Abstract
The FBN1 gene encodes profibrillin protein that is cleaved by the enzyme furin to release fibrillin-1 and a glucogenic hormone, asprosin. Asprosin is implicated in diverse metabolic functions as well as pathological conditions in mammals. However, till date, there are no studies on asprosin in any non-mammalian vertebrate. In this study, we have retrieved the spotted snakehead Channa punctata fbn1 gene (ss fbn1) from the testicular transcriptome data and validated it. The transcript is predicted to encode 2817 amino acid long putative profibrillin protein. Amino acid sequence alignment of deduced ss profibrillin with human profibrillin revealed that the furin cleavage site in profibrillin is well conserved in C. punctata. Further, differential expression of ss fbn1 was observed in various tissues with the highest expression in gonads. Prominent expression of furin was also observed in the gonads suggesting the possibility of proteolytic cleavage of profibrillin protein and secretion of asprosin in C. punctata. In addition, the C-terminal of the fbn1 gene of C. punctata that codes for asprosin protein has been cloned. Using in silico approach, physicochemical properties of the putative ss asprosin were characterized and post-translational changes were predicted. The putative ss asprosin protein sequence is predicted to consist of 142 amino acid residues, with conserved glycosylation sites. Further, the 3D model of ss asprosin was predicted followed by MD (molecular dynamics) simulation for energy minimization. Thus, the current study, for the first time in non-mammalian vertebrates, predicts and characterizes the novel protein asprosin using in silico approach.
Similar content being viewed by others
Introduction
The FBN1 gene in mammals is reported to encode profibrillin protein that is cleaved by activated protease, furin into fibrillin-1 and 140-amino acids-long asprosin1. Furin is a proprotein convertase belonging to a family of subtilisin serine proteases that cleaves at the R-X-K/R-R↓ or R/K-X-X-X-K/R-R↓ motif in the target protein and is ubiquitously expressed2. Fibrillin-1 is a major glycoprotein of extracellular matrix, belonging to the fibrillin family of proteins3. The C-terminal region of FBN1, specifically exons 65 and 66 are responsible for encoding the glucogenic hormone asprosin that was discovered by Romere et al. in 20161.
Asprosin is majorly expressed in the white adipose tissue in mammals1. It is shown to have diverse physiological effects acting through different receptors. This metabolic hormone regulates glucose release from the liver through interaction with G-protein coupled receptor (GPCR), olfactory receptor 734 (OLFR734)4. It also regulates the orexigenic effect via acting on the agouti-related peptide (AgRP) neurons in the brain5. In addition to this, asprosin regulates insulin secretion and inflammation in pancreas6. Not surprisingly, asprosin is implicated in various metabolic disorders including type 2 diabetes7, obesity8,9 and polycystic ovary syndrome10,11. In recent years, the profound effects of asprosin on reproductive functions have also been demonstrated12,13,14,15. However, all these studies are confined to mammals and till date the existence of asprosin in non-mammalian vertebrates is obscure.
In view of this lacuna, the present study was undertaken in fish Channa punctata. Teleost are the oldest and most diverse extant vertebrates. In this study, using various bioinformatics tools, fbn1 and furin genes were validated and the furin cleavage site was determined in the fbn1 encoded putative profibrillin protein in spotted snakehead. Further, tissue-dependent differential expression of the fbn1 and furin gene was demonstrated. In addition, an attempt has been made to overexpress ss asprosin in a heterologous bacterial system. Using in silico approach, the physicochemical properties and post-translational modifications have been predicted for ss asprosin. The 3D modelling of the deduced ss asprosin was carried out using I-TASSER followed by energy minimization through MD simulation.
Results
Identification and validation of fbn1 and furin sequence
The potential longest transcript of fbn1 obtained from testicular transcriptome data of C. punctata contained 8454 base pair long coding sequence and was partial from the 5′-end. Similarly, the best transcript of C. punctata furin containing 1869 base pair open reading frame (ORF) was retrieved and it was partial from the 5′-end.
Expression analysis of fbn1 and furin gene in Channa punctata
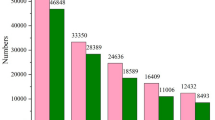
The expression analysis of fbn1 in different tissues of C. punctata revealed the highest expression in gonads followed by heart and different parts of the brain. Among the tissues that expressed fbn1 prominently (testis, ovaries, heart and brain), furin was highly expressed in gonads as observed by semi-quantitative PCR (Fig. 1; Supplementary Fig. S1).
