Abstract
Breast cancer is the commonest type of cancer in women worldwide and the leading cause of mortality for females. The aim of this research is to classify the alive and death status of breast cancer patients using the Surveillance, Epidemiology, and End Results dataset. Due to its capacity to handle enormous data sets systematically, machine learning and deep learning has been widely employed in biomedical research to answer diverse classification difficulties. Pre-processing the data enables its visualization and analysis for use in making important decisions. This research presents a feasible machine learning-based approach for categorizing SEER breast cancer dataset. Moreover, a two-step feature selection method based on Variance Threshold and Principal Component Analysis was employed to select the features from the SEER breast cancer dataset. After selecting the features, the classification of the breast cancer dataset is carried out using Supervised and Ensemble learning techniques such as Ada Boosting, XG Boosting, Gradient Boosting, Naive Bayes and Decision Tree. Utilizing the train-test split and k-fold cross-validation approaches, the performance of various machine learning algorithms is examined. The accuracy of Decision Tree for both train-test split and cross validation achieved as 98%. In this study, it is observed that the Decision Tree algorithm outperforms other supervised and ensemble learning approaches for the SEER Breast Cancer dataset.
Similar content being viewed by others
Introduction
According to factsheets by World Health Organization (WHO), breast cancer is the second foremost root of cancer death in women and it has a high mortality rate1. Breast cancer disease is a disorder in which the cells in the breast raise out of control. The Breast cancer manifests itself in a diversity of ways. Breast cancer type is resolute by which cells in the breast developed as cancerous. About ninety percentage of breast cancer disease are caused by genetic abnormalities that happen as an effect of the ageing process, and 5–10% of breast cancers are caused by an irregularity that is hereditary from the parents. Modern medical diagnoses are based on information gathered through clinical remark or other trials. Several researchers have emphasized the importance of Artificial Intelligence and Machine Learning in healthcare domains2. Correlation analysis and Principal Component Analysis (PCA) are used for the purpose of dimensionality reduction and to make the models perform well3. Supervised and Unsupervised learning methods are used for the Detection of Breast Cancer through Clinical Data4. Crystall algorithm is used to select the important features for the prediction of survival time for Breast Cancer Patients5. A combination of scaling and Principal Component Analysis (PCA) are used for feature selection in the breast tumor dataset. Both the supervised and unsupervised machine learning models are used for classifying the breast cancer dataset6. Minimal Redundancy Maximal Relevance and Chi-Square Algorithms are used to select the features from the breast cancer dataset7. Various feature selection and classification techniques based on Deep Learning have been assessed in the existing literature8. The main goal of this research work is to categorize and predict the alive and death status of cancer patients. The remaining section of this manuscript is prepared as follows- Section "Literature review" defines the Literature Review, Section "Materials and methods" describes the Materials and Methodology and Section "Results and discussion" illustrates the experimental results on the SEER breast cancer dataset and discusses the outcomes. Lastly, the conclusion and future enrichment are specified in Section "Conclusion and future enhancement".
Literature review
Feature selection techniques such as Recursive Feature Elimination, Forward Feature Selection, f-test and correlation are used with Wisconsin breast cancer data for extraction of important features9. Principal Component Analysis technique was used to indicate the genomic variants in rare genetic diseases10. Chi-Square, Singular Vector Decomposition and PCA are used to select the features from the breast cancer dataset11. PCA was used to extract the features from the Surface Enhanced Raman spectroscopy (SERS) and Raman Spectroscopy (RS) breast cancer serum12. Exploratory Data Analysis (EDA) of the breast cancer dataset was performed using PCA technique13. Receiver operating characteristic curve (ROC) and PCA method was used to visualize the prediction ability of various methods14. Random Forest and Principal Component Analysis methods are combined for attribute selection and accurate diagnosis of breast cancer patients15. Recent literatures for classifying breast cancer dataset have also been reviewed. Artificial Intelligence techniques such as Machine Learning and Deep Learning algorithms are used to perform the classification of breast cancer datasets16. Support Vector Machine (SVM) technique is employed for the classification of the Wisconsin breast cancer dataset17. An Improved Instance-Based K-Nearest Neighbour (IIBK) Classification was developed for solving the problem of Imbalanced Datasets with Enhanced Preprocessing18. Random Forest, KNN (k-Nearest-Neighbor) and Naive Bayes model are also used for the classification of the Wisconsin dataset19. MicroRNA regulated protein interaction pathways is predicted using fuzzy-based algorithms and also to rank Arabidopsis Thaliana20. SVM as well as K-Nearest Neighbor (KNN) algorithms are used to perform breast cancer prediction using tenfold cross-validation21. Four machine learning models such as Decision Tree, KNN, Binary SVM and AdaBoost are used to predict the stages of cancer22.
