Abstract
Ovarian cancer (OC) is an aggressive disease, primarily diagnosed in late stages with only 20% of patients surviving more than 5 years. Liquid biopsy markers have great potential to improve current diagnostic and prognostic methods. Here, we compared miRNAs and DNA methylation in matched plasma, whole blood and tissues as a surrogate marker for OC. We found that while both cfDNA and cf-miRNAs levels were upregulated in OC compared to patients with benign lesions or healthy controls, only cf-miRNA levels were an independent prognosticator of survival. Following on our previous work, we found members of the miR-200 family, miR-200c and miR-141 to be upregulated in both plasma and matched tissues of OC patients which correlated with adverse clinical features. We could also show that the upregulation of miR-200c and -141 correlated with promoter DNA hypomethylation in tissues, but not in plasma or matched whole blood samples. As cf-miRNAs are more easily obtained and very stable in blood, we conclude that they might serve as a more appropriate surrogate liquid biopsy marker than cfDNA for OC.
Similar content being viewed by others
Introduction
Ovarian cancer (OC) is a highly malignant disease with a 10-year survival rate of less than 30%, responsible for more than 200,000 deaths worldwide in 20201. Unfortunately, the vast majority of patients present with incurable advanced OC, with a dismal 5-years survival rate of < 20%2. In contrast, women diagnosed with early stage disease show an OS of > 90%. The current standard of care for OC is tumour cytoreductive surgery followed by mainly platinum-based chemotherapeutic regimens. However, around half of the patients will develop resistance to chemotherapy or relapse3. Therefore, there is a pending need to identify effective biomarkers for early screening, treatment response and prognosis in OC.
The serum tumour marker cancer antigen 125 (CA-125), with a sensitivity of less than 60% in early stage OC and up to 80% in advanced stages4 is the current gold standard biomarker for OC diagnosis and monitoring. While CA-125 is an attractive non-invasive marker for OC, the sensitivity and specificity must be improved to implement it as a marker for screening, particularly in early stage disease. Recently, our group has shown that the combination of CA-125 with a panel of seven circulating cell-free microRNAs (cf-miRNAs) could distinguish OC from healthy controls with an AUC of 0.975.
Other studies have also described cf-miRNAs as a promising minimally invasive clinical biomarker for profiling of cancer patients6,7,8. MicroRNAs are non-coding RNAs of 20–25 nucleotides long with the ability to regulate protein coding genes by repressing translation or mRNA degradation. They are transcribed in the nucleus and exported to the cytoplasm, resulting in a mature miRNA9. Cf-miRNAs are remarkably stable in body fluids such as plasma, thus making them an ideal non-invasive diagnostic tool for early cancer detection10,11,12,13. Our group has recently shown, for the first time, that total cf-miRNA levels are an independent prognostic marker for risk stratification in breast cancer14.
Likewise, the total amount of cfDNA has been explored as a potential liquid biopsy marker, and has been proposed to stem in varying ratios from DNA released from tumor cells together with DNA fragments from normal cells15. With cfDNA, one has the potential to analyze specific alterations coming from the tumor site such as mutations, copy number alterations or aberrant DNA methylation patterns. Since epigenetic aberrations occur early on in tumorigenesis16, tumor specific methylation of cfDNA might also be detectable in early stage cancer. However, for early detection, the low abundance of cancer specific cfDNA and corresponding high abundance of background DNA in circulation poses a huge challenge. Until now, only one methylation marker in cfDNA, SEPT9, has been translated to clinical screening with a specificity of 79% and sensitivity of 68% for the detection of colorectal cancer17.
Rational combinations of molecular markers in the blood might increase their specificity and sensitivity in diagnostics. Recently, a combination of a panel of DNA mutations and proteins in plasma was able to detect five different cancer types with a sensitivity of 69–98% and a specificity of > 99%18. Since miRNA expression can be regulated by aberrant DNA methylation of miRNA promoter sequences in OC19, probing for cfDNA promoter methylation and corresponding cf-miRNA abundance may have complementing prognostic value in liquid biopsies.
In this explorative case–control study, we sought out to assess the combination of cf-miRNA and corresponding promoter methylation in OC plasma and tissue samples as potential liquid biopsy markers. Our findings indicate that cf-miRNAs have more potential as a surrogate marker for OC than cfDNA and combinations of other markers such as proteins or mutations should be explored in future.