(a) Tissue distribution of fbn1 gene in different tissues of C. punctata. Real-time quantitative PCR was used to quantify the gene expression in each tissue. Each data point represents the mean ± SEM of tissues collected from 6 fish (N = 6). Two technical replicates for each sample were used. (b) Tissue distribution of furin using semi quantitative PCR. The gene expression of furin was studied in tissues which exhibited prominent fbn1 expression (testis, ovary, heart, forebrain midbrain and hindbrain) using semi-quantitative PCR followed by resolving into the 1% agarose gel.
Identification of furin cleavage site
The ss fbn1 transcript encoded 2817 amino acid long putative protein. The multiple sequence alignment of deduced profibrillin of C. punctata with profibrillin of H. sapiens showed the conserved furin cleavage site (R/K-X-X-X-K/R-R↓) (Fig. 2). On the basis of the presence of furin cleavage site, amino acid sequence of asprosin protein was deduced in C. punctata.
Amino acid alignment of C-terminal region of profibrillin of human and snakehead. The furin cleavage site has been highlighted in green colour and the predicted mature ss asprosin has been highlighted in yellow colour. The ‘*’ represents amino acids that are conserved, ‘:’ indicates amino acids with strongly similar properties, ‘.’ represents amino acids with less similar properties whereas the gaps represent mismatch residues.
Cloning and characterization of asprosin
Cloning and expression of ss recombinant asprosin
The 429 base pair sequence of fbn1 gene with one stop codon was cloned in the pProEx-HTc vector, transfected into DH5ɑ and validated through sequencing. After transformation in the BL21 cells, induction using IPTG was carried out. Proteins isolated from the uninduced pellet, induced pellet, uninduced supernatant and induced supernatant samples were analyzed using coomassie staining for presence of recombinant asprosin protein (Supplementary Fig. S2). Significant expression of recombinant asprosin (~ 19 kDa) having six histidine tag was observed in the supernatant of induced sample as compared to uninduced supernatant sample. However, in case of pellet, no protein band was detected around 19 kDa in both uninduced as well as induced sample. Further, using anti-His antibody, recombinant asprosin protein with histidine tag was validated in the induced supernatant sample (Fig. 3, Supplementary Fig. S3).
Expression analysis of recombinant ss asprosin. (a) 15% SDS-PAGE showing the protein expression in the uninduced pellet, induced pellet, uninduced supernatant and induced supernatant samples observed using coomassie staining. (b) Western blotting using anti-His antibody for validation of recombinant asprosin in supernatant of induced and uninduced samples.
Physicochemical properties
The putative ss asprosin and human asprosin comprises 142 and 140 residues, respectively with approximately 16 kDa molecular weight. The instability index of putative ss asprosin was 33.89 and human asprosin was 37.84. The aliphatic index of the putative ss asprosin and human asprosin was 94.72 and 89.86, respectively. The GRAVY values of putative ss asprosin was -0.585 and human asprosin was − 0.549 (Table 1). The percent identity matrix of the primary sequence of putative asprosin of C. punctata with asprosin of H. sapiens revealed 57.86% similarity. The post-translational modifications such as N-linked glycosylation and phosphorylation were predicted using Motif scan software (Fig. 4). Two conserved N-linked glycosylation sites at position 3 and 37 were predicted in ss asprosin. Phosphorylation sites at 5, 16, 42, 56, 88 and 89 positions were predicted in asprosin of C. punctata.
Protein modelling, quality assessment and validation of asprosin
The secondary and tertiary structure of ss asprosin showed the presence of β-strand. In the present study, MD simulation of the 3D structure of ss asprosin protein was carried out for the refinement and conformational dynamics. RMSD analysis showed that the model attained a stable plateau after 85 ns with an average value of 0.95 nm. Fluctuation of Cɑ atoms of every amino acid during energy minimization assessed through RMSF showed no major fluctuations. However, residues at position 5-20 and 115-125 fluctuated with RMSF value of approximately 1 nm. During the initial time period between 1 and 10 ns, Rg pattern was unstable, but after 90 ns simulation time, Rg value showed stable behaviour with an average value of ~ 1.73 nm. The Ramachandran plot for geometry evaluation of ss asprosin protein after MD simulation using PROCHECK module revealed that 81.5% residues lie in the most favoured region, 17.7% residues in additional allowed region, 0.8% residues in generously allowed region and 0% residues in disallowed region (Fig. 5).
Structural representation and stability parameters of ss asprosin after stimulation. (a) 3D model of ss asprosin in which arrow represent the β-sheets. (b) Ramachandran plot of amino acid residues of ss asprosin. (c) RMSD (Root mean square deviation) of the backbone Cɑ atoms of ss asprosin vs time. (d) RMSF (Root mean square fluctuation) of each residue of ss asprosin vs time. (e) Rg (Radius of gyration) vs time.