The time complexity of Naïve Bayes, logistic regression and decision tree is analysed using the breast cancer dataset. Logistic regression performs better than the other classifiers with the highest accuracy23. The dynamic ensemble learning algorithm is used to automatically identify the number of neural networks and their architecture24. The Bacterial Foraging Optimization—Genetic Algorithm (BFO-GA) is developed for solving the problem of Multiple Sequence Alignment (MSA)25. Support Vector Machine, Random Forest and Bayesian Networks are used to classify the Wisconsin dataset26. Enhanced Artificial Neural Network is used for predicting Protein Fold Recognition and Structural Class Prediction27. Protein sequence prediction and analysis are performed using a hybrid Knuth-Morris Pratt (KMP) and Boyer-Moore (BM) method28. Decision Tree based model evaluation is performed for breast cancer dataset using data mining approaches29. The Particle Swarm Optimization (PSO) algorithm was used to identify the cancer specific gene selection30. Deep Convolution Neural Networks with multi scale kernels is used to automate the diagnosis of breast ultrasonography images31. Convolutional Neural Network based diagnosis method was used to detect the early stage of breast cancer using image dataset32. An Improved Convolution Neural Network was developed to classify the brain tumors using Magnetic Resonance Image (MRI) data33. There are various metrics to evaluate the machine learning models. Accuracy, precision and recall are used to evaluate the models such as Logistic Regression, Nearest Neighbor and Support Vector Machines34. Propensity score matching was used to compare the survival outcomes in breast cancer patients, based on the axillary surgery35. The global burden of breast cancer in 2020 and the burden breast cancer in the year of 2040 was predicted36. Methods based on machine learning can assist physicians in reducing the number of false positive and false negative decisions. Based on the existing literatures, this research work focused on classifying the SEER breast cancer dataset using Machine Learning models such as Supervised and Ensemble Learning. In the exiting literature29, the features were chosen according to previously published sources and the features were chosen at random that were influenced by clinical and statistical significance. The current work focuses primarily on the features that were chosen from the SEER dataset using advanced feature selection techniques like Variance Threshold and PCA methods. These features were strongly correlated with the features chosen at random in the earlier work. All machine learning algorithms that performed the classification used the chosen features as input.
Materials and methods
Dataset description
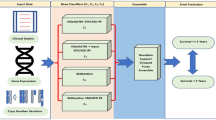
Cancer incidence data for all types of cancer can be found in the Surveillance, Epidemiology, and End Results (SEER) database (1972–2012). The SEER dataset consists of 7,12,319 breast cancer patient records with 149 features and this database37 is sustained by the National Cancer Institute (NCI) that comprises data on cancer incidence, prevalence, survival, and mortality in the United States. It was created by the United States government to collect data on cancer patients across the country. By law, all hospitals, clinics, laboratories, surgery sections, and organizations involved in the diagnosis and treatment of cancer must report information to this institute, which will be reviewed before being entered into the SEER database. The pseudocode for the proposed classification framework is shown in Fig. 1 and the overall architecture for this research work is shown in Fig. 2.
Handling missing values
The dataset contains more missing values. Hence, the features which have missing values of more than 20% are removed. The categorical features are imputed using the Random Forest classifier and continuous features are imputed using Random Forest Regressor. The parameter for the Random Forest Classifier technique is configured as the number of estimators is set to 100, criterion is set to gini with bootstrapping. The parameter for the Random Forest Regressor technique is configured as the number of estimators is set to 100, criterion is set to squared_error with bootstrapping.
Feature selection
Feature selection aims to discover the finest set of features that can be used to build models for the phenomena being studied. Because it is very hard to use more features and it may cause overfitting. In this research, a few feature selection techniques such as Variance Threshold and Principal Component Analysis (PCA) have been used to improve the model performance.
