. More in-depth work is needed to account for chronologies of these conditions. Moreover, comorbid conditions were established via IDC-10 codes based on Quan et al.55. At our institution, diagnoses with associated ICD-10 codes are typically first input into the EHR on admission and later confirmed retrospectively. However, there may be administrative delays in the coding of these conditions after a patient's initial encounter, both in our study and broadly at other institutions. This may require adequate alternatives for comorbidity identification, particularly for those conditions included in our final model. For example, the patients in our study population who were broadly indicated for fluid and electrolyte disorders had imbalances that span the range of sodium and potassium, acidosis, alkalosis, and volume depletion which coincide with both each other and worsened COVID-19 outcomes. An alternative strategy for identifying such conditions would be in flagging patients based on abnormal blood and urine laboratory values, e.g., increased urine sodium and/or osmolality, which may be more readily available. As the current method stands, however, this is a potential limitation for the immediate useability of the method in other practical clinical settings.
Lastly, mortality is often a key endpoint for identifying patients who are at high risk of adverse events or who may need closer monitoring and more aggressive interventions. However, as COVID-19 has a wide range of presentation, bearing many clinical abnormalities, mortality should not be used in isolation, and additional patient centered outcomes, such as the patient-specific quality of life and care, should be considered when making clinical decisions56,57.
Conclusions
In summary, portable chest X-ray is a valuable tool for monitoring and guiding the care of patients with COVID-19. This study found that patterns of COVID-19 lung disease identified on chest X-ray are predictive of, and significantly associated with, the survival outcomes of patients hospitalized with COVID-19. Multimodal prediction models may provide modest improvements in prognostic value over clinical risk factor alone, and further research into understanding the clinical and biological underpinnings of these improvement are necessary to provide additional information when guiding the management of patients with COVID-19.
Methods
Experimental design
This was a prognostic analysis of patients who (1) were admitted to Michigan Medicine between March 10, 2020 (the date of the first case in this state) and March 31, 2022 (the cutoff date of the released EHR data), (2) tested positive for COVID-19 or transferred in carrying a positive diagnosis, and (3) had at least one COVID-related chest X-ray image taken. We focused on patients with X-rays because patients without imaging were in general much younger and healthier, and images are valuable in triaging patients and managing resources58. Our outcome was the time from admission until in-hospital death, censored by discharge or the end of the study. Discharge was regarded as a censoring event, except for discharge to hospice, because the median survival for these patients was less than 30 days post-discharge. As it was a strong precursor to death, we considered both in-hospital death and discharge to hospice as failure events (see Supplement A).
From the EHR database, we extracted and created a set of demographic, socioeconomic, and clinical risk factors (see Supplement B) identified as being related to COVID-19 in the literature59,60,61,62,63,64,65,66,67,68,69,70,71,72. Patient demographics included age, sex, race (Black or non-Black), ethnicity (Hispanic or non-Hispanic), smoking status, alcohol use, and drug use. As patient-level socioeconomic factors were unavailable, we created four composite socioeconomic measures at the US census tract-level based on patient residences. These composites, measuring affluence, disadvantage, ethnic immigrant concentration, and education, were defined to be the proportion of adults meeting the corresponding criterion within a census tract73,74,75, and were further categorized by quartiles. For each of twenty-nine prevalent comorbidity conditions commonly used in literature55,76,77,78, we defined a binary indicator to flag whether the patient had any associated ICD-10 code at admission. Lastly, we obtained physiologic measurements within 24 h of admission, including body mass index (kg/m2), oxygen saturation, body temperature, respiratory rate, diastolic and systolic blood pressure, and heart rate.
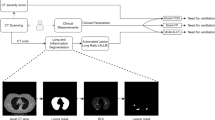
With multiple X-rays potentially taken for one patient, we chose the one closest to the time of admission and examined its role in predicting patient survival. We first pre-processed each image according to the pipeline depicted in Fig. 2. First, prior to feature extraction and selection, we retained only those images taken from the anterior–posterior or posterior-anterior positions so that the orientation of the images would be comparable. We then normalized these images so that the pixel intensities of each image conformed to a standard range of 0 (black) to 255 (white) units. We further used histogram equalization to enhance the contrast of the images79.
