Abstract
Hutchinson-Gilford Progeria Syndrome (HGPS) is a rare, fatal genetic disease that accelerates atherosclerosis. With a limited pool of HGPS patients, clinical trials face unique challenges and require reliable preclinical testing. We previously reported a 3D tissue engineered blood vessel (TEBV) microphysiological system fabricated with iPSC-derived vascular cells from HGPS patients. HGPS TEBVs exhibit features of HGPS atherosclerosis including loss of smooth muscle cells, reduced vasoactivity, excess extracellular matrix (ECM) deposition, inflammatory marker expression, and calcification. We tested the effects of HGPS therapeutics Lonafarnib and Everolimus separately and together, currently in Phase I/II clinical trial, on HGPS TEBVs. Everolimus decreased reactive oxygen species levels, increased proliferation, reduced DNA damage in HGPS vascular cells, and improved vasoconstriction in HGPS TEBVs. Lonafarnib improved shear stress response of HGPS iPSC-derived endothelial cells (viECs) and reduced ECM deposition, inflammation, and calcification in HGPS TEBVs. Combination treatment with Lonafarnib and Everolimus produced additional benefits such as improved endothelial and smooth muscle marker expression and reduced apoptosis, as well as increased TEBV vasoconstriction and vasodilation. These results suggest that a combined trial of both drugs may provide cardiovascular benefits beyond Lonafarnib, if the Everolimus dose can be tolerated.
Similar content being viewed by others
Introduction
Hutchinson-Gilford Progeria Syndrome (HGPS) is a rare, fatal genetic disease that accelerates atherosclerosis in children1. It is caused by a de novo point mutation in the LMNA gene that results in production of progerin, an aberrantly spliced version of prelamin A that remains permanently farnesylated and methylated at its C-terminus1. The farnesyl group anchors progerin to the nuclear lamina where it accumulates and causes DNA repair defects, altered gene expression, disorganized heterochromatin, telomere shortening, and cell senescence1,2. Patient symptoms include subcutaneous fat loss, progressive joint contracture, alopecia, bony dysplasia, and failure to thrive1,3. Fatality is caused by progressive atherosclerosis leading to heart attack or stroke by 15 years of age4.
Many HGPS children exhibit hypertension and altered ankle-brachial index, similar to that found in atherosclerosis of older non-HGPS individuals, although endothelial induced vasodilation is normal5. While the atherosclerotic lesions are similar to those found in older, non-HGPS individuals with atherosclerosis, in HGPS, cholesterol does not accumulate and adventitial thickening and fibrosis are observed4,6. Vascular SMC loss occurs throughout the HGPS vessel wall as SMCs are replaced by a dense matrix of proteoglycans and collagen, resulting in vascular stiffening that causes hypertension4,6. HGPS fibrotic plaques incorporate excessively expressed ECM proteins such as collagens I, III, IV, V, and VI7.
With a limited pool of ~ 350 HGPS patients worldwide, testing potential therapeutics for the disease faces unique and significant challenges8. Only one or two new approaches can be studied at a time, clinical trials require global recruitment but still suffer from low subject numbers, and historical controls must be used without placebo groups8. These challenges drive the need for accurate in vitro models that offer reliable preclinical testing of potential therapeutics before they progress into clinical trials. To date, in vitro HGPS preclinical studies rely on 2D cell culture which does not accurately model the 3D physiological microenvironment. Further, very few HGPS murine models exhibit cardiovascular pathology which appears after many months9. Since atherosclerosis is the fatal symptom of HGPS, a biomimetic model of HGPS cardiovascular pathology is an ideal platform for preclinical testing of therapeutics. Preclinical testing models that accurately predict drug responses can better identify candidates for clinical trials and determine the maximum tolerated dose (MTD) of therapeutics to potentially avoid toxicities that commonly occur during the initial stages of clinical trials.
To provide a human cell in vitro vascular model of HGPS, we developed 3D tissue engineered blood vessels (TEBVs) using induced pluripotent stem cell (iPSC)-derived vascular smooth muscle cells (viSMCs) with a luminal monolayer of iPSC-derived endothelial cells (viECs)10. TEBVs are fabricated within a few hours and perfused at physiological shear stress for several weeks. We differentiated iPSCs from healthy and HGPS patients into endothelial cells and smooth muscle cells that exhibit key markers and functional features of these cells. We then used the HGPS cells to create an HGPS TEBV model that replicates vascular disease characteristics after 3 weeks, including progerin expression, vascular calcification, loss of medial smooth muscle cells, reduced vasoactivity, and excess extracellular matrix (ECM) deposition10. Our model indicated a specific role of the endothelium in disease progression as viECs from HGPS patients exhibited blunted response to physiological shear stress and inflammatory marker expression10. Further, TEBVs can model early steps in atherosclerosis in straight11 and branched vessels12. This patient-specific 3D tissue engineered blood vessel platform offers the opportunity to study disease mechanisms and evaluate therapeutics in a controlled, biomimetic microenvironment.
Permanent farnesylation of progerin anchors the protein to the nuclear lamina, causing nuclear blebbing or misshapen nuclei, thickened nuclear lamina, and impaired mechanotransduction13. Cell and mouse models of HGPS have indicated that the farnesyl group of progerin is partly responsible for the HGPS phenotype1,14. Lonafarnib is a farnesyltransferase inhibitor (FTI) that inhibits post-translational farnesylation of progerin, reducing progerin intercalation into the nuclear envelope1,15,16. FTI treatment improves nuclear morphology in HGPS cells17,18,19,20, extends lifespan, improves skeletal properties, increases body weight and adipose tissue, and improves cardiovascular phenotype in animal models2,21,22,23,24. A clinical trial testing Lonafarnib indicated treatment increased body weight, reduced cardiovascular stiffness, increased bone density, and decreased mortality of trial participants3,15,25. Lonafarnib is currently the only FDA-approved therapy for HGPS.
There is currently a Phase I/II clinical trial to test the combination of Lonafarnib and the rapamycin analog Everolimus for HGPS (ClinicalTrials.gov Identifier: NCT02579044). Treatment of HGPS cells with rapamycin or its analogues increases autophagy-mediated progerin clearance, resulting in reduced nuclear abnormalities, senescence, and DNA damage levels in HGPS fibroblasts26,27. While rapamycin or its analogues have not been used in animal models of HGPS, genetic reduction of mTOR in a mouse progeria model improved lifespan, although progerin remained in cells28. Treating HGPS patients with the two drugs might enhance the effectiveness of Lonafarnib, although there are no published studies testing Lonafarnib and rapamycin combination treatment in HGPS mouse models. The objective of this study was to test the effect of Lonafarnib and Everolimus, separately and together, to assess if the beneficial effects of the drug combination on HGPS TEBV structure and function. Results of this study might inform clinical trial results.
Results
Everolimus treatment reduces progerin expression and misshapen nuclei in HGPS viSMCs and viECs
Treatment of viSMCs and viECs for 1 week with either 1 µM Lonafarnib or 0.1 µM Everolimus leads to minimal cell death and high cell viability, comparable to untreated and vehicle controls (Fig. S1). In SMCs, treatment with 1 µM Lonafarnib and 0.05 µM Everolimus does not reduce cell viability, while doubling the dose to 2 µM Lonafarnib and 0.1 µM Everolimus significantly decreases cell viability. For subsequent experiments, cells and TEBVs were treated with 1 µM Lonafarnib alone, 0.1 µM Everolimus alone, or 1 µM Lonafarnib combined with 0.05 µM Everolimus.
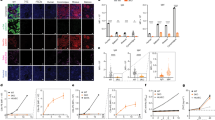
Next, progerin levels were measured after treating viSMCs and viECs for 7 days with 0.001–0.1 µM Everolimus, based on prior published studies29 and our own initial testing (Fig. 1a,b). Quantifying Western blots (Fig. 1c,d) indicated that by 0.05 µM Everolimus, progerin levels were significantly below vehicle levels. (The entire individual Western blots are shown in Fig. S2).
Everolimus reduces progerin expression and misshapen nuclei in HGPS viSMCs and viECs. (a, b) Cropped images of Western blot gels of progerin and (c, d) quantification in HGPS 167 CL2 viSMCs (a) and viECs (b) treated with Everolimus (Ev). Entire gels for each of the experiments performed with viSMCs and viECs are shown in Fig. S2. Data presented as fold change progerin expression compared with untreated HGPS control. ‘V’ indicates DMSO vehicle control. (e, f) Misshapen nuclei in HGPS 167 CL2 viSMCs (e) and viECs (f) treated with Everolimus for 7 days. Representative images are shown in Fig. S4. ‘V’ indicates DMSO vehicle control. Data presented as mean ± SD. N = 2–6 experiments per group.