Methods
Sample collection
Before surgery and chemotherapy, women filled in a questionnaire on sociodemographic information and whole blood samples were collected. The cohort is summarised in Supplementary Table 1. Three EDTA tubes (Sarstedt S-Monovette K3E, 1.6 mg EDTA/ml) with 9-ml whole blood were taken from all participants.
Plasma preparation
Whole blood samples were centrifuged at 1300G for 20 min. The plasma fraction was further processed by high-speed centrifugation at 12,000g for 10 min. Samples were immediately stored at − 80.
Tissue processing and nucleic acid extraction
Fresh frozen tissue sections were obtained from the NCT biobank. Tumor cell content was verified by a pathologist at the Pathology Department. Up to 25 mg of tissue was processed with the Quick-DNA/RNA FFPE Kit (Zymo Research, Freiburg, Germany) for simultaneous extraction of genomic DNA and total RNA, including miRNAs. Nucleic acid concentration and purity was confirmed with the NanoDrop™ 1000 UV/Vis-Spectralphotometer 3.3 (peqLab, Erlangen, Germany).
miRNA isolation from plasma
Circulating miRNAs were isolated from 300 µl thawed plasma using the NucleoSpin miRNA Plasma kit (Macherey–Nagel, Düren, Germany) according to the manufacturer’s procotol. Total miRNAs were quantified using the Qubit microRNA Assay Kit and the Qubit Fluorometer 3.0 (Thermo Fisher Scientific, Massachusetts, USA).
qRT-PCR
For miRNA extracted from plasma, 2 µl was synthesized to cDNA using the LNA miRNA RT kit (Qiagen). For total RNA extracted from tissue, 10 ng was synthesized to cDNA using the LNA miRNA RT kit. Individual miRNAs were amplified and quantified using LNA specific primers and the primaQUANT 2 × qPCR-CYBR-Green-Blue-MasterMix (Steinbrenner, Germany) according to the manufacturer’s protocol on the qTOWER instrument (Analytical Jena, Germany). Two replicates were performed for each sample. For plasma samples, the geometric mean across the cohort was used to calculate miRNA expression20. For tissue samples, relative expression was calculated using the U6 snoRNA as a reference.
cfDNA isolation
Plasma was thawn and cfDNA was isolated from up to 2 ml using the NucleoSnap cfDNA kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s procotol. cfDNA was quantified using the Qubit dsDNA HS Assay kit and the Qubit Fluorometer 3.0 (Thermo Fisher Scientific). DNA size distribution was assessed on the Bioanalyzer instrument using the DNA High Sensitivity Kit (Bioanalyzer, CA, USA).
DNA methylation analysis
For bisulfite conversion, 500 ng of tissue genomic DNA or 5–100 ng cfDNA was converted using the EZ DNA Methylation-Gold™ kit (Zymo Research, Freiburg; Germany). Subsequent PCR amplification was performed using HotStarTaq Plus DNA Polymerase kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions incorporating the T7 promoter sequence as listed in Supplementary Table 1. PCR products were verified by electrophoresis with a 1% agarose gel. According to the instructions of Sequenom MassARRAY EpiTyper Assay the PCR products were subjected to alkaline phosphatase treatment followed by in vitro transcription and RNaseA cleavage to result in specific fragmentation. The obtained fragments were subjected to matrix-assisted laser desorption/ionization time-of-flight mass-spectrometry (MALDI-TOF MS). Results were exported from the MassARRAY instrument with EpiTyper v1.0 software. Unless otherwise stated, DNA methylation values were calculated as the average methylation of all CpG sites within each PCR product.
Statistical analysis
Significance between groups was calculated by a nonparametric Mann Whitney test using GraphPad prism version 8.0. Kaplan–Meier survival plots for single markers were also computed with GraphPad prism, using the Log-rank model for significance. All other analyses were performed using R version 4.1.2. In order to compute univariate hazard ratios, a Cox proportional hazards regression model21 was fitted for each parameter separately with the respective overall or progression-free survival time as a dependent variable and the respective marker as a single covariate. Multivariate hazard ratios are stemming from a multivariate Cox proportional hazards regression model, where all parameters simultaneously were used as covariates when fitting the Cox models for overall or progression-free survival. For all Cox models, p values for the null hypothesis that the hazard ratio equals to 1 were derived by means of a standard Wald test. The REMARK (Reporting Recommendations for Tumor Marker Prognostic Studies) guidelines were implemented to report results22.