Broadly, there are two potential approaches for feature extraction, namely (1) artificial intelligence methods, which learn feature representations automatically from the data, and (2) engineered texture features. While deep learning has been shown to have high prognostic accuracy, learned features are difficult to interpret, not standardized, and often not reproducible, which may impact their reliability80. Thus, we extracted a standard panel of engineered texture features according to the PyRadiomics workflow47,81. Specifically, we applied six different filters (e.g., different transformations) to the pre-processed images to acquire additional information (e.g., at edges or boundaries) and derive different image types (e.g., shape)47. From the seven image filters (original + six transformations), we extracted seven classes of features from each image47,82,83,84, resulting in 1311 candidate image features. To obtain a short list of predictive clinical and image features, we performed feature screening by fitting Cox proportional hazards models21 on each feature one at a time and retaining those significant at the 0.05 level. Finally, we selected the features with the highest feature importance, and obtained a final Cox model, quantifying the adjusted associations of important clinical and radiomic features with in-hospital mortality85. We used the concordance index (C-index) to assess the predictiveness of models27,28 (see Supplement C). This study was approved by the Michigan Medicine Institutional Review Board (HUM00192931), which waived informed consent based on secondary analysis of deidentified datasets. All analysis was conducted in accordance with relevant guidelines and regulations.
Statistical analysis
We implemented five risk prediction algorithms, namely, the Cox proportional hazards model21, survival support vector machines22, random survival forests23,86, survival gradient boosting24, and ensemble averaging of the first four algorithms25. The Cox model, the most widely used method in survival analysis, assumes a risk function that is linear in the predictors. Survival support vector machines22 can account for non-linear relationships. Both random survival forests and survival gradient boosting combine multiple predictions from individual survival trees to achieve a more powerful prediction23,24,86,87. Ensemble averaging combines predictions from multiple models to produce a desired output and often performs better than individual models by averaging out their errors25. Supplement D details these methods.
We used cross-validation to unbiasedly estimate the predictiveness of each method. We randomly split the data into 80% training and 20% testing samples, maintaining the proportion of events in the full sample within each split. We then trained the various predictive models by using the training samples and computed the C-index by using the testing samples. We repeated the same procedure one hundred times and took an average of the C-index to obtain an unbiased estimate of the C-index for each method88,89. We applied each method with the demographic and clinical predictors, followed by the addition of radiomic features to assess their incremental prognostic utility via the C-index. Using ensemble averaging, which was the most predictive (see the section of “Results”), we developed a risk score to predict in-hospital mortality and classified patients into low- and high-risk groups using the median score as the cutoff.
Lastly, we detail the variable selection process for building a final Cox model. We selected clinical and image features based on their importance in prediction, defined by the absolute decrease in C-index with the “removal” of the concerned feature in the data90. To do so, we randomly split the data into 80% training and 20% testing samples, fit the model on the training data and calculated the feature importance using the testing data (Supplement D.6). We repeated the same procedure one hundred times, selected the features that were most important (on average) among these one hundred experiments, and included them in a multivariable Cox regression to assess their statistical associations with in-hospital mortality. All data processing and analysis was carried out with Python (version 3.8.8), NumPy (version 1.20.1), and scikit-survival (version 0.17.2).
We examined different subgroups to gauge how the prediction performance of the model improved with the added radiomic features. Because age and comorbidity burden were the most relevant to survival among the clinical factors, we considered patient subgroups defined by age (> versus ≤ 65 years old) and number of comorbidities at admission (> versus ≤ median seven comorbidities), respectively. We compared the change in prediction performance with the addition of the radiomic features between different subgroups.
Ethics approval
This study was approved by the Michigan Medicine Institutional Review Board (HUM00192931), which waived informed consent based on secondary analysis of deidentified datasets. All analysis was conducted in accordance with relevant guidelines and regulations.
Data availability
The datasets used in this study are not publicly available due to the need for institutional review board approval as a University of Michigan-affiliated researcher through the University of Michigan Health System (i.e., Michigan Medicine) Precision Health Initiative. For more information, please contact PHDataHelp@umich.edu.
Code availability
All code used to produce the results found in this work have been made publicly available at https://github.com/YumingSun/COVID_Imaging_Prediction.