Rapamycin treatment of HGPS fibroblasts does not affect lamin A/C29. Likewise, using a lamin A/C antibody, we found that one-week treatment of viSMCs or viECs with 0.1 µM Everolimus did not cause a statistically significant decline in lamin A or lamin C in viSMCs (Fig. S3a,b), but did reduce lamin C in viECs (Fig. S3c,d). This antibody also recognizes progerin. After treatment of viSMCs with 0.1 µM Everolimus progerin decreased relative to untreated or DMSO treated viSMCs, as was seen with the progerin-specific antibody (Fig. 1a,c). In viECs, Everolimus caused a decline in progerin, but progerin declined after DSMO treatment, which differed from the anti-progerin antibody (Fig. 1b,d).
Nuclei shape in HGPS is distorted, often showing blebs or non-elliptical shape30 (Fig. S4). We previously showed that HGPS viSMCs and viECs exhibit a higher fraction of misshapen nuclei than healthy cells10. iPSC-derived HGPS viSMCs and viECs treated with Everolimus for 7 days exhibited reduced levels of misshapen nuclei compared with no treatment and vehicle controls, reaching significance at 0.05 µM in both viSMCs and viECs (Fig. 1e,f).
Everolimus reduces ROS levels, increases proliferation, and decreases DNA damage in HGPS viSMCs and viECs
HGPS viSMCs from two donors (167 CL2 and 003 CL1D) exhibited significantly increased ROS levels, indicated by higher level of DCFDA mean fluorescence intensity, and reduced proliferation, marked by decreased Ki67 expression, compared with healthy viSMCs (Fig. 2a,b for 167 CL2 and Fig. S5a,b for 003 CL1D). HGPS viSMCs had significantly increased levels of DNA double-stranded breaks (DSBs), marked by a higher percentage of nuclei staining positively for ɣH2A.X foci (Fig. 2c, Fig. S5c) and more ɣH2A.X foci per cell (Fig. 2d, Fig. S5d). viECs from both HGPS donors displayed similar results with higher ROS levels (Fig. 2e, Fig. S5e), reduced proliferation (Fig. 2f, Fig. S5f), and more DNA DSBs (Fig. 2g,h, Fig. S5g,h). Representative images of DCFDA, Ki67, and γH2A.X staining are shown in Figs. S6, S7, and S8.
Everolimus reduces ROS levels, increases proliferation, and decreases DNA damage in HGPS 167 CL2 viSMCs and viECs. Healthy viSMCs (a–d) and viECs (e–h) and HGPS 167 CL2 viSMCs and viECs were treated with different combinations of Lonafarnib (LF) and Everolimus (Ev). ‘V’ indicates DMSO vehicle control. viECs and viSMCs from healthy donors are shown as gray bars and HGPS viECs and viSMCs are shown as white bars. Fold change of DCFDA mean fluorescence intensity compared with healthy in viSMCs (a) and viECs (e). Percent positive Ki67 nuclei in viSMCs (b) and viECs (f). Percent nuclei with ɣH2A.X foci in viSMCs (c) and viECs (g). Number of ɣH2A.X foci per total cells in viSMCs (d) and viECs (h). Data presented as mean ± SD. N = 3 experiments per group. Groups connected by the same letter are not significantly different from each other (p > 0.05). Groups labeled with different letters are significantly different from each other (p < 0.05). Exact p-values for significant differences are provided in Table S1.
For viSMCs from both HGPS donors, Lonafarnib partially reduced ROS levels, but to a level not significantly different from untreated HGPS or vehicle control (Fig. 2a, Fig. S5a). In contrast, Everolimus treatment reduced ROS to a level not significantly different than healthy viSMCs (Fig. 2a, Fig. S5a). Lonafarnib treatment did not improve HGPS viSMC proliferation for either donor (Fig. 2b, Fig. S5b). Everolimus and combination treatments significantly improved proliferation in HGPS viSMCs from donor 167 (Fig. 2b) but not in the 003 donor, which was at a higher level without drug treatment (Fig. S5b). Lonafarnib reduced the percentage of nuclei with DNA DSBs in viSMCs from donor 003 (Fig. S5c) but not in viSMCs from donor 167 (Fig. 2c). Everolimus treatment significantly reduced the percentage of nuclei with DNA DSBs to healthy levels in viSMCs from both HGPS donors (Fig. 2c, Fig. S5c).
In viECs, Lonafarnib partially reduced ROS levels in only the 003 line (Fig. 2e, Fig. S5e). Everolimus reduced ROS in HGPS viECs from both donors to a level that was not significantly different than healthy cells (Fig. 2e, Fig. S5e). Combination treatment with Lonafarnib and Everolimus reduced ROS levels to healthy levels in viECs from both donors (Fig. 2e, Fig. S5e). Lonafarnib only improved viEC proliferation in donor 167 (Fig. 2f). Everolimus and combination treatment significantly improved proliferation in viECs from both HGPS donors (Fig. 2f, Fig. S5f). In viECs, Lonafarnib had no significant effect on DNA DSBs for either donor (Fig. 2g,h, Fig. S5g,h). In contrast, Everolimus reduced percentage of nuclei with DNA DSBs and DNA DSBs per nucleus in viECs from both donors (Fig. 2g,h, Fig. S5g,h). Combination treatment resulted in similar DNA DSB as treatment with Everolimus alone in viECs from both donors (Fig. 2g,h, Fig. S5g,h).
Lonafarnib and Everolimus improve NO production and shear stress-sensitive gene expression in viECs exposed to physiological shear stress
To test the response of healthy and HGPS viECs to physiological shear stress which establishes normal endothelial cell function in vivo, we applied 12 dynes/cm2 shear stress for 24 h after 6-day static culture with or without treatment. When used, the drugs were applied during the 6-day static period and during the 24-h flow period. As marked by an increase in the relative DAF-FM diacetate mean fluorescence intensity, healthy viECs produced higher nitric oxide (NO) levels when exposed to shear stress compared with the same viECs under static conditions (Fig. 3a,b). In contrast, HGPS viECs from both donors exhibited reduced levels of NO production after exposure to shear stress, consistent with a dysfunctional endothelial phenotype (Fig. 3a,b)31. Lonafarnib treatment, Everolimus treatment, and combination treatment all restored NO production under shear stress to healthy levels in HGPS viECs from both donors (Fig. 3a,b). (Representative images are shown in Fig. S9).
Lonafarnib and Everolimus improve NO production and flow-mediated gene expression in HGPS viECs exposed to physiological shear stress. (a, b) DAF-FM diacetate (DA) mean fluorescence intensity (MFI) in healthy (gray), HGPS 167 CL2 (a), and HGPS 003 CL1D (b) viECs exposed to 12 dynes/cm2 for 24 h. Data presented as fold change DAF-FM DA MFI under shear stress compared with static condition from the same group. Gray bar represents healthy viECs. (c-i) Gene expression of (c) NOS3, (d) KLF2, (e) NRF2, (f) NQO1, (g) TXNRD1, (h) GCLM, and (i) GCLC in HGPS 167 CL2 viECs exposed to 12 dynes/cm2. Data were normalized to GAPDH expression and gene expression in viECs exposed to shear stress was normalized to respective viECs under static culture from the same group. Dashed line indicates normalized static control. Data presented as mean ± SD. N = 3 experiments per group. Asterisks indicate significant differences between each shear stress value and its own static control: *p < 0.05, **p < 0.01, ***p < 0.001, #p < 0.0001. Letters indicate significant differences between treatment groups under shear stress: groups labeled with different letters are significantly different from each other (p < 0.05). Exact p-values for significant differences for comparisons between treatment groups under shear stress are provided in Table S2.
We tested the expression of several shear stress-sensitive genes in healthy viECs and HGPS viECs with or without Lonafarnib and Everolimus. NO production is regulated by the expression of endothelial NO synthase which is encoded in the NOS3 gene. KLF2 (Kruppel-like factor 2) is a major anti-inflammatory and anti-thrombotic flow-responsive transcription factor which is upregulated by EC exposure to laminar flow and regulates ∼ 46% of EC flow-sensitive genes. KLF2 activates antioxidant transcription factor nuclear- erythroid 2-related factor 2 (NRF2) and both of these genes promote EC quiescence32. Since NRF2 transcription and nuclear location are affected by progerin33, we evaluated several NRF2 downstream targets: NAD(P)H-quinone oxidoreductase 1 (NQO1), thioredoxin reductase 1 (TXNRD1), glutamate-cysteine ligase modifier subunit (GCLM), and glutamate-cysteine ligase catalytic subunit (GCLC)34. While NOS3 expression did not significantly change in healthy viECs exposed to shear stress compared with viECs under static conditions, the expression of KLF2, NRF2, and NRF2 targets NQO1, TXNRD1, GCLM, and GCLC were upregulated (Fig. 3c–i). In contrast, after being exposed to 12 dynes/cm2 for 24 h, HGPS viECs from donor 167 downregulated NOS3 (Fig. 3c) while NRF2 and GCLC expression did not significantly change (Fig. 3e,i). NQO1, TXNRD1, and GCLM were upregulated by HGPS viECs to a significantly lesser extent than healthy viECs under the same shear stress conditions (Fig. 3f–h). KLF2 expression in HGPS viECs was reduced compared with healthy viECs but was not statistically significant (p = 0.0503, Fig. 3d).