Variance threshold
For feature selection, the variance threshold method is applied. It eliminates all attributes with variances below a predetermined level. By default, it removes all attributes with zero variance, or attributes having the same value across all instances. The relationship between features and the target variable is ignored by the variance threshold. A simple baseline method called Variance Threshold (VR) eliminates all features with zero variance. Nine features in the SEER dataset show too little variation (less than or equal to 0%), according to the variance threshold technique. We currently have 50 features. Table 1 displays the significant risk factors from the SEER breast cancer dataset.
Principal component analysis (PCA)
The Principal Component Technique was used to solve the problem of multicollinearity and the number of principal components was discovered using Variance Inflation Factor (VIF). This model used 13 components out of a total of 50.
Methodology
Decision tree classifier
Decision tree classifier is used to choose whether to split a node into two or more sub-nodes. For constructing decision trees, we can employ a diversity of machine learning models. The similarity of the resultant sub-nodes enhances with the creation of sub-nodes. The purity of the node expands as the target variable is increased. The decision tree splits the nodes into sub-nodes based on the input features, then selects the split that produces the maximum similar sub-nodes. This technique tries to divide the input dataset into the smallest subset possible at each split. The aim of Decision Tree algorithm is to reduce the loss metric value as much as possible. The loss functions such as Gini Impurity and Entropy are used to collate the class distribution beforehand and after the split. The loss metric named Gini Impurity is used to measure the variation between different classes. The parameter for the Decision Tree method is configured as the criterion is set to gini, splitter as best, minimum sample split as 2 and minimum sample leaf as 1.
Naive Bayes (NB) classifier
This Naïve Bayes model has newly gained popularity and is being used more frequently. It’s a statistical pattern recognition technique that makes a reasonable assumption about how data is generated. The parameters of NB are estimated using training samples in this model. This is a simple classifier, based on the assumption that all sample attributes are independent. Once the hypothesis is false, Naïve Bayes classifies the data in a perfect manner, because the classification hypothesis is only a symbol of function approximation, and the function estimate is achieved with low accuracy, whereas the classifier's accuracy is high. The parameter for the Naïve Bayes method is configured with the var smoothing as 1e-9. The conditional probability of individual variable Xk assumed the class label C is learned by Nave Bayes using training data and the conditional probability of individual variable Xk is specified the class label C. The Bayes rule is used to calculate the probability of C specified a particular instance, \({X}_{1}\),…\({X}_{n}\), using Eq. (1):
Because this classifier is based on the hypothesis that variables are conditionally independent. Equation (2) is used to calculate the posterior probability of the class:
The class with the highest posterior probability Eq. (3) is the classification result.
AdaBoost (AB) classifier
Freund and Schapire invented the adaptive boosting machine learning algorithm38, which is abbreviated as AB. AB is a meta-algorithm that works in aggregation with other learning algorithms to enhance the performance. AdaBoost is a training method for boosted classifiers, which are classifiers that have the form Eq. (4):
where individual ft is a poor learner that receipts input and yields a real-valued outcome that indicates the sample's class. The predicted sample class is identified by the weak learner outcome, and the value designates the level of sureness in that classification. Likewise, if the data is thought to be in a positive class, the T-layer classifier will be positive, else it will be negative. For each sample in the training set, individual weak learner model produces an output, hypothesis h(xi). Weak learner is elected and assumed a coefficient at respective iteration, t, so that the sum training error of the resulting t-stage boost classifier is minimized (Eq. (5)).
Ft − 1(xi) denotes the boosted classifier, E(F) denotes error function, and \({f}_{t}\)(x) =\({ \alpha }_{t}h\) (\({x}_{i}\)) denotes the weak learner for inclusion in the final classifier. In Adaboost, each new stage's classification is built on samples that have been incorrectly classified. Although AB is sensitive to noise and outliers data and it outperforms other learning algorithms in terms of overfitting. Random classification is the algorithm's base classifier (50 percent). The parameter for the AdaBoost method is configured as the number of estimators is set to 50, estimator is set to none, learning rate as 1.0 and the SAMME.R algorithm is used.
XG boost classifier
XGBoost (XGB) is classified as a boosting technique in Ensemble Learning. To improve prediction accuracy, ensemble learning combines multiple models into a collection of predictors. In the boosting technique, previous models' errors are attempted to be corrected by subsequent models by adding weights to the models. Gradient Boosted algorithms, unlike other boosting algorithms, optimise the loss function rather than increasing the weights of misclassified branches. With some regularisation factors, XGBoost is a more advanced gradient boosting implementation. The parameter for the XGBoost method is configured as the verbosity is set to 1 and the gbtree is used as booster.