References
Centers for Disease Control and Prevention. CDC covid data tracker. https://covid.cdc.gov/covid-data-tracker/ (Accessed 13 June 2022) (2022).
Karim, S. S. A. & Karim, Q. A. Omicron sars-cov-2 variant: A new chapter in the covid-19 pandemic. Lancet 398(10317), 2126–2128 (2021).
Kupferschmidt, K. & Wadman, M. Delta variant triggers new phase in the pandemic. Science 372(6549), 1375–1376 (2021).
McCue, C. et al. Long term outcomes of critically ill covid-19 pneumonia patients: Early learning. Intensive Care Med. 47(2), 240–241 (2021).
Michelen, M. et al. Characterising long term covid-19: A living systematic review. BMJ Glob. Health 6(9), e005427 (2021).
Jacobi, A. et al. Portable chest x-ray in coronavirus disease-19 (covid-19): A pictorial review. Clin. Imaging 64, 35–42 (2020).
Kim, H. W. et al. The role of initial chest x-ray in triaging patients with suspected covid-19 during the pandemic. Emerg. Radiol. 27(6), 617–621 (2020).
Akl, E. A. et al. Use of chest imaging in the diagnosis and management of covid-19: A who rapid advice guide. Radiology 298(2), E63–E69 (2021).
Borkowski, A. A. et al. Using artificial intelligence for covid-19 chest x-ray diagnosis. Fed. Pract. 37(9), 398–404 (2020).
Balbi, M. et al. Chest x-ray for predicting mortality and the need for ventilatory support in covid-19 patients presenting to the emergency department. Eur. Radiol. 31(4), 1999–2012 (2021).
Maroldi, R. et al. Which role for chest x-ray score in predicting the outcome in covid-19 pneumonia?. Eur. Radiol. 31(6), 4016–4022 (2021).
Monaco, C. G. et al. Chest x-ray severity score in covid-19 patients on emergency department admission: A two-centre study. Eur. Radiol. Exp. 4(1), 68 (2020).
Hussain, L. et al. Machine-learning classification of texture features of portable chest x-ray accurately classifies covid-19 lung infection. Biomed. Eng. Online 19(1), 88 (2020).
Ismael, A. M. & Şengür, A. Deep learning approaches for covid-19 detection based on chest x-ray images. Expert Syst. Appl. 164(114), 054 (2021).
Salvatore, M. et al. A phenome-wide association study (phewas) of covid-19 outcomes by race using the electronic health records data in michigan medicine. J. Clin. Med. 10(7), 1351 (2021).
Spector-Bagdady, K. et al. Coronavirus disease 2019 (covid-19) clinical trial oversight at a major academic medical center: Approach of michigan medicine. Clin. Infect. Dis. 71(16), 2187–2190 (2020).
Nypaver, M. et al. The michigan emergency department improvement collaborative: A novel model for implementing large scale practice change in pediatric emergency care. Pediatrics 142(1 MeetingAbstract), 105 (2018).
Abbas, A., Abdelsamea, M. M. & Gaber, M. M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl. Intell. 51, 854–864 (2021).
Gupta, A. et al. Association between antecedent statin use and decreased mortality in hospitalized patients with COVID-19. Nat. Commun. 12(1), 1325 (2021).
Cox, D. R. Regression models and life tables (with discussion). J. R. Stat. Soc. B 34(2), 187–220 (1972).
Therneau, T. M. & Grambsch, P. M. Modeling survival data: Extending the Cox model. In The Cox Model 39–77 (Springer, 2000).
Pölsterl, S., Navab, N. & Katouzian, A. An efficient training algorithm for kernel survival support vector machines. https://doi.org/10.48550/arXiv.1611.07054 (Preprint posted online November 21, 2016).
Ishwaran, H. et al. Random survival forests. Ann. Appl. Stat. 2(3), 841–860 (2008).
Hothorn, T. et al. Survival ensembles. Biostatistics 7(3), 355–373 (2006).
Zhou, Z. H. Ensemble Methods: Foundations and Algorithms (CRC Press, 2012).
Zwanenburg, A. et al. Image biomarker standardisation initiative. https://doi.org/10.48550/arXiv.1612.07003 (Preprint posted online December 21, 2016)
Harrell, F. E. et al. Evaluating the yield of medical tests. JAMA 247(18), 2543–2546 (1982).