Lonafarnib treatment did not affect NOS3 expression by HGPS viECs (Fig. 3c). In HGPS viECs treated with Everolimus and combination treatment, NOS3 was restored to a level not significantly different from static culture, similar to healthy viECs (Fig. 3c). Lonafarnib did significantly increase expression of KLF2, NRF2, NQO1, TXNRD1, GCLM, and GCLC in HGPS viECs from donor 167 to levels similar to or higher than healthy expression levels (Fig. 3d–i). Everolimus significantly increased expression of KLF2, NQO1, TXNRD1, and GCLM but to levels significantly lower than Lonafarnib treatment (Fig. 3d,f–h). Everolimus treatment had no significant effect on NRF2 or GCLC expression (Fig. 3e,i). Combination treatment did not offer any additional improvement over Lonafarnib monotherapy for KLF2, NRF2, NQO1, TXNRD1, GCLM, or GCLC.
Lonafarnib and Everolimus improve HGPS TEBV function and expression of SMC and EC markers
HGPS TEBVs were perfused for 3 weeks without treatment to develop a disease phenotype and then were left untreated or were treated with 1 µM Lonafarnib alone, 0.1 µM Everolimus alone, or a combination of 1 µM Lonafarnib and 0.05 µM Everolimus for one week. We previously showed that compared with TEBVs made with healthy cells, HGPS TEBVs exhibit significantly reduced vasoconstriction when exposed to 1 µM phenylephrine and significantly reduced vasodilation when exposed to 1 µM acetylcholine10. Lonafarnib treatment did not affect vasoconstriction in response to phenylephrine but resulted in significantly increased dilation in response to acetylcholine in HGPS TEBVs compared to untreated and vehicle controls (Fig. 4a,b). In contrast, HGPS TEBVs treated with Everolimus alone showed significantly greater constriction compared to untreated and vehicle controls, but no significant effect on dilation (Fig. 4a,b). Combination treatment with 1 µM Lonafarnib and 0.05 µM Everolimus showed a statistically significant additive effect upon dilation and constriction. The maximum level of vasoconstriction and vasodilation is comparable to TEBVs made with viECs and viSMCs from non-HGPS donors10. Treatment with the toxic combination dose of 2 µM Lonafarnib and 0.1 µM Everolimus did not improve vasodilation or vasoconstriction. TEBV treatment with Lonafarnib or Everolimus for two weeks did not offer any additional benefit in vasoactivity over 1-week treatment (Fig. S10a,b).
Lonafarnib and Everolimus treatment improves HGPS TEBV vasoactivity. HGPS 167 CL2 TEBVs were matured for 3 weeks, treated with Lonafarnib (LF) and Everolimus (Ev) for 1 week, then exposed to 1 µM phenylephrine followed by 1 µM acetylcholine for 5 min each and diameter change measured. Data presented as mean ± SD. N = 3 experiments per group. Results are compared to the reference condition without Lonafarnib and Everolimus.
We previously showed that HGPS TEBVs express decreased levels of SMC and EC markers compared with healthy TEBVs10. Lonafarnib significantly increased the expression of contractile SMC proteins calponin and myosin heavy chain-11 (MHC11) in HGPS TEBVs, while Everolimus significantly increased α-smooth muscle actin (αSMA) and calponin expression (Fig. 5a–c). Treatment with a combination of 1 µM Lonafarnib and 0.05 µM Everolimus combined the benefits of both single-drug treatments, resulting in significantly increased levels of αSMA, calponin, and MHC11 (Fig. 5a–c). Neither Lonafarnib alone nor Everolimus alone significantly improved endothelial cell markers vascular endothelial cadherin (VE-Cadherin), platelet endothelial cell adhesion molecule (PECAM), or von Willebrand factor (vWF) (Fig. 5d–f). While combination treatment with 1 µM Lonafarnib and 0.05 µM Everolimus did not affect VE-Cadherin or PECAM expression (Fig. 5d,e), this treatment did significantly elevate vWF expression (Fig. 5f). Treatment with the toxic combination of 2 µM Lonafarnib and 0.1 µM Everolimus resulted in low expression levels of all 6 proteins which did not differ from untreated HGPS TEBVs (Fig. 5a–f). Representative images of αSMA, calponin, MHC11, VE-Cadherin, PECAM, and vWF expression with Lonafarnib treatment, Everolimus treatment, and the toxic combination are shown in Fig. S11.
Lonafarnib and Everolimus treatment improves expression of contractile proteins in HGPS TEBVs. HGPS 167 CL2 TEBVs were matured for 3 weeks, treated with Lonafarnib (LF) and Everolimus (Ev) for 1 week and then processed for immunostaining. Representative images and quantification of SMC markers α-smooth muscle actin (αSMA, a), calponin (b), and myosin heavy chain-11 (MHC11, c) and endothelial markers vascular endothelial cadherin (VE-Cadherin, d), platelet endothelial cell adhesion molecule (PECAM, e), and von Willebrand factor (vWF, f). Scale bar: 50 µm. Data presented as mean ± SD. N = 3 experiments per group. Results are compared to the reference condition without Lonafarnib and Everolimus using Dunnett’s test. Representative images for the 1 µM Lonafarnib treatment, 0.1 µM Everolimus treatment, and 2 µM Lonafarnib + 0.1 µM Everolimus treatment conditions are shown in Fig. S11.
Lonafarnib and Everolimus mitigate disease pathology in HGPS TEBVs
We assessed the effects of Lonafarnib and Everolimus on disease pathology using the HGPS TEBV model10,35. HGPS TEBVs express progerin, stain strongly for extracellular matrix proteins fibronectin and collagen IV, and express the inflammatory marker vascular cell adhesion molecule 1 (VCAM-1) (Fig. 6a). Based on comparison of immunostaining images, treatment with Lonafarnib alone did not affect progerin expression, whereas treatment with Everolimus alone and Everolimus in combination with Lonafarnib reduced progerin expression in HGPS TEBVs (Fig. 6a, Fig. S12a). Additionally, Everolimus alone was insufficient to reduce fibronectin, collagen IV, or VCAM-1 expression, while Lonafarnib alone or in combination with Everolimus reduced expression of all 3 proteins (Fig. 6a, Fig. S12a,b). Treatment with the toxic combination of 2 µM Lonafarnib and 0.1 µM Everolimus did not reduce expression of fibronectin, collagen IV, or VCAM-1 and resembled the untreated HGPS case (Fig. S12a,b).
Lonafarnib and Everolimus treatment mitigates HGPS TEBV pathology. Representative immunofluorescence images of (a) progerin, fibronectin (FN), collagen IV (Col-IV), vascular cell adhesion molecule-1 (VCAM-1), and (b) H&E staining, alizarin red staining, and TUNEL staining in HGPS 167 CL2 TEBVs matured for 3 weeks then treated with 1 µM Lonafarnib (LF) + 0.05 µM Everolimus (Ev) for 1 week. Scale bar 50 µm for progerin, FN/Col-IV, and VCAM-1, and 200 µm for H&E, alizarin red, and TUNEL. (c) Quantification of alizarin red-positive area in untreated and treated HGPS 167 Cl2 TEBVs. (d) Nuclei density and (e) TEBV thickness in untreated and treated HGPS 167 CL2 TEBVs. Data presented as mean ± SD. N = 3 TEBVs per group for alizarin red and 4 TEBVs per group for cell density. *p < 0.05, **p < 0.01 compared with untreated case. Representative images for the 1 µM Lonafarnib treatment, 0.1 µM Everolimus treatment, and 2 µM Lonafarnib + 0.1 µM Everolimus treatment conditions are shown in Figs. S12 and S13.
Compared with healthy TEBVs35, HGPS TEBVs displayed lower medial cell density, stained strongly for Alizarin Red, and showed increased TUNEL staining (Fig. 6b–d and Fig. S13). Treatment with Lonafarnib alone reduced calcification as indicated by Alizarin Red staining and increased medial cell density but did not reduce apoptosis marked by TUNEL staining (Fig. 6c,d, Fig. S13b). Treatment with Everolimus alone showed little improvement in calcification (Alizarin Red) or in TUNEL staining (Fig. 6c, Fig. S13b). Treatment with the toxic combination of 2 µM Lonafarnib and 0.1 µM Everolimus did not significantly reduce Alizarin Red staining and resulted in increased TUNEL staining (Fig. 6c, Fig. S13b). Everolimus alone was also not sufficient to significantly increase medial cell density in the TEBVs (Fig. 6d). Treatment with a combination of 1 µM Lonafarnib and 0.05 µM Everolimus significantly decreased Alizarin Red staining, increased cell density, and reduced TUNEL staining (Fig. 6c,d, Fig. S13b). H&E staining indicated that all treatment conditions resulted in reduced wall thickness in HGPS TEBVs (Fig. 6e) and comparable to levels reported for TEBVs made with non-HGPS viECs and viSMCs10.