Gradient boosting algorithm
Gradient boosting (GB) is a boosting algorithm based on the ensemble techniques. In this model, each predictor alters the error of the previous model. The training sample weights are not adjusted in Adaboost. As an alternative, each model is trained using the ancestor's residual errors as labels. Gradient Boosting technique use CART (Classification and Regression Trees) as the base learner. The Gradient Boosting is an ensemble model that can be made up of N number of trees. The first tree model is trained using the feature matrix X and labels y. The residual errors (r1) in the first tree training set are considered using the predictions labelled y1 (hat). The second tree is trained using Tree1's feature matrix X and residual errors r1 as labels. Using the predicted results r1, the residual r2 is calculated (hat). This procedure is repetitive until all N trees in the ensemble have been trained. The parameter for the Gradient Boosting method is configured as the number of estimators is set to 100, criterion is set to friedman_mse, the learning rate as 0.1 and log loss is used as loss metric.
Shrinkage occurs when the prediction of each model in the ensemble is grown by the learning rate (lr), which ranges from 0 to 1. All the trees have been trained and each tree predicts a label with Eq. (6) providing the ultimate prediction. The mathematical notations which are used in this research work in shown in Supplementary Table S1.
Results and discussion
Machine learning models that are supervised and ensemble predict breast cancer survival. The proposed method to predict breast cancer survival included five machine learning models, including NB, Decision tree classifier, Ada Boost, XG Boost, and Gradient Boosting classifier. The experiments are performed using an Intel(R) Core (TM) i5-1235U 1.30 GHz CPU with 8 GB of RAM and Windows 11 as the operating system. Python 3.8 was used to develop the proposed framework.
Performance metrics
The Performance metrics which are used in this research work are discussed below.
Accuracy
Accuracy refers to the correctly classified instances by the total amount of instances present in the SEER dataset (Eq. 7).
where TP = True Positive, FP = False Positive, TN = True Negative, FN = False Negative, TP = Dead persons correctly known as dead. TN = Alive persons correctly recognized as dead. FP = Alive persons wrongly recognized as dead. FN = Dead persons wrongly recognized as alive.
TP rate
It is used to find the high true-positive rate using the Eq. (8). The true-positive rate is also known as sensitivity and it measures the part of actual positives which are appropriately recognized.
FP rate
The False Positive rate (Eq. 9) refers to the probability of falsely refusing the null hypothesis for a specific test. It usually refers to the anticipation of the false positive ratio.
F-measure
F-Measure is the mixture of both precision and recall (Eq. 10), which is used to calculate the score. This kind of measure is often used in the field of Information Retrieval to estimate the query classification performance.
where, Precision = \(\frac{\mathrm{TP}}{\mathrm{TP}+\mathrm{FP}}\) and Recall = \(\frac{\mathrm{TP}}{\mathrm{TP}+\mathrm{FN}}\)
Performance of the proposed model
The SEER breast cancer data contains 149 features with 712,319 records. In the SEER data, six categorical features such as 'siteo2v', 'eod13', 'eod2', 'icdot10v', 'plc_brth_cntry' and 'plc_brth_state' which will not contribute to the model as we want. Hence, the six features are dropped. Then we found that the SEER data has some features which have more null values. Around 84 features have null values of more than 20%. Even if we try to impute them, it may impact the model in a bad way. So, we dropped those features as well. Now we are left with 58 features. Among 58 features we have 13 features that have null values of less than 20% (Table 2) and 45 features which don’t have null values.
The missing values are imputed using Random Forest Classifier for categorical features and Random Forest Regressor for continuous features. After imputing the missing values, the important features are selected using the Variance Threshold method. By using this method, 50 features are selected among 58 features. For finding the multicollinearity, the Variance Inflation Factor (VIF) value is calculated for the 50 features and it is shown in Table 3.
After finding the VIF values, the dataset is performed with the Standard Scaler method and then it is split into training and testing records. The Xtrain consists of 498,623 records with 50 features and Xtest consists of 213,696 records with 50 features. To solve the problem of multicollinearity, the Principal Component Analysis (PCA) dimensionality reduction technique is used to reduce the feature dimensions. For achieving this, the Principal Explained Variance Ratio method is used to find the number of components. Now the features end up with 13 components and the Principal Explained Variance Ratio for the 13 features is shown in Table 4.