Ethics approval and consent to participate
Ovarian cancer (OC) patients (n = 72), women who were treated for unknown pelvic mass and healthy volunteers with no known conditions (n = 53) were recruited at the University Hospital Heidelberg, Germany and at the National Center for Tumor Diseases (NCT), Heidelberg, Germany, between May 2015 and August 2018. All participants provided written informed consent. The study was approved by the ethics committee of the University of Heidelberg (S-046/2018, S-266/2011, S-393/2019) in accordance with good clinical practice guidelines, national laws and the Declaration of Helsinki.
Results
Study population
Plasma and whole blood from EDTA tubes as well as fresh frozen tissues were used in this study as outlined in Fig. 1. Patients were recruited at the Heidelberg University Hospital between May 2015 and August 2018. The cohort characteristics are outlined in Supplementary Table 1.
Study overview. Plasma cf-miRNA and cf-DNA was isolated from 125 study participants and analysed for miR-200c expression and promoter methylation analysis. Genomic DNA was obtained from whole blood (n = 110). Genomic DNA and RNA were simultaneously isolated from matched tissues (n = 46). This figure was created with Biorender.com and exported under a paid subscription.
Total levels of circulating cfDNA and cf-miRNAs are globally upregulated in OC
Since circulating nucleic acids are proposed to be released from tumors and can serve as surrogate markers, we first assessed whether OC patients have more cfDNA and cf-miRNA amounts compared to patients with benign lesions or healthy women with no known malignancies. cfDNA and cf-miRNA was isolated from the matched plasma samples, using independent isolation methods. Because there was no difference between the healthy controls and benign group (Supp Fig. 1), we decided to pool these two groups hereby called “non-malignant”. We also log transformed the data due to abnormal distribution and found a significant increase in the amounts of both cfDNA and cf-miRNA in OC cases (Fig. 2A,B). Remarkably, the total amount of circulating cfDNA and miRNA was positively correlating in matched plasma samples (Fig. 2C).
Circulating cfDNA and cf-miRNAs are upregulated in OC. Total cfDNA and cf-miRNAs were measured in plasma. Dot plots depict the total levels (log transformed) of cfDNA (A) and cf-miRNAs (B) in a group of healthy controls and patients with benign lesions (non-malignant) and OC cases. Significance is calculated by a Mann Whitney test.
Circulating cf-miRNA but not cfDNA is an independent marker of survival in OC
Our group has recently shown that total levels of cf-miRNAs have prognostic value in both breast14 and OC5. Because cfDNA levels were similarly increased as cf-miRNA levels in OC, we hypothesized that the levels of cfDNA might also be associated with patient outcome. To this end, we dichotomized the patients according to the median levels of circulating DNA or miRNA and could reproduce in the reduced cohort, where plasma was available for both analyses (n = 72), that cf-miRNA levels significantly associated with survival (Fig. 3A; P = 0.005). However, levels of cfDNA were not associated with survival using the same approach of dichotomization (Supp Fig. 2A) but only when we split the data into 3 sub-groups in a reduced number of patients (Fig. 3B; P = 0.03). Next, we performed univariate and multivariate cox regression analysis using Grade, CA-125, R status, FIGO stage and age as categorical variables. Strikingly, only levels of cfDNA and cf-miRNA were significant predictors of survival in univariate analysis. Finally, in multivariate analysis including FIGO stage, cfDNA and cf-miRNA levels, only cf-miRNA remained as a significant independent predictor of survival in this cohort (Fig. 3C,D). Neither cfDNA nor cf-miRNA were significantly associated with progression-free survival (Supp Fig. 2).
Circulating miR-200c and miR-141 are surrogate markers for OC and correlate with adverse clinical features
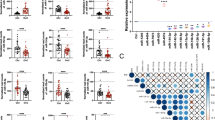
Next, we were interested to know which miRNAs were contributing to the increase of total miRNA levels in OC patients. Previously, our group identified a signature of 7 miRNAs which could serve as diagnostic markers for OC5. One of these candidates, miR-200c has already been documented to be upregulated in OC23. miR-200c belongs to the miR-200 miRNA family which have repeatedly been described as modulators of the metastatic cascade in cancer. Since miR-200c is transcribed together and generally co-expressed with miR-141, we analysed the expression of both miRNAs in OC plasma and a subset of matched tissues by qRT-PCR. Both miR-200c and miR-141 were significantly increased in both plasma and tissues in OC (Fig. 4A,B). Additionally, increased circulating levels of miR-200c and -141 significantly associated with a number of adverse clinical features, highlighting their potential as surrogate markers for OC (Fig. 4C).