Discussion
Since the discovery of asprosin in 20161, there are at present no reports on asprosin in any of the other vertebrate groups. This is surprising as it is shown to play a critical role in metabolism, immunity and reproduction in mammalian species1,6,13,14,15,16. In the current study, the partial cDNA sequence for spotted snakehead fbn1 and furin was obtained. ss fbn1 comprises a 8454 bp long coding sequence that is predicted to code for 2817 amino acid long profibrillin protein with conserved furin cleavage site. The ss furin transcript retrieved from the transcriptome of C. punctata had 1869 bp ORF. The constitutive and ubiquitous expression of ss fbn1 was observed in many tissues of C. punctata. Unlike humans, in which the maximum expression of FBN1 has been reported in the white adipose tissue1, the gonads of C. punctata expressed the maximum fbn1. The gonads also show prominent gene expression of the protease furin, thereby indicating the potential production of asprosin in teleosts. Similar expression of FBN1 and FURIN was also observed in the ovaries of beef heifers wherein it has been suggested to play an important role in ovarian functions13. The importance of asprosin in regulation of gonadal activities is further corroborated by a recent report in mice where Leydig and Sertoli cells are found to be immune-positive for asprosin and intratesticular administration of asprosin has been shown to promote steroidogenesis as well as spermatogenesis17. Based on our observations, and studies in mammals, it can be hypothesized that asprosin might be implicated in reproduction in teleosts also.
For the first time in a teleostean model, we have successfully cloned the fbn1 region encoding asprosin protein of C. punctata and expressed it in the bacterial system. The IPTG-induced recombinant ss asprosin protein observed in the supernatant had an approximate molecular weight of 19 kDa. Intriguingly, the recombinant protein was not detected in the pellet which might be due to either the absence, or extremely low amount of inclusion bodies formed of recombinant ss asprosin. Recombinant ss asprosin was validated using anti-His antibody. In the case of human, the constructed recombinant protein has molecular weight of ~ 17 kDa with six histidine residues, however the molecular weight of asprosin found in human plasma is 30 kDa1. The discrepancy in size has been attributed to the lack of several post-translational changes in the recombinant asprosin expressed in the bacterial system as opposed to the asprosin found in the plasma. Indeed, it was shown that when recombinant asprosin is expressed in mammalian cell line, then the molecular weight is almost similar to that found in plasma1. Based on this observation, it can be suggested that the molecular weight of asprosin in Channa punctata might be more than 19 kDa and needs to be confirmed in further studies.
The percent identity matrix revealed that putative ss asprosin is more than 55% similar with the human asprosin and comprises 142 amino acids, whereas mammalian asprosin consists of 140 residues1. Interestingly, alignment of ss asprosin with human asprosin reveals that two amino acids at position 13th (Met) and 94th (Ser) exist in ss asprosin but are lost in due course of evolution in human. In silico analysis revealed that the putative ss asprosin and human asprosin were stable as its instability index, a parameter to measure the stability of protein under ex vivo conditions, were lower than the threshold value of 4018. The predicted ss asprosin and human asprosin had a high aliphatic index, thereby indicating its thermostability. The aliphatic index represents the relative volume of the aliphatic side chain (valine, alanine, leucine and isoleucine) occupied in a protein and determines the thermostability of a globular protein with values ranging between 71.13 and 143.54. Higher aliphatic value denotes a thermostable protein19. In addition to this, based on the negative grand average of hydropathicity index (GRAVY), the deduced ss asprosin and human asprosin seems to be hydrophilic in nature. The negative GRAVY value represents that a protein is hydrophilic and globular in nature, while positive GRAVY value implies a hydrophobic and membrane-bound protein20. The hydrophilic nature of ss asprosin implies that it is a soluble protein and might not require binding protein for its transport.
Post-translational analysis revealed the presence of glycosylation sites. Glycosylation is reported to increase the stability, as well as, half-life of proteins21,22. In case of other glycoproteins such as thyroid stimulating hormone, luteinizing hormone and follicle-stimulating hormone, glycosylation in these protein hormones helps in their interaction with the cognate receptor by increasing binding affinity and effecting the signalling23,24,25,26,27,28. The N-linked glycosylation sites in ss asprosin at position 3 and 37 correspond to the glycosylation sites in human asprosin1 and hence seem to be well conserved and might play a crucial role in the functional aspect of asprosin. Moreover, in ss asprosin, six phosphorylation sites are also predicted. Although phosphorylation plays a critical role in proteins involved in cell signalling, the implication of phosphorylation sites in ss asprosin is yet to be understood.