We assessed expression of ATG7, LC3-I/LC3-II and p62 which are all associated with autophagy and are upregulated after Everolimus treatment in progerin-expressing cells to help clear progerin26. Immunohistochemical staining was done as described previously36 and shows that HGPS TEBVs treated with Everolimus alone or in combination with Lonafarnib at both therapeutic and toxic doses express all three autophagy proteins at more abundant levels compared to untreated controls (Fig. S14). These results are consistent with studies of progerin clearance in 2D fibroblast cultures from HGPS patients37. The presence of these autophagy associated proteins along with reduced level of progerin after Everolimus treatment in HGPS TEBVs suggests that Everolimus helps clear progerin through autophagy.
Discussion
In this study, we showed that a TEBV model of HGPS, together with 2D cell culture, can assess the effect of a combination of drugs on cell and blood vessel function. In HGPS viSMCs and viECs, Everolimus treatment reduced progerin expression and misshapen nuclei in a dose-dependent manner. Everolimus treatment also improved proliferation in one donor and reduced ROS levels and DNA double stranded breaks in both donors. Lonafarnib had a more limited effect on these phenotypes which varied between donors. These results are consistent with studies that showed that rapamycin decreases DNA damage in HGPS fibroblasts while Lonafarnib does not, and that rapamycin also decreases ROS levels in HGPS fibroblasts38,39. Treatment with a combination of Lonafarnib and Everolimus showed a beneficial effect upon SMC proteins and vasoactivity.
Progerin expression varies among cell and tissue types and SMCs are among the highest progerin-expressing cells, making them a major target of study in HGPS13,14. However, less is known about the role of the endothelium in the disease, despite the well-established role of EC senescence and dysfunction in non-HGPS atherosclerosis13. Over 85% of in vitro pre-clinical studies of HGPS therapeutics between 2010 and 2020 were performed with fibroblasts from HGPS patients40. HGPS fibroblasts accumulate double stranded DNA breaks over several passages and have elevated ROS levels41,42,43. Further, NRF2 transcription and nuclear location are affected by progerin expression44 and antioxidant treatment of HGPS fibroblasts alleviates characteristic HGPS nuclear abnormalities45. HGPS ECs show reduced NO production, impaired mechanosensitivity, and increased inflammation13,46,47.
Mice in which only ECs express the LMNAG609G/G609G mutation exhibit cardiac fibrosis13,46 and a pro-inflammatory senescence associated secretory phenotype (SASP)48. Mice in which only the endothelial cells express progerin exhibit reduced vasodilation47 as well as increased collagen and adventitial thickening, which is found in HGPS patients13. Further, in the murine model in which only ECs express the LMNAG609G/G609G mutation, the myocardin-related transcription factor A (MRTFA) binds to NOS3, which inhibits NO release. Blocking MRFTA restores NO activity48. Likewise, in this murine HGPS model, progerin reduces SIRT7 levels in ECs and vascular EC specific SIRT7 overexpression significantly reduced inflammation and promoted survival47. Thus, restoring EC function significantly improves vascular function and could improve overall survival of individuals with HGPS47. A limitation of these animal studies is that most do not replicate the human vascular pathology.
The HGPS TEBV system models key functional features of arteries under physiological conditions and replicates pathology observed in the disease. As we showed previously10, iPSC-derived viSMCs express high levels of αSMA, calponin and MHC11, a terminal marker of SMC differentiation. The viSMCs also exhibit contractile potential that is increased compared with a previous generation of the HGPS TEBV model we reported, resulting in increased vasoconstriction and vasodilation in healthy TEBVs and a larger detectable difference in vasoactivity between healthy and HGPS TEBVs35. The viECs express CD31, CD144 in 99% of the cells, as well as VEGF receptor 2 and von Willebrand Factor10. In healthy TEBVs, viECs produce nitric oxide which results in TEBV vasodilation. Limitations of the model are that the iPSC-derived cells are not as mature as primary cells and TEBVs do not incorporate elastin. Our experiments only tested the effects of Lonafarnib and Everolimus at a limited timescale of 7 and 14 days, but the TEBV model can show the response to therapeutics.
Despite the large influence of shear stress on EC phenotype, only one other study explored HGPS ECs under shear stress conditions and showed reduced NO production in HGPS ECs compared with healthy cells49. We previously showed that HGPS viECs exhibit abnormal nuclear morphology, impaired tube formation, and blunted shear stress-responsive gene expression10. We also reported a specific role of the endothelium in HGPS TEBV vasoactivity reduction and inflammatory protein expression10. Here we showed that HGPS viECs from two donors exhibit characteristics of endothelial dysfunction including elevated ROS levels, DNA damage, and reduced NO production when exposed to physiological shear stress. These results support the growing body of data that indicate that ECs may play a role in mediating HGPS atherosclerotic development.
Limited results exist on the effect of both Lonafarnib and Everolimus on ECs, SMCs, or the vessel wall. Human coronary ECs exogenously expressing progerin exhibit increased oxidative stress and DNA damage48. Treatment of these cells with zoledronate and pravastatin, which respectively block farnesylation and geranylation, reduces the abundance of the pathogenic progerin protein and consequently decreases oxidative stress. Human umbilical vein endothelial cells overexpressing progerin exhibited increased cell loss after 72 h exposure to 12 dynes/cm2 which was reversed after addition of Lonafarnib50.
In the current study, Lonafarnib and Everolimus treatment rescued NO production under shear stress conditions to healthy levels both when administered alone and in combination. Lonafarnib also significantly increased expression of shear stress-responsive genes to healthy levels or higher while Everolimus had a more limited effect. These results are consistent with a recent study that showed Lonafarnib improves shear stress adaptability in progerin-expressing ECs50. In accordance with reduced NO levels after exposure to shear stress, HGPS viECs also exhibited reduced expression of the NOS3 gene. However, while NO levels increased under shear stress in healthy viECs and treated HGPS viECs, this did not correlate with a significant increase in NOS3 levels. This indicates that the changes in NO production may be due to causes other than gene regulation such as post-translational modifications of the NOS3 protein (eNOS) which affects its activity. HGPS iPSC-derived ECs express lower levels of total eNOS protein as well as increased abundance of phosphorylated threonine 495 eNOS, which inhibits eNOS activity49.
HGPS patients develop severe atherosclerosis with vascular dysfunction, stiffening, calcification, inflammation, fibrotic plaque formation, and medial SMC depletion51. We previously showed that compared with healthy TEBVs, HGPS TEBVs exhibit reduced vasoactive response to phenylephrine and acetylcholine, decreased contractile markers indicating SMC loss, progerin expression, inflammatory marker expression, and excess ECM deposition10. Lonafarnib alone was sufficient to improve endothelium-dependent vasodilation in response to acetylcholine, contractile marker expression, ECM deposition, calcification, inflammatory marker expression, and cell retention. However, Lonafarnib alone did not significantly affect vasoconstriction in response to phenylephrine, progerin expression, or TUNEL expression. In contrast, Everolimus alone was only sufficient to decrease progerin expression, improve vasoconstriction in response to phenylephrine, and increase some contractile markers, but did not affect ECM deposition, inflammatory marker expression, or calcification. These results are consistent with our previous report that Everolimus did not improve calcification or apoptosis in a previous HGPS TEBV model35. Treatment with both 1 µM Lonafarnib and 0.05 µM Everolimus combined the beneficial effects of both therapeutics and resulted in a significant increase in endothelial marker vWF and decrease in apoptosis (TUNEL expression), which were not improved by either drug alone. Treatment with the toxic combination of 2 µM Lonafarnib and 0.1 µM Everolimus did not improve disease characteristics and exacerbated TUNEL expression, indicating the importance of proper dosing when combining the two therapies to avoid toxicity. Since the treatment was administered after maturation of the HGPS TEBVs, these results indicate that drug treatment can reverse the phenotypes we tested after the onset of symptoms.
Our results indicate that Everolimus decreases progerin expression, leading to a reduction in ROS levels and DNA damage and improvement in vascular cell proliferation. Everolimus treatment also improves contractile marker expression in HGPS TEBVs and strengthens vasoconstriction. While Everolimus does enhance NO production in HGPS viECs exposed to shear stress, this did not translate to a significant improvement in EC-dependent vasodilation in HGPS TEBVs. The beneficial effects of Everolimus on TEBV structure and function are due, in part, to increased autophagy (Fig. S14). In contrast, Lonafarnib not only improves NO production in HGPS viECs exposed to shear stress, but also significantly increases expression of shear stress-responsive genes. This correlates with a significant improvement in vasodilation in Lonafarnib-treated HGPS TEBVs. Lonafarnib also dominates the positive effect on ECM deposition, cell retention, inflammatory marker expression, and calcification. Lonafarnib was effective at increasing viSMC proliferation.