Circulating miR-200c and miR-141 are potential surrogate markers for OC. Expression levels of circulating miR-200c and -141 are significantly upregulated in OC cases compared to controls, as depicted by box plot with SEM (A). Both miR-200c and miR-141 expression is upregulated in a panel of OC tissues compared to benign tissues (B). Significance is calculated by a Mann Whitney test and depicted as ***P < 0.001 and ****P < 0.0001. The table below shows the correlation of circulating miRNA expression with clinical features of OC.
Promoter methylation of miR-200c and miR-141 correlates with expression in tissues but not in plasma
Since the bulk of circulating cfDNA is likely not tumor specific, we sought to look for a more specific parameter that could be used in combination with cf-miRNAs as a surrogate marker for OC. We hypothesized that the miR-200 family might be epigenetically regulated in OC, as concluded by a recent meta-analysis24. The miR-200 family consists of five miRNAs, miR-200a, miR-200b and miR-429; transcribed on chromosome 1 and miR-200c and miR-141; transcribed on chromosome 12. With a difference of one base pair in the seed region, the miR-200 family have the potential to target thousands of mRNAs which can have a profound impact in disease25. We utilized the quantitative MassArray technology26 to analyse DNA methylation of two regions: 200c_2 and 200c_5 in the upstream promoter region of miR-200c on chromosome 12 (Supplementary Fig. 3a) in the same set of tissues where we had measured miRNA expression (Fig. 4B). We found a highly significant promoter hypomethylation in genomic DNA of tumor tissues compared to benign in both regions (Fig. 5A) which significantly correlated with miR-200c (over)expression in the matched tissue samples (Fig. 5B). Next, we sought out to investigate DNA methylation in cfDNA samples. 110 samples of 125 had sufficient quality to undergo MassArray analysis. In contrast to tumor tissues, we observed no significant differences in miR-200c promoter methylation in plasma samples (Fig. 5C). Overall, the variation in methylation in plasma was much broader compared to the tissues. Of interest, a mild but significant correlation between matched plasma cfDNA methylation and miRNA expression was observed (Fig. 5D), indicating that at least in part, the cfDNA methylation profile reflected that of primary tumors. Direct comparison of matched tissue and cfDNA revealed that the majority of patients with hypomethylated DNA from tissue had hypermethylated cfDNA (Supplementary Fig. 3B), indicating a strong influence of background signal in the plasma. Promoter methylation analysis of genomic DNA from whole blood (n = 143) confirmed this observation, however, in contrast to cfDNA (Supplementary Fig. 3C) and there was no correlation between whole blood methylation and miRNA expression (data not shown). In conclusion, methylation of the miR-200c promoter in cfDNA was partly reflective of tumor tissue methylation in OC.
miR-200c expression correlates with promoter methylation in OC tissues but not in cfDNA. Quantitative DNA methylation analysis of the miR-200 promoter in a panel of non-malignant and cancer tissues is depicted as percentage where ****P < 0.0001 (A). DNA methylation correlated with matched miRNA expression, plotted as raw Ct values, in corresponding tissues (n = 38) (B). Quantitative DNA methylation analysis of the miR-200 promoter in cfDNA of non-malignant and malignant OC (C) and one representative CpG site *P < 0.05 (D). The correlation between cfDNA methylation and miRNA expression is shown (E).
Discussion
In summary, we presented one of the only studies in OC where cf-miRNA, cfDNA, whole blood and matched tissue samples have been simultaneously investigated. We have again demonstrated the potential of cf-miRNA levels as an independent prognostic marker for survival compared to cfDNA. We have also shown that two circulating miRNAs have potential as surrogate markers in OC. We verified one possible biological mechanism of miRNA regulation by DNA methylation in matched tumor tissues and partly in cfDNA. Our results indicate that circulating miRNAs might be a more suitable liquid biopsy marker than cfDNA in OC, and combinations with other markers such as proteins or mutations should be explored in the future to evaluate options for disease diagnosis and therapy monitoring.