Currently, lack of NMR/crystal experimental structure of asprosin is a major constraint in understanding the structural aspect of the protein. This study provides the first comprehensive description of structural parameters using in silico approach via prediction of tertiary structure of asprosin in C. punctata. The presence of β-strands in the predicted 3D model of ss asprosin might be involved in strengthening the backbone of the protein and thereby enhancing the stability29. Similarly, tertiary structure of human asprosin predicted using bioinformatics tools also showed the presence of several β-strands30. Molecular dynamics (MD) simulation and energy minimization represent an important tool for the optimization of 3D models and relaxation of geometric chains with the unfavourable bond angle, bond length and torsion angles31,32. MD simulation of 3D model of ss asprosin was carried out for 100 ns and stability parameters revealed that it is stable in the physiological system. The RMSD value determines the protein stability on the basis of conformational changes in the Cα backbone of complete structure from the initial to the final position. The conformation of a protein is relative to the fluctuations occurring during the simulation process. These fluctuations are plotted as RMSD value vs the time during which simulation was in process. The smaller deviations in RMSD values indicate a stable structure33. After 80 s of simulation, the ss asprosin showed stable RMSD value of approximately 0.95 nm. RMSF value determines the fluctuations in the Cα atom of individual residue during MD simulation. The amino acid residues between position 5-20 and 115-125 in the 3D model of ss asprosin include coils and hence show more fluctuations in the RMSF value. Residues involved in the formation of loops, coils and turns and therefore more exposed to solvents are known to have high fluctuations in the RMSF value34. Loops, coils and turns are flexible random structures in the model and play crucial roles in protein function and folding35,36. The ss asprosin showed stable Rg value which is an indicator of globularity and compactness of the protein37. Further, the Ramachandran plot analysis revealed more than 80% residues of ss asprosin in the most favoured region and 0% residues in disallowed regions. Thus, the geometrical evaluation of ss asprosin along with RMSD, RMSF and Rg value attests the robustness of the predicted 3D model of putative ss asprosin protein.
Conclusion
Asprosin is an important metabolic hormone, playing diverse roles in mammals and implicated in various metabolic disorders. The hormone is encoded by the C-terminal of the FBN1 gene. Interestingly, since its discovery in 2016, there have been no studies on asprosin in any other vertebrate group. Since, teleost are being the most abundant extant vertebrates, we undertook the current study to explore the possibility of the presence of asprosin in a teleost model, Channa punctata. The expression of fbn1 gene and the enzyme furin responsible for cleavage of the profibrillin into fibrillin-1 and asprosin, in various tissues of C. punctata suggest the presence of asprosin in teleost. The conserved furin cleavage site in the profibrillin helped in determining the putative primary ss asprosin sequence. Understanding the physicochemical properties of ss asprosin helped in determining the nature of the putative protein. Although, N-linked glycosylation sites in ss asprosin were found similar to human asprosin, the exact role of glycosylation and phosphorylation in asprosin are yet to be elucidated. The cloning and expression of ss recombinant asprosin has been carried out. In future, the purified ss recombinant asprosin could be used to understand the role played by this hormone in fish physiology, especially in reproductive functions.
Material and methods
Identification and validation of fbn1 and furin gene transcript
The transcript of fbn1 and furin gene were retrieved from the testicular transcriptome of Channa punctata, previously annotated with Takifugu rubripes, Oreochromis niloticus and Rattus norvegicus reference protein sequences38. The transcript with longest open reading frame (ORF) and maximum percentage identity was selected from several fbn1 and furin transcripts with alternate ORF (Gene runner version 3.05, Hastings Software, Inc., USA) and using Blastx (http://blast.ncbi.nlm.nih.gov/Blast.cgi), the nucleotide sequence was verified.
Reverse transcriptase polymerase chain reaction (RT-PCR) was used to validate the transcript encoded sequence. Using Clustal omega, multiple sequence alignment was constructed to determine the conserved region for designing gene specific primers for fbn1 and furin (http://www.ebi.ac.uk/Tools/msa/clustalo)39. Table 2 includes the details of the primers. For RT-PCR procedure, the following steps were employed: initial denaturation (95 °C for 5 min), 35 cycles of denaturation (95 °C for 30 s), annealing (60 °C for 30 s) and extension (72 °C for 45 s), followed by final extension (72 °C for 10 min). 1% agarose gel (Himedia, India) was used to resolve the amplified product of desired length with ethidium bromide staining. For elution, Wizard SV Gel and PCR Clean-Up System (Gel extraction kit Cat. No. A9281, Promega, USA) was used and Sanger sequencing was employed for fbn1 and furin nucleotide sequence. Using Clustal Omega, the obtained partial sequence was aligned with the transcript encoded sequence and it exhibited complete similarity. The same partial sequence of fbn1 and furin were submitted to the NCBI GenBank and accession number OP271666 and OP921042 has been assigned.