Harrell, F. E. Jr., Lee, K. L. & Mark, D. B. Multivariable prognostic models: Issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat. Med. 15(4), 361–387 (1996).
Holste, G. et al. End-to-end learning of fused image and non-image features for improved breast cancer classification from mri. In Proceedings of the IEEE/CVF International Conference on Computer Vision, 3294–3303 (2021).
Zhou, H. et al. Diagnosis of distant metastasis of lung cancer: Based on clinical and radiomic features. Transl. Oncol. 11(1), 31–36 (2018).
Militello, C. et al. CT Radiomic Features and Clinical Biomarkers for Predicting Coronary Artery Disease. Cogn. Comput. 15(1), 238–253 (2023).
Huang, S. C. et al. Multimodal fusion with deep neural networks for leveraging CT imaging and electronic health record: A case-study in pulmonary embolism detection. Sci. Rep. 10(1), 1–9 (2020).
Liu, Z. et al. Imaging genomics for accurate diagnosis and treatment of tumors: A cutting edge overview. Biomed. Pharmacother. 135, 111173 (2021).
Tomaszewski, M. R. & Gillies, R. J. The biological meaning of radiomic features. Radiology 298(3), 505–516 (2021).
Brouqui, P. et al. Asymptomatic hypoxia in COVID-19 is associated with poor outcome. Int. J. Infect. Dis. 102, 233–238 (2021).
Struyf, T. et al. Cochrane COVID-19 Diagnostic Test Accuracy Group. Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID‐19. Cochrane Database Syst. Rev. (5) (2022).
Garrafa, E. et al. Early prediction of in-hospital death of covid-19 patients: A machine-learning model based on age, blood analyses, and chest x-ray score. Elife 10, e70640 (2021).
Schalekamp, S. et al. Model-based prediction of critical illness in hospitalized patients with covid-19. Radiology 298(1), E46–E54 (2021).
Soda, P. et al. Aiforcovid: Predicting the clinical outcomes in patients with covid-19 applying ai to chest-x-rays. An Italian multicentre study. Med. Image Anal. 74, 102216 (2021).
Shen, B. et al. Initial chest radiograph scores inform covid-19 status, intensive care unit admission and need for mechanical ventilation. Clin. Radiol. 76(6), 473.e1-473.e7 (2021).
Liu, Y. et al. Tumor heterogeneity assessed by texture analysis on contrast-enhanced CT in lung adenocarcinoma: Association with pathologic grade. Oncotarget 8(32), 53664–53674 (2017).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. Imagenet classification with deep convolutional neural networks. Adv. Neural. Inf. Process. Syst. 25, 1–9 (2012).
He, K. et al. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 770–778 (2016).
Chandra, T. B. et al. Coronavirus disease (covid19) detection in chest x-ray images using majority voting based classifier ensemble. Expert Syst. Appl. 165(113), 909 (2021).
Johri, S. et al. A novel machine learning-based analytical framework for automatic detection of covid-19 using chest x-ray images. Int. J. Imaging Syst. Technol. 31(3), 1105–1119 (2021).
Selvi, J. T., Subhashini, K. & Methini, M. Investigation of covid-19 chest x-ray images using texture features—A comprehensive approach. Computational 1, 45–58 (2021).
van Griethuysen, J. J. M. et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77(21), e104–e107 (2017).
Zhang, Q., Wu, Y. N. & Zhu, S. C. Interpretable convolutional neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 8827–8836 (2018).
Varghese, B. A. et al. Predicting clinical outcomes in covid-19 using radiomics on chest radiographs. Br. J. Radiol. 94(1126), 20210221 (2021).
Iori, M. et al. Mortality prediction of COVID-19 patients using radiomic and neural network features extracted from a wide chest X-ray sample size: A robust approach for different medical imbalanced scenarios. Appl. Sci. 12(8), 3903 (2022).
Blain, M. et al. Determination of disease severity in covid-19 patients using deep learning in chest x-ray images. Diagn. Interv. Radiol. 27(1), 20–27 (2021).
Liu, X. et al. Temporal radiographic changes in covid-19 patients: Relationship to disease severity and viral clearance. Sci. Rep. 10(1), 10263 (2020).