While levels of circulating cfDNA and mutational load have been readily uptaken in the clinic for disease monitoring, circulating cf-miRNAs are still far from implementation. We believe that cf-miRNAs are more appropriate as surrogate markers for OC and an ideal plasma biomarker in comparison to total cfDNA for a number of reasons. First of all, previous work from our lab demonstrated that miRNAs, including miR-200c, are highly stable in blood13 and can be obtained from as little as 200 µl plasma. They are easily detectable in both healthy and diseased individuals by simple amplification methods such as qPCR. On the contrary, cfDNA is hardly detectable in healthy individuals and the amount of blood required for analysis is at least 2 ml. We have shown, for the first time, that cf-miRNA is more representative of corresponding tissues compared to cfDNA methylation. The miR-200 promoter was significantly hypomethylated in a panel of tissues which strongly correlated with miRNA expression. On the other hand, no differences on general methylation were observed in cfDNA, and in a subset of patients with matched tissues, we observed a general increased methylation of corresponding cfDNA. One explanation for this may be that the signal is diluted due to the influence of normal circulation, probably coming from blood cells. Indeed, whole blood methylation analysis confirmed the results in cfDNA. Nevertheless, we did see a trend toward hypomethylation at specific CG sites, which partly correlated with miRNA expression in corresponding plasma.
Overexpression of the miRNA-200 family has already been well described in OC27. In line with our study, miR-200a/b/c were elevated in OC patient serum (n = 70) which associated with disease progression, advanced stage and metastasis28. Another recent study of ascites, the fluid released from the surrounding pelvic region in OC patients, also found upregulation of the miR-200 family29. We have demonstrated that DNA methylation of the upstream miRNA promoter region associates with miR-200c expression in OC tissues and partly in plasma. Similarly, a recent meta-analysis on the TCGA cohort revealed a correlation between promoter methylation and miR-200 expression in more than 500 OC patients24. While most studies describe the miR-200 family as tumor suppressive miRNAs, downregulated in cancer, some studies have had similar findings to ours. For example in pancreatic cancer, hypomethylation and upregulation of miR-200a and 200b led to an epithelial-mesenchymal transition (EMT) transition30. While we did not directly characterize EMT in our study, we also observed higher levels of circulating miR-200c and -141 in patients with lymph node infiltration and metastatic disease. In addition to EMT, epigenetic regulation of the miR-200 family has been attributed to drug resistance in cancer. Shindo and colleagues demonstrated that miR-200b,a and miR-429 were downregulated in cisplatin resistant bladder cancer which associated with CpG island hypermethylation31. As platinum resistance is also a major contributor in OC progression32 circulating miR-200 might also have potential in disease monitoring.
While, the field of liquid biopsy is currently placing a large emphasis on cfDNA, we could show that a small panel of cf-miRNAs are more likely to reflect the tumor status. Few studies until now have been able to show a significant impact of cfDNA methylation in OC. Recently, Giannopoulou et al. found a moderate correlation between ESR1 methylation in OC primary tumors and matched cfDNA33. The technology used to assess cfDNA methylation should also be considered. Most studies until now have used PCR based methylation assays which are not very accurate and introduce PCR bias. We have utilized the quantitative MassARRAY technology based on mass spectrometry26 which has the major advantage of accurately quantifying methylation levels. Another technical challenge with methylation analysis is that most methods require a bisulfite conversion step, which can degrade up to 96% of DNA34. A limitation of our work is that we focused on just one miRNA promoter. A whole genome methylation screening should be implemented to identify novel markers. Although this is difficult due to the low amount of cfDNA available, one study until now has performed methylome screening in a large panel of tumor tissues and serum from OC. They found that methylation of just three genes could identify 90% of cases and was predictive of therapy response35. In future, we may look to analyse the same set of markers in corresponding tissues and plasma. Similarly to our results, the authors also reported a discrepancy between cfDNA and tumor tissue methylation, possibly due to contamination of blood cells in the plasma. Another angle could be to combine miRNA markers with mutation status in cfDNA. Recently, this approach in combination with protein markers has shown great promise in OC18.