In this study, five machine learning algorithms are used to predict the survival of breast cancer such as Naïve Bayes, Decision tree classifier, Ada Boost, XG Boost, and Gradient Boosting classifier. In the Decision Tree, the criterion for determining the quality of a split is entropy, which is calculated using information gain given by entropy, and the random state is 0 for generating random states. When building an NB classifier with zero training instances, the default precision for numeric attributes is 0.1. In Adaboost, the Decision Stump algorithm is chosen as the base classifier. The number of iterations to be accomplished is set to 10 and the weight pruning threshold is set to 100. In the Gradient Boosting Classifier log loss function was used and the learning rate was set to 0.1, the criterion is friedman_mse. In the XG Boost classifier gbtree booster was used and the learning rate is 0.3. These machine learning models have been implemented, and the comparison results are summarized in Tables 5 and 6. The alive and death count of breast cancer patients predicted by machine learning models is shown in Table 7. The comparison of machine learning models (percent) by train test split and cross-validation strategy, including NB, Decision tree classifier, Ada Boost, XG Boost, and Gradient Boosting classifier is shown in Tables 5 and 6.
Figures 3 and 4 shows the comparison of Accuracy for the various machine learning techniques such as Naïve Bayes, AdaBoost, Decision Tree, Gradient Boosting and XG Boosting algorithms using Train-Test Split and Cross Validation Methods. From Figs. 3 and 4, it is inferred that the Decision Tree algorithm performs better than the other algorithms in terms of Accuracy. Figure 5 shows the comparison of performance metrics values for the various machine learning algorithms using the Train-Test Split method. From Fig. 5, it is inferred that the Decision Tree algorithm provides better results compared to other machine learning models. The Fig. 6, shows the comparison of performance metrics values for the various machine learning algorithms using the Cross-Validation method. From Fig. 6, it is inferred that the Decision Tree algorithm provides better results compared to other machine learning algorithms.
These machine learning models are associated in terms of precision, recall, F1 score, and accuracy using train test split and cross-validation strategies. From the experimental results, it is inferred that the decision tree model achieved 98% accuracy which is the highest among those other machine learning models. For the SEER breast cancer dataset, it is inferred that the Decision Tree classifier algorithm performs 6.12% better than the NB algorithm, 1.02% better than the Adaboost algorithm and 8.16% better than the GB and XGB algorithms using the train test method. For the cross-validation method, it is inferred that the Decision Tree classifier algorithm performs 5.1% better than the NB algorithm, 1.02% better than the Adaboost algorithm, 9.18% better than the GB and 7.14% better than the XGB algorithm. From the experimental results it is inferred that the Decision Tree outperforms the other machine learning models. As shown in Tables 5 and 6, the Decision Tree machine learning model is the best model for classifying the SEER breast cancer disease dataset.
Conclusion and future enhancement
Given that breast cancer is one of the most common causes of death for women, early detection is crucial. The burden on doctors can be decreased by using automatic classification systems as diagnostic tools. Modern machine learning classifiers make it possible to identify breast cancer tumours early. Even while false positive and false negative results are frequently acknowledged to be significant in medical research, the majority of past studies have primarily focused on accuracy. As a result, we looked at various performance metrics in addition to accuracy, precision, and recall. In this work, variance threshold and principal component analysis were used to determine the features. Then, the chosen features are fed into the machine learning classifiers as input to carry out the classification task. This study evaluates the effectiveness of different machine learning classification methods for predicting breast cancer survival, including Naive Bayes, Decision Tree, Ada Boost, XG Boost, and Gradient Boosting classifiers. The decision tree approach was the most successful, according to the comparative results. In the future, several machine-learning techniques might be used to classify datasets pertaining to the breast cancer disease.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author upon reasonable request.
References
https://www.who.int/news-room/fact-sheets/detail/breast-cancer.
Bi, W. L. et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J. Clin. 69, 127–157 (2019).
Ibrahim, S., Nazir, S. & Velastin, S. A. Feature selection using correlation analysis and principal component analysis for accurate breast cancer diagnosis. J. Imaging. 7(11), 225. https://doi.org/10.3390/jimaging7110225 (2021).
Haq, A. et al. Detection of breast cancer through clinical data using supervised and unsupervised feature selection techniques. IEEE Access. 1, 1–1. https://doi.org/10.1109/ACCESS.2021.3055806 (2021).