The Lonafarnib dose in this study (1 µM) is below the typical mean plasma levels of HGPS patients on this drug (1.7–2.8 µM)15, but is still effective in improving cell and TEBV function. The dose of Everolimus that led to significant effects in 2D and TEBV studies, 0.05 µM, is slightly less than that used in other in vitro studies26,29. For the ongoing clinical trial with Lonafarnib and Everolimus, plasma levels of Everolimus are reported to be comparable to values in other studies with this drug52. Reported mean levels in cancer studies53 are as high as 6.26–12.52 nM, which is on the lower level of effectiveness in this study. Plasma Everolimus levels between 18 and 27 nM are reported to cause oral mucosal ulcers in cancer patients54,55. Everolimus does not appear to cause any vascular problems56 and may have some additional beneficial anti-inflammatory effects57. Lonafarnib is used to treat infections and side effects include nausea and diarhea58. Other than the results of this study, there is no information on adverse effects of the combination of Lonafarnib and Everolimus. While normal TEBVs may be a way to test drug toxicity, the HGPS TEBVs appear to be more sensitive than the healthy TEBVs. Thus, we did not observe toxicity with 0.05 µM EV and 1 µM LF for HGPS TEBVs for this time period.
Overall, our results show that combining Lonafarnib and Everolimus resulted in additional benefits over either therapy alone (Table S4). However, proper dosing of the drugs is essential to identify the therapeutic window. Since calcification would affect vessel stiffness, the most likely vascular response detected would be a decrease in pulse wave velocity. While such changes have been observed with Lonafarnib15, detecting an additional beneficial effect of the drug combination may be challenging. Additional study is warranted to better understand the long-term effects of these therapies and how they might interact in different tissues.
Materials and methods
Experimental design
We tested phenotypes previously identified in HGPS fibroblasts or mice but not tested in vascular cells or human-derived blood vessels, including reactive oxygen species levels, proliferation, and DNA damage, endothelial shear stress response, blood vessel vasoactivity, progerin expression, extracellular matrix deposition, inflammation, calcification, and apoptosis. To test the effects of Lonafarnib and Everolimus on these phenotypes, we performed studies in iPSC-derived smooth muscle cells (viSMCs), endothelial cells (viECs), and tissue engineered blood vessels. All experiments were independently performed at least 3 times and 2D cell culture experiments were repeated in two HGPS donors.
Cell culture and isolation
Primary skin fibroblasts with the classic LMNA Exon 11 1824 C>T HGPS mutation (167 CL2) and control primary skin fibroblasts (168 CL2) were provided by the Progeria Research Foundation and converted to iPSCs by the Duke iPSC Shared Resource Facility. iPSCs with the classic LMNA Exon 11 1824 C>T HGPS mutation (003 CL1D) were provided by the Progeria Research Foundation. The Duke University Health System Institutional Review Board has determined that the protocol using these cells meets the definition of research not involving human subjects as described in 45 CFR 46.102(f), 21 CFR 56.102(e) and 21 CFR 812.3(p) and satisfies the Privacy Rule as described in 45CFR164.514. iPSCs were maintained in feeder-free conditions on hESC-qualified Matrigel (BD biosciences) in mTeSR Plus (StemCell Technologies). Cells were passaged at 80–90% confluency with 0.5 mM EDTA (Invitrogen) for maintenance culture or with Accutase (StemCell Technologies) and 10 µM ROCK inhibitor Y-27632 (Tocris Bioscience) for differentiation.
viSMC and viEC differentiation
viSMCs and viECs were differentiated using a modified protocol as previously described10,59. iPSCs were dissociated on day 0 with Accutase and replated on Matrigel-coated plates at 37,000 cells/cm2 for viSMC differentiation or 47,000 cells/cm2 for viEC differentiation. On day 1, the medium was replaced with mesoderm induction medium consisting of N2B27 medium (1:1 mix of Neurobasal medium and DMEM/F12 with HEPES supplemented with N2 and B27 minus vitamin A, all Gibco) with 25 ng/mL BMP4 (PeproTech) and 8 µM CHIR99021 (Cayman Chemical). The media was not changed for 3 days to induce a mesoderm state.
For viSMC differentiation, media was changed on day 4 to viSMC induction medium consisting of N2B27 medium, 10 ng/mL PDGF-BB (PeproTech), and 2 ng/mL Activin A (PeproTech). The viSMC induction medium was changed daily. On day 6, cells were dissociated with Accutase and re-plated on collagen-coated plates in viSMC media comprised of N2B27 media supplemented with 2 ng/mL Activin A and 2 µg/mL heparin (Sigma-Aldrich) to induce a contractile SMC phenotype. Media was changed every other day and viSMCs were routinely passaged at 80–90% confluency using Accutase onto collagen-coated plates. viSMCs were used between passages 4–6.
For viEC differentiation, on day 4 the media was changed to viEC induction media consisting of Stempro-34 SFM medium (Thermo Fisher), 200 ng/mL VEGF165 (Genscript), and 2 µM forskolin (Sigma-Aldrich). The viEC induction media was changed daily and conditioned media was collected on days 5, 6, and 7. On day 7, viECs were dissociated with Accutase and MACS separated for CD31+CD144+, CD31−CD144+ and CD31+CD144− cells. viECs were fed with viEC conditioned media consisting of 1:1 fresh Stempro-34 SFM and conditioned medium supplemented with 2 µg/mL heparin until the first passage, then cultured in viEC media consisting of Stempro34-SFM supplemented with 50 ng/mL VEGF and 2 µg/mL heparin.
viEC magnetic activated cell sorting (MACS)
On day 7 of the viEC differentiation, cells were dissociated with Accutase and neutralized with cold Stempro-34 SFM medium. Cells were spun at 1000 rpm for 5 min and then washed with MACS buffer consisting of PBS with 0.5% BSA and 2 mM EDTA. Cells were resuspended in 8 µL buffer per million cells and co-stained for 15 min on ice with 2 µL per million cells each FCR blocking reagent (Miltenyi Biotec), CD31 microbeads (Miltenyi Biotec), and CD144 microbeads (Miltenyi Biotec) antibody. Cells were rinsed once with MACS buffer then resuspended in 1 mL MACS buffer and ran through an LS column attached to a MACS separator (Miltenyi Biotec) to discard CD31-CD144- cells.
Cell and TEBV drug treatment
Cells and TEBVs were treated with Lonafarnib, Everolimus, or a combination of the two drugs for a total of 7 days. Drugs were added every 2 days with media change for a total of 3 times throughout the treatment. The control “no treatment” condition cells were cultured for 7 days along with the treated cells with media change every 2 days but without any addition of drug to the media.
viEC parallel plate flow studies
The effect of physiological shear stresses on healthy and HGPS viECs was evaluated as described previously10,60. viECs were seeded at a density of 33,000 cells/cm2 onto slide flasks (Nunc) or collagen-coated glass slides and allowed to attach and develop a confluent monolayer in the flasks. Once fully confluent, flasks were incorporated in a parallel plate flow chamber connected to a closed circular flow loop60. Steady laminar flow was applied using a pre-conditioning treatment as described previously60,61 to maintain cell attachment. Once 12 dynes/cm2 was reached, cells were maintained in the flow circuit for 24 h. After 24 h under flow, viECs were removed from the circuit and tested for NO production or shear stress-sensitive gene expression.
RT-PCR and primers
Total RNA was extracted from normal and HGPS viECs using the RNeasy Mini Kit (Qiagen). RNA was reverse transcribed into cDNA using the iScript cDNA Synthesis Kit (Bio-Rad). RT-PCR was performed in triplicate using the iQ SYBR Green Supermix (Bio-Rad) and the CFX Connect Real-Time PCR Detection System (Bio-Rad). Human GAPDH served as an internal control (VHPS-3541, Real-time primers). Primer sequences for human NOS3, KLF2, NRF2, NQO1, TXNRD1, GCLM and GCLC are as follows: NOS3-F: 5′‐CAT CTT CAG CCC CAA ACG GA‐3′, NOS3-R: 5′‐AGC GGA TTG TAG CCT GCA AC‐3′; KLF2-F: 5′‐GCA CGC ACA CAG GTG AGA AG‐3′, KLF2-R: 5′‐ACC AGT CAC AGT TTG GGA GGG‐3′; NRF2-F: 5′‐AAA CCA CCC TGA AAG CAC AG‐3′, NRF2-R: 5′‐AGT GTT CTG GTG ATG CCA CA‐3′; NQO1-F: 5′‐GGC ATT CTG CAT TTC TGT G‐3′, NQO1-R: 5′‐GGC GTT TCT TCC ATC CTT C‐3′; TXNRD1-F: 5′‐CTG CGG GAG AAA AAA GTC G‐3′, TXNRD1-R: 5′‐CAG GGA TGC CCA AGT AAC G‐3′; GCLM-F: 5′‐TCA GGG AGT TTC CAG ATG TC‐3′, GCLM-R: 5′‐CAA TAG GAG GTG AAG CAA TG‐3′; GCLC-F: 5′‐CAA GAG AAG GGG GAA AGG AC‐3′, GCLC-R: 5′‐GAC CTC GGG CAG TGT GAA C‐3′.
To account for variability among experiments, a normalization factor was calculated by dividing the global average of all conditions within each group by the average of each experiment10. Normalized values were calculated for each experiment by scaling each data point by the normalization factor of the experiment.