Expression analysis of fbn1 and furin gene in C. punctata
Fish procurement and maintenance
Adult spotted snakehead C. punctata weighing 80–100 g was obtained from the wild populations in and around the National Capital Region of Delhi, India and stocked in dechlorinated fresh water tanks (15 fish per tank containing 45 L water) having dimension: 74 cm × 34 cm × 32 cm (L × B × H) under light regimen of 12 L:12 D at 25 ± 2 °C. The water was changed on alternate days and the water temperature was maintained at 24–26 °C. The fish were acclimatized for 3 weeks. The protocol has been approved by the Institutional Animal Ethics Committee (DU/ZOOL/IAEC-R/2021/6), Department of Zoology, University of Delhi and experiment was carried out following the relevant guidelines and regulations of the IAEC. The studies involving the live animals follows the recommendations in the ARRIVE guidelines.
Tissue-specific expression of fbn1 and furin
Healthy spotted snakeheads (N = 6) were sacrificed with an overdose of 2-phenoxyethanol (5 mL/L water, Sisco Research Laboratories, Mumbai, India). Different tissues including brain (forebrain, hindbrain and midbrain), heart, liver, adipose tissue, gut, gonads and lymphoid organs including head kidney, spleen, skin, gills and trunk kidney were dissected out and stored at − 80 °C until RNA extraction after washing with 1×PBS.
RNA extraction and cDNA synthesis
Using the TRIzol reagent, total RNA was extracted from tissues following the manufacturer's protocol (Invitrogen). For assessment of RNA integrity, 1% agarose gel was used to observe the band intensities of 28S and 18S rRNA and RNA levels were quantified using a Nanodrop (ND-1000, Nanodrop Technologies, USA). The RNA samples having 1.8–2.0 absorbance ratio at A260/280 were selected for the cDNA preparation. In order to remove genomic contamination, RNA was treated with the enzyme DNase I (Thermo Scientific, USA) in 0.2 mL PCR tubes in a standard thermocycler at 37 °C for 30 min, and to terminate the DNase activity, EDTA (Thermo Scientific) was added and reaction was run at 65 °C for 10 min and then 4 °C hold. Using avian myeloblastosis virus (AMV) reverse transcriptase, the treated samples were processed for reverse transcriptional synthesis of single-strand cDNA following manufacturer’s specifications (Cat. No. K1622, Thermo Scientific, USA) in a thermocycler at 65 °C (for 10 min), 25 °C (for 5 min), 42 °C (for 60 min), 72 °C (for 10 min) and 4 °C hold. Through amplification of the 18S rRNA housekeeping gene, the synthesis of cDNA was confirmed.
Real-time quantitative PCR (qPCR) for fbn1
The partially validated sequence of fbn1 was used for qPCR primers design in order to quantitate the target mRNA transcript. For validation of primers, melt curve analysis was employed. 1% agarose was used to resolve the amplified products of fbn1 gene and using ethidium bromide staining method, single bands were visualized. The qPCR primers percentage efficiency was checked by a standard curve using two-fold serial dilutions of ovarian cDNA. Similarly, using specific primers, 18S rRNA was quantified as a reference gene in each sample. The reaction was carried out in qPCR machine having standard cycle mode: 50 °C (for 2 min), 95 °C (for 2 min), 95 °C (for 15 s), 59 °C (for 15 s) and 72 °C (for 1 min) using power SYBR Green (Cat. No. 4367659, Applied Biosystems, USA) and finally a dissociation step: 95 °C (for 15 s), 60 °C (for 1 min) and 95 °C (for 15 s) for melt-curve analysis.
Semi-quantitative PCR (RT-PCR) for furin
Tissues expressing high levels of fbn1 such as different parts of the brain (forebrain, midbrain and hindbrain), heart, testis and ovary were selected for studying the expression of furin. Using semi-quantitative primers of furin and 18S rRNA, RT-PCR procedure was employed with the following steps: initial denaturation (95 °C for 5 min), 35 cycles of denaturation (95 °C for 30 s), annealing (60 °C for 30 s) and extension (72 °C for 45 s), followed by final extension (72 °C for 10 min). 1% agarose gel (Himedia, India) was used to resolve the amplified products of desired length with ethidium bromide staining. The gel was observed under ChemiDocTM XRS + Imaging system (Bio-rad).
Statistical analysis
18S rRNA expression was used to normalize the relative expression of fbn1. In order to calculate the relative fold change, 2− ∆∆CT method40 was employed, wherein the tissue that showed lowest expression, i.e., head kidney was considered as a reference for tissue distribution. For the statistical analysis GraphPad Prism 8.0.1 software (GraphPad Software, La Jolla, CA) was used.