Liu, S. et al. Survival time prediction of breast cancer patients using feature selection algorithm crystall. IEEE Access 9, 24433–24445. https://doi.org/10.1109/ACCESS.2021.3054823 (2021).
Nguyen, Q.H., Do, T.T., Wang, Y., Heng, S.S., Chen, K., Ang, W.H.M., Philip, C.E., Singh, M., Pham, H.N., & Nguyen B.P., et al. Breast cancer prediction using feature selection and ensemble voting. In Proceedings of the 2019 International Conference on System Science and Engineering (ICSSE); Dong Hoi City, Vietnam. pp. 250–254 (2019).
Haq, A. U., Li, J., Memon, M. H., Khan, J. & Din, S. U. A novel integrated diagnosis method for breast cancer detection. J. Intell. Fuzzy Syst. 38(2), 2383–2398. https://doi.org/10.3233/JIFS-191461 (2020).
Haq, A. et al. A survey of deep learning techniques-based Parkinson’s disease recognition methods employing clinical data. Expert Syst. Appl. 208, 8045. https://doi.org/10.1016/j.eswa.2022.118045 (2022).
Dhanya, R., Paul, I. R., Sindhu Akula, S., Sivakumar, M., & Nair, J. J. A comparative study for breast cancer prediction using machine learning and feature selection. In 2019 International Conference on Intelligent Computing and Control Systems (ICCS), pp. 1049–1055. https://doi.org/10.1109/ICCS45141.2019.9065563 (2019).
Zhou, Y. et al. Genetic determinants and absence of breast cancer in Xavante Indians in Sangradouro Reserve Brazil. Sci. Rep. 13, 1452 (2023).
Shafique, R. et al. Breast cancer prediction using fine needle aspiration features and upsampling with supervised machine learning. Cancers 15(3), 681 (2023).
Cheng, Z. et al. Application of serum SERS technology based on thermally annealed silver nanoparticle composite substrate in breast cancer. Photodiagn. Photodyn. Ther. 1, 103284 (2023).
Pereira de Souza, N. M. et al. Rapid and low-cost liquid biopsy with ATR-FTIR spectroscopy to discriminate the molecular subtypes of breast cancer. Talanta 254, 123858 (2023).
Pan, Y. et al. Prognostic and immune microenvironment analysis of cuproptosis-related LncRNAs in breast cancer. Funct. Integr. Genomics 23, 38 (2023).
Bian, K., Zhou, M., Hu, F. & Lai, W. RF-PCA: A new solution for rapid identification of breast cancer categorical data based on attribute selection and feature extraction. Front. Genet. 11, 566. https://doi.org/10.3389/fgene.2020.566057 (2020).
Hasan, S., Sagheer, A. & Veisi, H. Breast cancer classification using machine learning techniques: A review. Turk. J. Comput. Math. Educ. (TURCOMAT). 12, 1970–1979 (2021).
Telsang V. A., & Hegde, K. Breast cancer prediction analysis using machine learning algorithms. In: 2020 International Conference on Communication, Computing and Industry 4.0 (C2I4), pp. 1–5. https://doi.org/10.1109/C2I451079.2020.9368911 (2020).
Manikandan, P., Ramyachitra, D., Kalaivani, S. & Ranjani, R. An improved instance based K-nearest neighbor (IIBK) classification of imbalanced datasets with enhanced preprocessing. Int. J. Appl. Eng. Res. 11, 642–649 (2016).
Sharma, S., Aggarwal, A., & Choudhury, T. Breast cancer detection using machine learning algorithms. In 2018 International Conference on Computational Techniques, Electronics and Mechanical Systems (CTEMS), pp. 114–118. https://doi.org/10.1109/CTEMS.2018.8769187 (2018).
Manikandan, P., Ramyachitra, D. & Nandhini, R. Fuzzy based algorithms to predict MicroRNA regulated protein interaction pathways and ranking estimation in Arabidopsis thaliana. Gene 692, 170–175 (2019).
Islam, M.M., Iqbal, H., Haque, M. R., & Hasan, M.K. Prediction of breast cancer using support vector machine and K-Nearest neighbors. In 2017 IEEE Region 10 Humanitarian Technology Conference (R10-HTC), pp. 226–229. https://doi.org/10.1109/R10-HTC.2017.8288944 (2017).