Yasin, R. & Gouda, W. Chest x-ray findings monitoring covid-19 disease course and severity. Egypt. J. Radiol. Nucl. Med. 51(1), 193 (2020).
Castelli, G. et al. Brief communication: Chest radiography score in young covid-19 patients: Does one size fit all?. PLoS ONE 17(2), e0264172 (2022).
Quan, H. et al. Coding algorithms for defining comorbidities in icd-9-cm and icd-10 administrative data. Med. Care 43(11), 1130–1139 (2005).
Cau, R. et al. Long-COVID diagnosis: From diagnostic to advanced AI-driven models. Eur. J. Radiol. 148, 110164 (2022).
Sudre, C. H. et al. Attributes and predictors of long COVID. Nat. Med. 27(4), 626–631 (2021).
Jiao, Z. et al. Prognostication of patients with covid-19 using artificial intelligence based on chest x-rays and clinical data: A retrospective study. Lancet Digit. Health 3(5), e286–e294 (2021).
Alqahtani, J. S. et al. Prevalence, severity and mortality associated with copd and smoking in patients with covid-19: A rapid systematic review and meta-analysis. PLoS ONE 15(5), e0233147 (2020).
Centers for Disease Control and Prevention. Scientific evidence for conditions associated with higher risk for severe covid-19. https://www.cdc.gov/coronavirus/2019-ncov/science/sciencebriefs/underlying-evidence-table.html (Accessed 13 June 2022) (2022).
Ebinger, J. E. et al. Pre-existing traits associated with covid-19 illness severity. PLoS ONE 15(7), e0236240 (2020).
Jordan, R. E., Adab, P. & Cheng, K. K. Covid-19: Risk factors for severe disease and death. BMJ 368, m1198 (2020).
Khan, M. M. A. et al. Effects of underlying morbidities on the occurrence of deaths in covid-19 patients: A systematic review and meta-analysis. J. Glob. Health 10(2), 020503 (2020).
Kim, L. et al. Risk factors for intensive care unit admission and in-hospital mortality among hospitalized adults identified through the us coronavirus disease 2019 (covid-19)-associated hospitalization surveillance network (covid-net). Clin. Infect. Dis. 72(9), e206–e214 (2021).
Mikami, T. et al. Risk factors for mortality in patients with covid-19 in new york city. J. Gen. Intern. Med. 36(1), 17–26 (2021).
Rod, J. E., Oviedo-Trespalacios, O. & Cortes-Ramirez, J. A brief-review of the risk factors for COVID-19 severity. Rev. Saude Publica 54, 60 (2020).
Salerno, S. et al. Comprehensive evaluation of covid-19 patient short-and long-term outcomes: Disparities in healthcare utilization and post-hospitalization outcomes. PLoS ONE 16(10), e0258278 (2021).
Ssentongo, P. et al. Association of cardiovascular disease and 10 other pre-existing comorbidities with covid-19 mortality: A systematic review and meta-analysis. PLoS ONE 15(8), e0238215 (2020).
Wang, B. et al. Does comorbidity increase the risk of patients with covid-19: Evidence from meta-analysis. Aging 12(7), 6049–6057 (2020).
Williamson, E. J. et al. Factors associated with covid-19-related death using opensafely. Nature 584(7821), 430–436 (2020).
Wu, Z. & McGoogan, J. M. Characteristics of and important lessons from the coronavirus disease 2019 (covid-19) outbreak in China: Summary of a report of 72314 cases from the chinese center for disease control and prevention. JAMA 323(13), 1239–1242 (2020).
Yang, J. et al. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: A systematic review and meta-analysis. Int. J. Infect. Dis. 94, 91–95 (2020).
Clarke, P. & Melendez, R. (Ann Arbor, MI) National neighborhood data archive (nanda): Neighborhood socioeconomic and demographic characteristics of census tracts, united states, 2000–2010. Inter-university Consortium for Political and Social Research.
Gu, T. et al. Characteristics associated with racial/ethnic disparities in covid-19 outcomes in an academic health care system. JAMA Netw. Open 3(10), e2025197 (2020).
Salerno, S. et al. Patterns of repeated diagnostic testing for covid-19 in relation to patient characteristics and outcomes. J. Intern. Med. 289(5), 726–737 (2021).