Identification of furin cleavage site
For the prediction and analysis of putative peptide sequence encoded by fbn1 gene, ExPASy server (http://ca.expasy.org/)41 was used. The sequence alignment was constructed for the putative partial sequence of profibrillin of C. punctata with the profibrillin of Homo sapiens using Clustal omega. Furin cleavage site (R-X-K/R-R↓ or R/K-X-X-X-K/R-R↓; where ↓ represents cleavage site)2 was manually identified in the aligned sequences near the C-terminal region. The amino acid sequence towards the C-terminus after the furin cleavage site represented the putative ss asprosin protein sequence as mentioned by the Romere et al.1.
Molecular cloning and characterization of putative ss asprosin protein
Cloning
The C-terminal region of ss fbn1 that is predicted to code for asprosin (8026–8454 bp) was cloned in the pProEx-HTc vector, having N-terminal His-tag. To clone the gene region of interest, the sequences of forward and reverse primers were 5′-CCTGGATCCTAAGCACTAACGCAACACACGATGAGC-3′ (carrying a BamHI site) and 5′-CGCAAGCTTTTAATGGAGGATGATCTGCACCCTC-3′ (containing a HindIII site). The amplified product using these primers was digested with the BamHI and HindIII restriction enzymes and the resulting fragment was ligated into the pProEx-HTc plasmid, which was also previously digested with the same restriction enzymes. After ligation, the plasmid containing gene of interest was transformed into the DH5α cells and grown overnight on 100 µg/ml ampicillin resistance agar plates at 37 °C. The white colonies from the plate were picked, grown and maintained in the LB medium supplemented with 100 µg/ml ampicillin at 37 °C overnight with constant shaking (220 rpm). The DH5α cells containing plasmid were pellet down and DNA was extracted using WizardⓇ Plus Minipreps DNA purification system (Cat. No. A7660, Promega, USA). The integrity of plasmid construct was validated through Sanger sequencing.
Expression
BL21(DE3) cells were transformed with pProEx-HTc vector derivative expressing asprosin protein and inoculated into LB medium containing 25 µg/ml chloramphenicol and 50 µg/ml ampicillin followed by incubation at 37 °C with constant shaking (220 rpm) until A600 reached at OD 0.6. 1 mM Isopropyl 1-thio-d-galactopyranoside (IPTG) was added in the culture for the induction of recombinant protein and culture was grown for an additional 2 h at 37 °C with constant shaking (220 rpm). The cells were pellet down and resuspended in cell lysis buffer containing 50 mM Tris–Cl, pH 8.0, 300 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 10 mg/ml lysozyme, 0.1% SDS, 1× protease inhibitor cocktail and 1 mM phenylmethylsulfonyl fluoride and lysed by sonication. The cell lysate was then centrifuged at 12,000 rpm at 4 °C for 30 min. The supernatant and pellet were collected separately and stored at − 80 °C. Further, to check the induction of asprosin protein, protein samples from induced and uninduced cells were mixed with 2× SDS loading dye and loaded on 15% SDS-PAGE for gel electrophoresis and observed using coomassie brilliant blue staining. To validate the recombinant asprosin protein, western blotting was done using anti-His antibody (Anti-PolyHistidine-Peroxidase antibody, mouse monoclonal, Cat No. A7058, Sigma-Aldrich). The uninduced supernatant sample was taken as control. Briefly, for western blotting, protein was resolved in the 15% SDS-PAGE and transferred onto nitrocellulose membrane in transfer buffer (193 mM glycine, 25 mM Tris and 20% methanol, pH 8.5). After transfer, blocking of nitrocellulose membrane was done in 5% BSA for 1 h and subsequently incubated in the primary anti-His antibody (dilution 1:2000) for 1 h. The membrane was washed three times with TBST (50 mM Tris, 150 mM NaCl, 0.1% Tween-20, pH 7.6) and bands were developed using Luminata™ Crescendo Western HRP substrate (Millipore Corporation) and the luminescent image analyzer amersham imager-600 (GE/Biosciences AB) was used for visualization.
Physicochemical properties and post-translational modifications
To determine the molecular weight, grand average of hydropathicity index (GRAVY) and other physicochemical properties, the deduced primary sequence of asprosin of C. punctata and H. sapiens asprosin were subjected to ProtParam tool (http://expasy.org/cgi-in/protparam)42. Clustal omega was employed to compute the percentage similarity of deduced asprosin of C. punctata with asprosin of H. sapiens. Motifscan software (https://myhits.sib.swiss/cgi-bin/motif_scan) was employed for determining the post-translational modifications such as glycosylation and phosphorylation in the deduced asprosin of C. punctata.