Laghmati, S., Cherradi, B., Tmiri, A., Daanouni, O., & Hamida, S. Classification of patients with breast cancer using neighbourhood component analysis and supervised machine learning techniques. In 2020 3rd International Conference on Advanced Communication Technologies and Networking (CommNet), pp. 1–6. https://doi.org/10.1109/CommNet49926.2020.9199633 (2020).
Mandal, S. K. Performance analysis of data mining algorithms for breast cancer cell detection using Naïve Bayes, logistic regression and decision tree. Int. J. Eng. Comput. Sci. 6, 20388–20391 (2017).
Alam, K. M. R., Siddique, N. & Adeli, H. A dynamic ensemble learning algorithm for neural networks. Neural. Comput. Appl. 1, 1–16. https://doi.org/10.1007/s00521-019-04359-7 (2019).
Manikandan, P. & Ramyachitra, D. Bacterial foraging optimization—genetic algorithm for multiple sequence alignment with multi-objectives. Sci. Rep. 7, 1 (2017).
Bazazeh, D., & Shubair, R. Comparative study of machine learning algorithms for breast cancer detection and diagnosis. In 2016 5th International Conference on Electronic Devices, Systems and Applications (ICEDSA), pp. 1–4. https://doi.org/10.1109/ICEDSA.2016.7818560 (2016).
Sudha, P., Ramyachitra, D. & Manikandan, P. Enhanced artificial neural network for protein fold recognition and structural class prediction. Gene Rep. 12, 261–275 (2018).
Manikandan, P. & Ramyachitra, D. PATSIM: Prediction and analysis of protein sequences using hybrid Knuth-Morris Pratt (KMP) and Boyer-Moore (BM) algorithm. Gene 657, 50–59 (2018).
Ponnuraja, C. Decision tree classification and model evaluation for breast cancer survivability: A data mining approach. Biomed. Pharmacol. J. 10, 281–289. https://doi.org/10.13005/bpj/1107 (2017).
Ramyachitra, D., Sofia, M. & Manikandan, P. Interval-value Based Particle Swarm Optimization algorithm for cancer-type specific gene selection and sample classification. Genom. Data 5, 46–50 (2015).
Qi, X. et al. Automated diagnosis of breast ultrasonography images using deep neural networks. Med. Image Anal. 52, 185–198 (2019).
Haq, A. U., et al. DEBCM: deep learning-based enhanced breast invasive ductal carcinoma classification model in IoMT healthcare systems. IEEE J. Biomed. Health Inf. https://doi.org/10.1109/JBHI.2022.3228577.
Haq, A. U. et al. DACBT: deep learning approach for classification of brain tumors using MRI data in IoT healthcare environment. Sci. Rep. 12, 15331. https://doi.org/10.1038/s41598-022-19465-1 (2022).
Sharma, A., Kulshrestha, S., & Daniel, S. Machine learning approaches for breast cancer diagnosis and prognosis. In 2017 International Conference on Soft Computing and its Engineering Applications (icSoftComp), pp. 1–5. https://doi.org/10.1109/ICSOFTCOMP.2017.8280082 (2017).
Cha, C. et al. Survival benefit from axillary surgery in patients aged 70 years or older with clinically node-negative breast cancer: A population-based propensity-score matched analysis. Eur. J. Surg. Oncol. 1, 1 (2022).
Arnold, M. et al. Soerjomataram I Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast 66, 15–23 (2022).
Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Research Data (1973–2013), National Cancer Institute, DCCPS, Surveillance Research Program, Surveillance Systems Branch, released April 2016, based on the November 2015 submission.
Freund, Y., & Schapire, R.E. A desicion-theoretic generalization of on-line learning and an application to boosting. In: Vitányi, P. (eds) Computational Learning Theory. EuroCOLT 1995. Lecture Notes in Computer Science, vol 904. Springer, Berlin, Heidelberg. https://doi.org/10.1007/3-540-59119-2_166 (1995).
Author information
Authors and Affiliations
Contributions
The conceptualization and design of this study involved input from all authors. P.M. and U.D. performed the analysis, interpreted the findings, and prepared the manuscript. The figures were prepared and the statistical analysis was carried out by P.M. and C.P. The final draught of this manuscript was approved by all authors after they had reviewed the findings.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Manikandan, P., Durga, U. & Ponnuraja, C. An integrative machine learning framework for classifying SEER breast cancer. Sci Rep 13, 5362 (2023). https://doi.org/10.1038/s41598-023-32029-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-32029-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.