One strength of our study is the same material (matched plasma miRNA, cfDNA, whole blood and tissue) was available from the same donors for analysis, albeit only from one cohort and not for all samples. For miRNA analysis, the number of individuals analysed across independent cohorts was considerably large for OC. No studies until now have utilized cf-miRNA levels as a biomarker, or compared this to cfDNA from the same donor. Even though the correlation of both nucleic acids was significant, miRNA levels surpassed cfDNA with the association of clinical features. In particular, miRNA levels were strongly associated with OS, which we have previously shown for both OC5 and breast cancer14 which was not the case for cfDNA. This finding suggests that cf-miRNAs may be even more informative than traditional biopsies as prognostic markers. A potential explanation for this may be due to the protection of miRNAs in circulation. While DNA fragments are released from tumors or blood cells, miRNAs are either packaged into extracellular vesicles or exported from cells in a protein complex. These mechanisms can protect cf-miRNAs from degradation and may explain why their specificity for cancer is better retained than for cfDNA. Further studies should be done to elaborate on this point.
The introduction of novel targeted therapies, namely the anti-angiogenic bevacizumab during the course of our sample collection may have also impacted the prognosis of our cohort36 and thus our conclusions related to cf-miRNA levels. One recent study in breast cancer tissues, found a number of changes in miRNA expression in patients undergoing neoadjuvant therapy with bevacizumab37. We are not aware of any such studies regarding cf-miRNA and we did not have access to longitudinal samples in our study to analyse this.
In conclusion, our data support practicability and clinical relevance of circulating miRNAs as liquid biopsy markers and propose prognostic potential of circulating miR-200c and miR-141 for OC. Ultimately, we describe a link between miR-200c overexpression and miR-200c promoter hypomethylation in OC tissue, which is partly reflected by cfDNA.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71(3), 209–249 (2021).
Jayson, G. C., Kohn, E. C., Kitchener, H. C. & Ledermann, J. A. Ovarian cancer. Lancet 384(9951), 1376–1388 (2014).
Armbruster, S., Coleman, R. L. & Rauh-Hain, J. A. Management and treatment of recurrent epithelial ovarian cancer. Hematol. Oncol. Clin. N. Am. 32(6), 965–982 (2018).
Soletormos, G. et al. Clinical use of cancer biomarkers in epithelial ovarian cancer: Updated guidelines from the European Group on tumor markers. Int. J. Gynecol. Cancer 26(1), 43–51 (2016).
Gahlawat, A. W., Witte, T., Haarhuis, L. & Schott, S. A novel circulating miRNA panel for non-invasive ovarian cancer diagnosis and prognosis. Br. J. Cancer 127(8), 1550–1556 (2022).
Patel, M., Verma, A., Aslam, I., Pringle, H. & Singh, B. Novel plasma microRNA biomarkers for the identification of colitis-associated carcinoma. Lancet 385(Suppl 1), S78 (2015).
Cuk, K. et al. Circulating microRNAs in plasma as early detection markers for breast cancer. Int. J. Cancer 132(7), 1602–1612 (2013).
Madhavan, D. et al. Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clin. Cancer Res. 18(21), 5972–5982 (2012).
O’Brien, J., Hayder, H., Zayed, Y. & Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne) 9, 402 (2018).
Turchinovich, A., Weiz, L., Langheinz, A. & Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 39(16), 7223–7233 (2011).
Mitchell, P. S. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 105(30), 10513–10518 (2008).
Jung, M. et al. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin. Chem. 56(6), 998–1006 (2010).
Ward Gahlawat, A. et al. Evaluation of storage tubes for combined analysis of circulating nucleic acids in liquid biopsies. Int. J. Mol. Sci. 20, 3 (2019).
Gahlawat, A. W., Fahed, L., Witte, T. & Schott, S. Total circulating microRNA level as an independent prognostic marker for risk stratification in breast cancer. Br. J. Cancer 20, 22 (2022).
De Mattos-Arruda, L. et al. Circulating tumour cells and cell-free DNA as tools for managing breast cancer. Nat. Rev. Clin. Oncol. 10(7), 377–389 (2013).
Feinberg, A. P., Ohlsson, R. & Henikoff, S. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 7(1), 21–33 (2006).
Potter, N. T. et al. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin. Chem. 60(9), 1183–1191 (2014).
Cohen, J. D. et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359(6378), 926–930 (2018).
Loginov, V. I. et al. Novel miRNA genes deregulated by aberrant methylation in ovarian carcinoma are involved in metastasis. Gene 662, 28–36 (2018).
Schwarzenbach, H., da Silva, A. M., Calin, G. & Pantel, K. Data normalization strategies for microRNA quantification. Clin. Chem. 61(11), 1333–1342 (2015).