Crabb, B. T. et al. Comparison of international classification of diseases and related health problems, tenth revision codes with electronic medical records among patients with symptoms of coronavirus disease 2019. JAMA Netw. Open 3(8), e2017703 (2020).
Elixhauser, A. et al. Comorbidity measures for use with administrative data. Med. Care 36(1), 8–27 (1998).
van Walraven, C. et al. A modification of the elixhauser comorbidity measures into a point system for hospital death using administrative data. Med. Care 47(6), 626–633 (2009).
Jain, A. K. Fundamentals of Digital Image Processing (Prentice-Hall Inc, 1989).
Yip, S. S. & Aerts, H. J. Applications and limitations of radiomics. Phys. Med. Biol. 61(13), R150 (2016).
Galloway, M. M. Texture analysis using gray level run lengths. Comput. Graph. Image Process. 4(2), 172–179 (1975).
Chu, A., Sehgal, C. M. & Greenleaf, J. F. Use of gray value distribution of run lengths for texture analysis. Pattern Recogn. Lett. 11(6), 415–419 (1990).
Haralick, R. M., Shanmugam, K. & Dinstein, I. Textural features for image classification. IEEE Trans. Syst. Man Cybern. SMC 3(6), 610–621 (1973).
Thibault, G. et al. Shape and texture indexes application to cell nuclei classification. Int. J. Pattern Recogn. Artif. Intell. 27(01), 1357002 (2013).
Fisher, A., Rudin, C. & Dominici, F. All models are wrong, but many are useful: Learning a variable’s importance by studying an entire class of prediction models simultaneously. J. Mach. Learn. Res. 20(177), 1–81 (2019).
Ishwaran, H. et al. Random survival forests for high-dimensional data. Stat. Anal. Data Min. ASA Data Sci. J. 4(1), 115–132 (2011).
Salerno, S. & Li, Y. High-dimensional survival analysis: Methods and applications. https://doi.org/10.48550/arXiv.2205.02948 (Preprint posted online May 5, 2022).
Uno, H. et al. Evaluating prediction rules for t-year survivors with censored regression models. J. Am. Stat. Assoc. 102(478), 527–537 (2007).
Uno, H. et al. On the c-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat. Med. 30(10), 1105–1117 (2011).
Breiman, L. Random forests. Mach. Learn. 45(1), 5–32 (2001).
Acknowledgements
We thank Dr. Brahmajee Nallamothu for leading the development and curation of DataDirect, a newly launched, GPU-based analytics platform through the Michigan Medicine Precision Health Initiative. We are grateful to Anisa Driscoll and Cinzia Smothers for their continued analytical support with respect to database management, data processing, and use of the DataDirect platform. Additionally, we would like to thank Dr. Hamid Usefi and two reviewers for their insightful comments and suggestions, which have benefited the revision of this manuscript.
Funding
National Institutes of Health, National Cancer Institute grant R01-CA249096-01A1 (YL).
Author information
Authors and Affiliations
Contributions
Conceptualization: Y.S., S.S., P.H., J.K., Y.L. Methodology: Y.S., S.S., X.W., P.H., J.K., M.W.S., S.J., D.C.C., Y.L. Investigation: Y.S., S.S., X.H., Z.P., E.Y., C.S., J.S., X.W. Visualization: Y.S., S.S. Supervision: X.W., P.H., J.K., M.W.S., S.J., D.C.C., Y.L. Writing—original draft: Y.S., S.S., X.H., Y.L. Writing—review and editing: Y.S., S.S., X.H., Z.P., E.Y., C.S., J.S., X.W., P.H., J.K., M.W.S., S.J., D.C.C., Y.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sun, Y., Salerno, S., He, X. et al. Use of machine learning to assess the prognostic utility of radiomic features for in-hospital COVID-19 mortality. Sci Rep 13, 7318 (2023). https://doi.org/10.1038/s41598-023-34559-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-34559-0
This article is cited by
-
Predicting oxygen needs in COVID-19 patients using chest radiography multi-region radiomics
Radiological Physics and Technology (2024)
-
MultiCOVID: a multi modal deep learning approach for COVID-19 diagnosis
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.