Modelling and MD simulation
Automated modelling program I-TASSER (Iterative-Threading ASSEmbly Refinement) server43 was employed for modelling 3D structure of putative ss asprosin protein in order to gain insight into asprosin protein. For validation of the obtained 3D model, MD simulation was carried out using GROMACS (Groningen Machine for Chemical Simulations) 2019.2 version for 100 ns with force field Charmm2744. Simulation system was solvated with the TIP4P water model in a triclinic box. 25 Na+ and 15 Cl− ions were added to the system for achievement of electroneutrality by steepest descent energy minimization. For the position restrained simulation, system equilibrium was done in two phases: NVT (constant number of particles, volume and temperature) and NPT (constant number of particles, pressure and temperature) ensemble for 500 ns each. System temperature and pressure was maintained at 300 K and 1 bar during MD runs via Berendsen and Parinello-Rahman methods. For long-range electrostatic interactions and bond length constrain, Particle Mesh Ewald (PME) and LINC algorithm were used45. GROMACS modules such as gmx rms, gmx rmsf and rmx gyrate were performed for protein analysis. After MD simulation, the 3D model of asprosin was subjected to the SAVES (Structural Analysis and Verification Server) and geometry evaluation was done using PROCHECK module (http://servicesn.mbi.ucla.edu/SAVES/)46.
Ethics declaration
The protocol has been approved by the Institutional Animal Ethics Committee (DU/ZOOL/IAEC-R/2021/6), Department of Zoology, University of Delhi and all the methods were performed in accordance with the relevant guidelines and regulations of the IAEC. The studies involving the live animals follows the recommendations in the ARRIVE guidelines.
Data availability
The datasets analysed during the current study are available in the National Centre for Biotechnology Information (NCBI) GenBank repository [Accession number OP271666 and OP921042].
References
Romere, C. et al. Asprosin, a fasting-induced glucogenic protein hormone. Cell 165, 566–579 (2016).
Molloy, S. S., Anderson, E. D., Jean, F. & Thomas, G. Bi-cycling the furin pathway: From TGN localization to pathogen activation and embryogenesis. Trends Cell Biol. 9, 28–35 (1999).
Dietz, H. C. & Pyeritz, R. E. Mutations in the human gene for fibrillin-1 (FBN1) in the Marfan syndrome and related disorders. Hum. Mol. Genet. 4, 1799–1809 (1995).
Li, E. et al. OLFR734 mediates glucose metabolism as a receptor of asprosin. Cell Metab. 30(2), 319–328 (2019).
Mishra, I. et al. Protein tyrosine phosphatase receptor δ serves as the orexigenic asprosin receptor. Cell Metab. 34, 549–563 (2022).
Lee, T., Yun, S., Jeong, J. H. & Jung, T. W. Asprosin impairs insulin secretion in response to glucose and viability through TLR4/JNK-mediated inflammation. Mol. Cell. Endocrinol. 486, 96–104 (2019).
Zhang, X., Jiang, H., Ma, X. & Wu, H. Increased serum level and impaired response to glucose fluctuation of asprosin is associated with type 2 diabetes mellitus. J. Diabetes Investig. 11, 349–355 (2020).
Long, W. et al. Decreased circulating levels of asprosin in obese children. Horm. Res. Paediatr. 91, 271–277 (2019).
Wang, M. et al. Serum asprosin concentrations are increased and associated with insulin resistance in children with obesity. Ann. Nutr. Metab. 75, 205–212 (2019).
Alan, M. et al. Asprosin: A novel peptide hormone related to insulin resistance in women with polycystic ovary syndrome. Gynecol. Endocrinol. 35, 220–223 (2018).
Jiang, Y., Liu, Y., Yu, Z., Yang, P. & Zhao, S. Serum asprosin level in different subtypes of polycystic ovary syndrome: A cross-sectional study. Rev. Assoc. Méd. Bras. 67, 590–596 (2021).
Wei, F., Long, A. & Wang, Y. The Asprosin-OLFR734 hormonal signaling axis modulates male fertility. Cell Discov. 5, 1–3 (2019).
Maylem, E. R. S., Spicer, L. J., Batalha, I. & Schutz, L. F. Discovery of a possible role of asprosin in ovarian follicular function. J. Mol. Endocrinol. 66, 35–44 (2021).
Keskin, T., Erden, Y. & Tekin, S. Intracerebroventricular asprosin administration strongly stimulates hypothalamic-pituitary-testicular axis in rats. Mol. Cell. Endocrinol. 538, 111451 (2021).
Maylem, E. R. S. et al. A potential role of fibrillin-1 (FBN1) mRNA and asprosin in follicular development in water buffalo. Theriogenology 178, 67–72 (2022).
Duerrschmid, C. et al. Asprosin is a centrally acting orexigenic hormone. Nat. Med. 23, 1444–1453 (2017).