Cox, D. R. Regression models and life-tables. J. R. Stat. Soc. B. 34(2), 187 (1972).
Altman, D. G., McShane, L. M., Sauerbrei, W. & Taube, S. E. Reporting recommendations for tumor marker prognostic studies (REMARK): Explanation and elaboration. BMC Med. 10, 51 (2012).
Koutsaki, M., Libra, M., Spandidos, D. A. & Zaravinos, A. The miR-200 family in ovarian cancer. Oncotarget 8(39), 66629–66640 (2017).
Agustriawan, D. et al. DNA methylation-regulated microRNA pathways in ovarian serous cystadenocarcinoma: A meta-analysis. Comput. Biol. Chem. 65, 154–164 (2016).
Uhlmann, S. et al. miR-200bc/429 cluster targets PLCgamma1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene 29(30), 4297–4306 (2010).
Ehrich, M. et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl. Acad. Sci. USA 102(44), 15785–15790 (2005).
Giannopoulou, L., Kasimir-Bauer, S. & Lianidou, E. S. Liquid biopsy in ovarian cancer: Recent advances on circulating tumor cells and circulating tumor DNA. Clin. Chem. Lab. Med. 56(2), 186–197 (2018).
Zuberi, M. et al. Expression of serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clin. Transl. Oncol. 17(10), 779–787 (2015).
Zavesky, L. et al. Ascites-derived extracellular microRNAs as potential biomarkers for ovarian cancer. Reprod. Sci. 2018, 1933719118776808 (2018).
Li, A. et al. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 70(13), 5226–5237 (2010).
Shindo, T. et al. Epigenetic silencing of miR-200b is associated with cisplatin resistance in bladder cancer. Oncotarget 9(36), 24457–24469 (2018).
Galluzzi, L. et al. Systems biology of cisplatin resistance: Past, present and future. Cell Death Dis. 5, e1257 (2014).
Giannopoulou, L. et al. ESR1 methylation in primary tumors and paired circulating tumor DNA of patients with high-grade serous ovarian cancer. Gynecol. Oncol. 150(2), 355–360 (2018).
Grunau, C., Clark, S. J. & Rosenthal, A. Bisulfite genomic sequencing: Systematic investigation of critical experimental parameters. Nucleic Acids Res. 29(13), E65–E75 (2001).
Widschwendter, M. et al. The potential of circulating tumor DNA methylation analysis for the early detection and management of ovarian cancer. Genome Med. 9(1), 116 (2017).
Garcia, A. & Singh, H. Bevacizumab and ovarian cancer. Ther. Adv. Med. Oncol. 5(2), 133–141 (2013).
Lindholm, E. M. et al. miRNA expression changes during the course of neoadjuvant bevacizumab and chemotherapy treatment in breast cancer. Mol. Oncol. 13(10), 2278–2296 (2019).
Acknowledgements
We would like to thank all patients who participated in this study. We are grateful to the liquid and tissue biobanks of the NCT, Heidelberg for providing samples. We would like to extend our thanks to the technical support of Denise Keitel, Kai Sona and Terence Osere and to all of the nurses and doctors who supported the study. We thank Lisa Haarhuis for retrieving patient information.
Funding
Open Access funding enabled and organized by Projekt DEAL. Our study was funded by the University Hospital of Heidelberg. For the publication fee, we acknowledge financial support by Deutsche Forschungsgemeinschaft within the funding programme “Open Access Publikationskosten” as well as by Heidelberg University.
Author information
Authors and Affiliations
Contributions
A.W.G. and T.W. designed experiments. A.W.G. analysed the data and wrote the manuscript. Tumor tissues were verified and provided by P.S. A.W.G., S.S. and T.W. conceptualised the project and all authors edited and revised the final manuscript.
Corresponding author
Ethics declarations
Competing interests
HeiScreen GmbH is a company shared by the University Hospital Heidelberg. SS is shareholder of Heiscreen and Heiscreen NKY GmbH. TW, AWG, SS have a patent Filing EPO EP19174483.8
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gahlawat, A.W., Witte, T., Sinn, P. et al. Circulating cf-miRNA as a more appropriate surrogate liquid biopsy marker than cfDNA for ovarian cancer. Sci Rep 13, 5503 (2023). https://doi.org/10.1038/s41598-023-32243-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-32243-x
This article is cited by
-
Liquid biopsy: from concept to clinical application
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.