DAF-FM DA NO assay
viECs were maintained in static culture in viEC media with or without therapeutics for 6 days. Media was changed every 2 days. On day 6, media was changed on static samples. Other samples were exposed to 12 dynes/cm2 for 24 h. On day 7, static and flow samples were exposed to 1 µM DAF-FM diacetate (Thermo Fisher) in serum free viEC media for 30 min at 37 °C. Cells were rinsed once with PBS and then maintained in fresh media for 15 min to allow for de-esterification of intracellular diacetates. Images were taken on a Zeiss 880 Airyscan inverted confocal microscope at 20 × magnification and analyzed for mean fluorescence intensity using FIJI.
Quantification of misshapen nuclei
The nuclei per field were quantified from Hoechst-stained nuclei using FIJI. The number of misshapen nuclei in each image were identified as those with abnormal protrusions or shape (Fig. S4a). To correct for differences in cell dispersion, the fraction of misshapen nuclei per total number of nuclei in each frame was determined. Three independent experiments were performed, and three image frames were quantified per experiment. Manual analysis of misshapen nuclei was performed with blinded images to prevent bias.
TEBV fabrication, treatment, and testing
TEBVs were fabricated as previously described10. Briefly, 1.5 × 106 viSMCs were dissociated with Accutase, resuspended in 300 µL of viSMC media, and incorporated in a 2.05 mg/mL rat tail collagen type 1 solution (Corning) in 0.6% acetic acid. Serum-free 10 × DMEM (Sigma-Aldrich) was added at a 1:10 ratio to the collagen solution. The pH of the solution was raised to 8.5 by adding 5 M NaOH, causing gelation. Prior to complete gelation, the solution was placed in a 3 mL syringe mold (BD Biosciences) with the stopper completely removed and a two-way luer-lock stopcock attached to the luer-lock end of the syringe. An 800 μm diameter steel mandrel was inserted in the middle of the solution in the syringe and held in the center with parafilm wrapped over the syringe opening. The solution gelled for 30 min at room temperature prior to compression. After gelation, the vessel construct was transferred onto 0.2 μm nylon filter paper (Whatman) on 10 KimWipes. Plastic compression was applied to the construct by suspension in the filter paper for seven minutes to remove over 90% of the water content. Once compressed, TEBVs were placed in custom perfusion chambers and sutured onto grips at both ends.
After gelation and mounting, TEBVs were endothelialized by resuspending 0.5 × 106 viECs in 0.5 mL viEC media. viECs were perfused through the TEBV lumen using 1 mL luer-lock syringes (BD) connected to the grips of the custom chambers. The TEBVs were evenly endothelialized by rotating the chambers on a rotator at a rate of 10 revolutions per hour for 30 min at 37ºC. Chambers were attached to a flow circuit containing a 25 mL media reservoir connected by tubing. Continuous, steady, laminar flow at 2 mL/min was applied to the TEBVs by attaching the perfusion circuit to a peristaltic pump that applied a physiological shear stress of 6.8 dynes/cm2 (Masterflex). The TEBVs were matured for up to 5 weeks in viSMC media and the media was changed 3 times per week.
HGPS TEBVs were matured for 3 weeks without treatment to develop the HGPS phenotype, then treated by adding different combinations of Lonafarnib (Progeria Research Foundation) and Everolimus (StemCell Technologies) in the media. Lonafarnib and Everolimus were replenished with each media change, every 2 days for 1–2 weeks. Vasoactivity was tested weekly to determine function before and after treatment.
TEBV vasoactivity was measured at room temperature in the perfusion circuit with the TEBVs and imaged using a stereoscope (AmScope) while being recorded with ISCapture software. After 30 s of normal perfusion, 1 μM phenylephrine was added to a syringe port (Ibidi) integrated in the flow circuit. Five minutes later, 1 μM acetylcholine was added to the same port. TEBV diameter was measured based on images taken prior to phenylephrine addition, 5 min after phenylephrine addition, and 5 min after acetylcholine addition. The diameter at each time point was determined by averaging 4 random widths along the length of the vessel using ImageJ. Vasoconstriction was calculated as the percent change in diameter from the initial diameter at baseline to the diameter after 5 min of exposure to phenylephrine. Vasodilation was calculated as the percent change in diameter from the constricted state to the diameter after 5 min of exposure to acetylcholine.
Immunostaining
For 2D viSMC and viEC immunostaining analysis, cells were fixed with 4% paraformaldehyde for 15 min and then washed 3 times with PBS. Cells were permeabilized with 0.1% Triton-X for 5 min at room temperature and then rinsed with PBS 3 times. Cells were blocked in 10% goat serum in PBS for 1 h at room temperature. Cells were then incubated with primary antibody at 1:200 in 10% goat serum overnight at 4ºC. Cells were then washed with PBS 3 times and incubated with secondary antibody at 1:500 in 10% goat serum for 2 h at room temperature. Cells were washed 3 times with DPBS and incubated with Hoechst 33342 at 1:1000 in DPBS for 5 min at room temperature. Samples were then washed 3 times with DPBS and imaged on a Zeiss 880 Airyscan inverted confocal microscope at 20X magnification. Images were analyzed using FIJI.
For 3D TEBV immunostaining analysis, TEBVs were fixed in 10% formalin for 10 min while still attached to the chamber grips. TEBVs were then placed in a well-plate and further fixed in 10% formalin for an additional 50 min. TEBVs were washed with DPBS 3 times and cut en face to expose the inner lumen. Sections were permeabilized with 0.1% Triton-X for 10 min and then washed 3 times with DPBS. Samples were blocked in 10% goat serum in PBS for 8 h at room temperature, then incubated with primary antibody (1:100) in 10% goat serum overnight at 4 °C. Samples were washed 3 times with PBS and then incubated with secondary antibody (1:500) in 10% goat serum for 1 h at room temperature. Samples were washed 3 times and incubated with Hoechst 33342 (1:1000) in DPBS for 5 min at room temperature. Samples were washed 3 times with DPBS and mounted onto glass slides with FluorSave (Millipore). TEBVs were imaged using a Zeiss 510 inverted confocal microscope at 20× or 40× magnification and analyzed using Zeiss LSM image browser.
Primary antibodies used were rabbit anti-ki67 (Abcam), rabbit anti-calponin (Abcam), rabbit anti-αSMA (Abcam), rabbit anti-MyHC11 (Abcam), rabbit anti-vWF (Abcam), mouse anti-fibronectin (Abcam), rabbit anti-collagen IV (Abcam), mouse anti-ɣH2A.X (Abcam), mouse anti-CD31 (BD biosciences), mouse anti-VE-cadherin (Santa Cruz), rabbit anti-VCAM-1 (Abcam), mouse anti-E-selectin (Santa Cruz), mouse anti LC3 (Axxora/Enzo), rabbit polyclonal anti-ATG7 (Sant Cruz), rabbit polyclonal anti-p62 (Enzo), mouse anti-lamin A/C (Abcam) and rabbit anti-progerin26. Anti-MyHC11, anti-VE-Cadherin, and anti-E-selectin were used at 1:100 dilution. Anti-LC3 was used at 1:20 dilution and anti-lamin A/C was used at 1:1000. Other primary antibodies were used at 1:200 dilution. Secondary antibodies used were Alexa Fluor 594 goat anti-rabbit and Alexa Fluor 488 goat anti-mouse (Thermo Fisher) at 1:500 dilution and Hoechst 33342 (Fischer Scientific) at 1:1000 dilution.
Immunohistochemistry imaging and analysis
TEBVs were fixed in 10% formalin for 10 min attached to the chamber grips to maintain structural integrity. TEBVs were then placed in a well-plate and fixed in 10% formalin for an additional 50 min for a total of one hour of fixation. TEBVs were maintained in DPBS or 70% ethanol until paraffin embedded. Paraffin embedded cross-sections were then stained with hematoxylin/eosin, Alizarin Red, and TUNEL as previously described35. Human bone was used as a positive Alizarin Red control and mouse liver was used as a positive TUNEL control. Images were taken on a Nikon Eclipse TE2000-U microscope at 20× magnification using the NIS Elements software and analyzed using ImageJ.
Live/dead staining
viSMCs and viECs were incubated in their respective media containing 2 μM Calcein AM and 1 μM Ethidium homodimer-1 (Life Technologies) for 1 h at room temperature. viSMCs were then imaged on a fluorescent microscope at 10× and analyzed using ImageJ. Live cells were quantified by determining the maxima of the FITC (green) channel and dead cell counts were quantified by determining the maxima of the TXRED (red) channel. The percent of live and dead cells was determined by dividing the live or dead cell count by the total number of live and dead cells.
Western blotting
Whole cell lysates were prepared by dissolving cells in Laemmli Sample Buffer containing 5% of 2-mercaptoethanol (Bio-Rad). Antibodies used for immunoblotting include: anti-progerin26 with 1:500 dilution, and mouse anti-GAPDH (sc-47724; Santa Cruz) with 1:3000 dilution. Protein samples were loaded on 12% polyacrylamide gels (Bio-Rad) and run at 100 V until the dye front reached the bottom of the gel. The proteins were transferred onto 0.45 μm pore-size nitrocellulose membranes (Bio-Rad) using Turboblot (Bio-Rad). Then, blots were probed with primary and secondary antibodies, and images were developed using ECL (Bio-Rad). Image Lab software (Bio-Rad) was used for image capture and analysis. For each experiment, all conditions (control, DMSO vehicle and Everolimus does) run on the same gel. For each gel, values are normalized to control and then the results of the different gels averaged. Complete gels are shown in Fig. S2.
DCFDA assay
viSMCs and viECs were cultured with or without therapeutics for 7 days, then washed once with 1X buffer (Abcam). Cells were incubated with 10 µM DCFDA solution (Abcam) diluted in 1× buffer for 5 min at room temperature. After staining, cells were washed with 1× buffer twice and then imaged on a Zeiss 880 Airyscan confocal microscope at 20× magnification. Images were analyzed in FIJI for mean fluorescence intensity.
Quantification of DCFDA, Ki67, and γH2A.X
For image analysis in FIJI, a threshold was set to identify positive staining for each marker analyzed (DCFDA, Ki67, γH2A.X) and kept standard across all the images. For DCFDA, mean fluorescence intensity was calculated and normalized by the healthy condition to report fold change. For Ki67, positive cells were automatically counted in FIJI based on the positive threshold and divided by the total number of cells to report percent Ki67-positive cells. For γH2A.X, positive foci were automatically counted using FIJI’s particle counting function based on the positive threshold and size and divided by the total number of nuclei to report γH2A.X foci per cell. To report percent γH2A.X-positive nuclei, the number of nuclei with foci was manually counted based on the automatic detection of foci by FIJI overlayed with the Hoechst staining, then this number was divided by the total number of nuclei.
Statistical analysis
Statistical analysis was performed using JMP Pro 14 (SAS) and GraphPad Prism. Data were analyzed using a one- or two-way ANOVA and post-hoc Tukey test to compare means across groups or Dunnett’s test when comparing means to one control. Repeated measures ANOVA was used for time-dependent assays. Data are represented as mean ± SD with N = number of independent experiments; p ≤ 0.05 was considered significant.
Data availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
References
Gordon, L. B., Brown, W. T. & Collins, F. S. in GeneReviews® (eds M. Adam, P. et al.) (University of Washington, 2019).
Strandgren, C., Revêchon, G., Carvajal, A. S. & Eriksson, M. Emerging candidate treatment strategies for Hutchinson-Gilford progeria syndrome. Biochem. Soc. Trans. 45, 1279–1293. https://doi.org/10.1042/BST20170141 (2017).
Gordon, L. B. et al. Association of lonafarnib treatment vs no treatment with mortality rate in patients with hutchinson-gilford progeria syndrome. JAMA 319, 1687–1695. https://doi.org/10.1001/jama.2018.3264 (2018).
Olive, M. et al. Cardiovascular pathology in hutchinson-gilford progeria: Correlation with the vascular pathology of aging. Arterioscler. Thromb. Vasc. Biol. 30, 2301–2309. https://doi.org/10.1161/atvbaha.110.209460 (2010).
Merideth, M. A. et al. Phenotype and course of hutchinson-gilford progeria syndrome. N. Engl. J. Med. 358, 592–604. https://doi.org/10.1056/NEJMoa0706898 (2008).
Stehbens, W. E., Wakefield, S. J., Gilbert-Barness, E., Olson, R. E. & Ackerman, J. Histological and ultrastructural features of atherosclerosis in progeria. Cardiovasc. Pathol. 8, 29–39. https://doi.org/10.1016/S1054-8807(98)00023-4 (1999).
Stehbens, W. E., Delahunt, B., Shozawa, T. & Gilbert-Barness, E. Smooth muscle cell depletion and collagen types in progeric arteries. Cardiovasc. Pathol. 10, 133–136. https://doi.org/10.1016/S1054-8807(01)00069-2 (2001).
Collins, F. S. Seeking a cure for one of the rarest diseases: Progeria. Circulation 134, 126–129. https://doi.org/10.1161/CIRCULATIONAHA.116.022965 (2016).
Osorio, F. G. et al. Splicing-directed therapy in a new mouse model of human accelerated aging. Sci. Transl. Med. 3, 106–107. https://doi.org/10.1126/scitranslmed.3002847 (2011).
Atchison, L. et al. iPSC-derived endothelial cells affect vascular function in a tissue-engineered blood vessel model of hutchinson-gilford progeria syndrome. Stem Cell Rep. 14, 325–337. https://doi.org/10.1016/j.stemcr.2020.01.005 (2020).
Zhang, X. et al. Modeling early stage atherosclerosis in a primary human vascular microphysiological system. Nat. Comm. 11, 5426. https://doi.org/10.1038/s41467-020-19197-8 (2020).
Lee, J. H. et al. Emulating early atherosclerosis in a vascular microphysiological system using branched tissue-engineered blood vessels. Adv. Biol. 5, 2000428. https://doi.org/10.1002/adbi.202000428 (2021).
Osmanagic-Myers, S. et al. Endothelial progerin expression causes cardiovascular pathology through an impaired mechanoresponse. J. Clin. Investig. 129, 531–545. https://doi.org/10.1172/JCI121297 (2019).
Zhang, J. et al. A human iPSC model of hutchinson gilford progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell 8, 31–45. https://doi.org/10.1016/j.stem.2010.12.002 (2011).
Gordon, L. B. et al. Clinical trial of a farnesyltransferase inhibitor in children with Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. 109, 16666–16671. https://doi.org/10.1073/pnas.1202529109 (2012).
Gordon, L. B., Harten, I. A., Patti, M. E. & Lichtenstein, A. H. Reduced adiponectin and HDL cholesterol without elevated C-reactive protein: Clues to the biology of premature atherosclerosis in Hutchinson-Gilford Progeria Syndrome. J. Pediatr. 146, 336–341. https://doi.org/10.1016/j.jpeds.2004.10.064 (2005).
Yang, S. H. et al. Blocking protein farnesyltransferase improves nuclear blebbing in mouse fibroblasts with a targeted Hutchinson-Gilford progeria syndrome mutation. Proc. Natl. Acad. Sci. 102, 10291–10296. https://doi.org/10.1073/pnas.0504641102 (2005).
Toth, J. I. et al. Blocking protein farnesyltransferase improves nuclear shape in fibroblasts from humans with progeroid syndromes. Proc. Natl. Acad. Sci. 102, 12873–12878. https://doi.org/10.1073/pnas.0505767102 (2005).
Glynn, M. W. & Glover, T. W. Incomplete processing of mutant lamin A in Hutchinson-Gilford progeria leads to nuclear abnormalities, which are reversed by farnesyltransferase inhibition. Hum. Mol. Genet. 14, 2959–2969. https://doi.org/10.1093/hmg/ddi326 (2005).
Capell, B. C. et al. Inhibiting farnesylation of progerin prevents the characteristic nuclear blebbing of Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 102, 12879–12884. https://doi.org/10.1073/pnas.0506001102 (2005).
Yang, S. H., Qiao, X., Fong, L. G. & Young, S. G. Treatment with a farnesyltransferase inhibitor improves survival in mice with a Hutchinson-Gilford progeria syndrome mutation. Biochimica et Biophysica Acta BBA Mol. Cell Biol. Lipids 1781, 36–39. https://doi.org/10.1016/j.bbalip.2007.11.003 (2008).
Yang, S. H. et al. A farnesyltransferase inhibitor improves disease phenotypes in mice with a Hutchinson-Gilford progeria syndrome mutation. J. Clin. Invest. 116, 2115–2121. https://doi.org/10.1172/JCI28968 (2006).
Fong, L. G. et al. Prelamin A and lamin A appear to be dispensable in the nuclear lamina. J. Clin. Invest. 116, 743–752. https://doi.org/10.1172/JCI27125 (2006).
Capell, B. C. et al. A farnesyltransferase inhibitor prevents both the onset and late progression of cardiovascular disease in a progeria mouse model. Proc. Natl. Acad. Sci. USA 105, 15902–15907 (2008).
Gordon, L. B. et al. Impact of farnesylation inhibitors on survival in hutchinson-gilford progeria syndrome. Circulation 130, 27–34. https://doi.org/10.1161/circulationaha.113.008285 (2014).
Cao, K. et al. Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in hutchinson-gilford progeria syndrome cells. Sci. Transl. Med. 3, 89ra58. https://doi.org/10.1126/scitranslmed.3002346 (2011).
Gabriel, D., Gordon, L. B. & Djabali, K. Temsirolimus partially rescues the hutchinson-gilford progeria cellular phenotype. PLoS ONE 11, e0168988. https://doi.org/10.1371/journal.pone.0168988 (2016).
Cabral, W. A. et al. Genetic reduction of mTOR extends lifespan in a mouse model of Hutchinson-Gilford Progeria syndrome. Aging Cell 20, e13457. https://doi.org/10.1111/acel.13457 (2021).
DuBose, A. J. et al. Everolimus rescues multiple cellular defects in laminopathy-patient fibroblasts. Proc. Natl. Acad. Sci. 115, 4206. https://doi.org/10.1073/pnas.1802811115 (2018).
Goldman, R. D. et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. 101, 8963–8968. https://doi.org/10.1073/pnas.0402943101 (2004).
Gimbrone, M. A. & García-Cardeña, G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ. Res. 118, 620–636. https://doi.org/10.1161/circresaha.115.306301 (2016).
Nakajima, H. & Mochizuki, N. Flow pattern-dependent endothelial cell responses through transcriptional regulation. Cell Cycle 16, 1893–1901. https://doi.org/10.1080/15384101.2017.1364324 (2017).
Gonzalo, S., Kreienkamp, R. & Askjaer, P. Hutchinson-Gilford Progeria Syndrome: A premature aging disease caused by LMNA gene mutations. Ageing Res. Rev. 33, 18–29. https://doi.org/10.1016/j.arr.2016.06.007 (2017).
Kim, M. et al. Laminar flow activation of ERK5 protein in vascular endothelium leads to atheroprotective effect via NF-E2-related factor 2 (Nrf2) activation. J. Biol. Chem. 287, 40722–40731. https://doi.org/10.1074/jbc.M112.381509 (2012).
Atchison, L., Zhang, H., Cao, K. & Truskey, G. A. A tissue engineered blood vessel model of hutchinson-gilford progeria syndrome using human iPSC-derived smooth muscle cells. Sci. Rep. 7, 8168. https://doi.org/10.1038/s41598-017-08632-4 (2017).
Rosenfeldt, M. T., Nixon, C., Liu, E., Mah, L. Y. & Ryan, K. M. Analysis of macroautophagy by immunohistochemistry. Autophagy 8, 963–969. https://doi.org/10.4161/auto.20186 (2012).
Graziotto, J. J., Cao, K., Collins, F. S. & Krainc, D. Rapamycin activates autophagy in Hutchinson-Gilford progeria syndrome. Autophagy 8, 147–151. https://doi.org/10.4161/auto.8.1.18331 (2012).
Mao, X., Bharti, P., Thaivalappil, A. & Cao, K. Peroxisomal abnormalities and catalase deficiency in Hutchinson-Gilford Progeria Syndrome. Aging 12, 5195–5208. https://doi.org/10.18632/aging.102941 (2020).
Bikkul, M. U. et al. Farnesyltransferase inhibitor and rapamycin correct aberrant genome organisation and decrease DNA damage respectively, in Hutchinson-Gilford progeria syndrome fibroblasts. Biogerontology 19, 579–602. https://doi.org/10.1007/s10522-018-9758-4 (2018).
Lai, W.-F. & Wong, W.-T. Progress and trends in the development of therapies for Hutchinson-Gilford progeria syndrome. Aging Cell 19, e13175. https://doi.org/10.1111/acel.13175 (2020).
Liu, Y. et al. Involvement of xeroderma pigmentosum group A (XPA) in progeria arising from defective maturation of prelamin A. FASEB J. 22, 603–611. https://doi.org/10.1096/fj.07-8598com (2008).
Liu, Y., Rusinol, A., Sinensky, M., Wang, Y. & Zou, Y. DNA damage responses in progeroid syndromes arise from defective maturation of prelamin A. J. Cell Sci. 119, 4644. https://doi.org/10.1242/jcs.03263 (2006).
Liu, B. et al. Genomic instability in laminopathy-based premature aging. Nat. Med. 11, 780–785. https://doi.org/10.1038/nm1266 (2005).
Kubben, N. et al. Repression of the antioxidant NRF2 pathway in premature aging. Cell 165, 1361–1374. https://doi.org/10.1016/j.cell.2016.05.017 (2016).
Xiong, Z.-M. et al. Methylene blue alleviates nuclear and mitochondrial abnormalities in progeria. Aging Cell 15, 279–290. https://doi.org/10.1111/acel.12434 (2016).
del Campo, L. et al. Vascular smooth muscle cell-specific progerin expression in a mouse model of Hutchinson-Gilford progeria syndrome promotes arterial stiffness: Therapeutic effect of dietary nitrite. Aging Cell 18, e12936. https://doi.org/10.1111/acel.12936 (2019).
Sun, S. et al. Vascular endothelium–targeted Sirt7 gene therapy rejuvenates blood vessels and extends life span in a Hutchinson-Gilford progeria model. Sci. Adv. 6, eaay5556. https://doi.org/10.1126/sciadv.aay5556 (2020).
Bidault, G. et al. Progerin expression induces inflammation, oxidative stress and senescence in human coronary endothelial cells. Cells 9, 1201–1217 (2020).
Gete, Y. G. et al. Mechanisms of angiogenic incompetence in Hutchinson-Gilford progeria via downregulation of endothelial NOS. Aging Cell 20, e13388. https://doi.org/10.1111/acel.13388 (2021).
Danielsson, B. E. et al. Progerin-expressing endothelial cells are unable to adapt to shear stress. Biophys. J. 121, 620–628. https://doi.org/10.1016/j.bpj.2022.01.004 (2022).
Scaffidi, P. & Misteli, T. Lamin A-dependent nuclear defects in human aging. Science 312, 1059–1063. https://doi.org/10.1126/science.1127168 (2006).
Gordon, L. B. et al. The progeria research foundation 10th international scientific workshop; researching possibilities, ExTENding lives—Webinar version scientific summary. Aging 13, 9143–9151. https://doi.org/10.18632/aging.202835 (2021).
Shipkova, M. et al. Therapeutic drug monitoring of everolimus: A consensus report. Therapeutic Drug Monitoring. 38, 143–169 (2016).
Fukudo, M., Ishibashi, K. & Kitada, M. Real-world pharmacokinetics and pharmacodynamics of everolimus in metastatic breast cancer. Invest. New Drugs 39, 1707–1715. https://doi.org/10.1007/s10637-021-01131-4 (2021).
Mueller-Schoell, A. et al. Therapeutic drug monitoring of oral targeted antineoplastic drugs. Eur. J. Clin. Pharmacol. 77, 441–464. https://doi.org/10.1007/s00228-020-03014-8 (2021).
Gomez-Lara, J. et al. Coronary endothelium-dependent vasomotor function after drug-eluting stent and bioresorbable scaffold implantation. J. Am. Heart Assoc. 10, e022123. https://doi.org/10.1161/JAHA.121.022123 (2021).
Jin, Y. P., Valenzuela, N. M., Ziegler, M. E., Rozengurt, E. & Reed, E. F. Everolimus inhibits anti-HLA I antibody-mediated endothelial cell signaling, migration and proliferation more potently than sirolimus. Am. J. Transpl. 14, 806–819. https://doi.org/10.1111/ajt.12669 (2014).
Koh, C. et al. Oral prenylation inhibition with lonafarnib in chronic hepatitis D infection: A proof-of-concept randomised, double-blind, placebo-controlled phase 2A trial. Lancet. Infect. Dis 15, 1167–1174. https://doi.org/10.1016/S1473-3099(15)00074-2 (2015).
Patsch, C. et al. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat. Cell Biol. 17, 994–1003. https://doi.org/10.1038/ncb3205 (2015).
Abutaleb, N. O. & Truskey, G. A. Differentiation and characterization of human iPSC-derived vascular endothelial cells under physiological shear stress. STAR Protocols 2, 100394. https://doi.org/10.1016/j.xpro.2021.100394 (2021).
Mathur, A. B., Truskey, G. A. & Reichert, W. M. Synergistic effect of high-affinity binding and flow preconditioning on endothelial cell adhesion. J. Biomed. Mater. Res. Part A 64A, 155–163. https://doi.org/10.1002/jbm.a.10403 (2003).
Acknowledgements
We thank Dr. Zheng-Mei Xiong of the Cao Lab (University of Maryland) for providing the progerin antibody. We thank the Progeria Research Foundation for providing fibroblasts and iPSCs. We also thank the Duke Pathology core for preparing the histological sections, particularly Dr. Zuowei Su for staining the histology samples. This work was support by National Institutes of Health Grants R01 HL138252-01 (GAT), UH3TR000505 (GAT), and UH3TR002142 (GAT); T32GM144291 (KS), and National Science Foundation Graduate Research Fellowship Grants DGE1644868 (NOA), 1106401 (LA).
Author information
Authors and Affiliations
Contributions
Conceptualization: G.A.T.; methodology: N.O.A., L.A., G.A.T.; performing experiments: N.O.A., L.A., L.C., A.B., K.S., Y.G.; visualization of results: N.O.A., G.A.T., K.S.; supervision: G.A.T.; resources: G.A.T., K.C.; writing—original draft: N.O.A., L.A.; writing—review and editing: N.O.A., G.A.T., K.S., Y.G., K.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Abutaleb, N.O., Atchison, L., Choi, L. et al. Lonafarnib and everolimus reduce pathology in iPSC-derived tissue engineered blood vessel model of Hutchinson-Gilford Progeria Syndrome. Sci Rep 13, 5032 (2023). https://doi.org/10.1038/s41598-023-32035-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-32035-3
This article is cited by
-
Molecular hallmarks of ageing in amyotrophic lateral sclerosis
Cellular and Molecular Life Sciences (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.