Maurya, S., Krishna, A., Lal, B. & Singh, A. Asprosin promotes steroidogenesis and spermatogenesis with improved glucose metabolism in adult mice testis. Andrologia 2022, e14579. https://doi.org/10.1111/and.14579 (2022).
Guruprasad, K., Reddy, B. B. & Pandit, M. W. Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. Des. Sel. 4, 155–161 (1990).
Ikai, A. Thermostability and aliphatic index of globular proteins. J. Biochem. 88, 1895–1898 (1980).
Kyte, J. & Doolittle, R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 (1982).
Perlman, S. et al. Glycosylation of an N-terminal extension prolongs the half-life and increases the in vivo activity of follicle stimulating hormone. J. Clin. Endocrinol. Metab. 88, 3227–3235 (2003).
Sinclair, A. M. & Elliott, S. Glycoengineering: The effect of glycosylation on the properties of therapeutic proteins. J. Pharm. Sci. 94, 1626–1635 (2005).
Smith, P. L., Kaetzel, D., Nilson, J. & Baenziger, J. U. The sialylated oligosaccharides of recombinant bovine lutropin modulate hormone bioactivity. J. Biol. Chem. 265, 874–881 (1990).
Bishop, L. A., Robertson, D. M., Cahir, N. & Schofield, P. R. Specific roles for the asparagine-linked carbohydrate residues of recombinant human follicle stimulating hormone in receptor binding and signal transduction. Mol. Endocrinol. 8, 722–731 (1994).
Ulloa-Aguirre, A., Midgley, A. R. Jr., Beitins, I. Z. & Padmanabhan, V. Follicle-stimulating isohormones: Characterization and physiological relevance. Endocr. Rev. 16, 765–787 (1995).
Grossmann, M. et al. Expression of human thyrotropin in cell lines with different glycosylation patterns combined with mutagenesis of specific glycosylation sites: Characterization of a novel role for the oligosaccharides in the in vitro and in vivo bioactivity. J. Biol. Chem. 270, 29378–29385 (1995).
Arey, B. J. et al. Induction of promiscuous G protein coupling of the follicle-stimulating hormone (FSH) receptor: A novel mechanism for transducing pleiotropic actions of FSH isoforms. Mol. Endocrinol. 11, 517–526 (1997).
Bousfield, G. R. et al. Differential effects of α subunit asparagine56 oligosaccharide structure on equine lutropin and follitropin hybrid conformation and receptor-binding activity. Biochemistry 43, 10817–10833 (2004).
Kellis, J. T., Nyberg, K. & Fersht, A. R. Contribution of hydrophobic interactions to protein stability. Nature 333, 784–786 (1988).
Liu, L. et al. The effects of asprosin on exercise-intervention in metabolic diseases. Front. Physiol. 13, 1285 (2022).
Han, Y. & Elliott, J. Molecular dynamics simulations of the elastic properties of polymer/carbon nanotube composites. Comput. Mater. Sci. 39, 315–323 (2007).
Chuphal, B., Rai, U., Kumar, R. & Roy, B. Molecular and functional characterization of spotted snakehead NOD1 with an emphasis on structural insights into iE-DAP binding motifs employing advanced bioinformatic tools. J. Biomol. Struct. Dyn. 40(16), 7483–7495 (2022).
Aier, I., Varadwaj, P. K. & Raj, U. Structural insights into conformational stability of both wild-type and mutant EZH2 receptor. Sci. Rep. 6, 1–10 (2016).
Bös, F. & Pleiss, J. Multiple molecular dynamics simulations of TEM β-lactamase: Dynamics and water binding of the Ω-loop. Biophys. J. 97, 2550–2558 (2009).
Leszczynski, J. F. & Rose, G. D. Loops in globular proteins: A novel category of secondary structure. Science 234, 849–855 (1986).
Yang, J. Comprehensive description of protein structures using protein folding shape code. Proteins Struct. Funct. Genet. 71, 1497–1518 (2008).
Lobanov, M. Y., Bogatyreva, N. S. & Galzitskaya, O. V. Radius of gyration as an indicator of protein structure compactness. Mol. Biol. 42, 623–628 (2008).
Roy, A., Basak, R. & Rai, U. D. novo sequencing and comparative analysis of testicular transcriptome from different reproductive phases in freshwater spotted snakehead Channa punctatus. PLoS ONE 12, e0173178 (2017).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Gasteiger, E. et al. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 (2003).
Gasteiger, E. et al. Protein identification and analysis tools on the ExPASy server. Proteom. Protocols Handb. 2005, 571–607 (2005).
Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 9, 1–8 (2008).
Brooks, B. R. et al. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–217 (1983).
Hess, B., Bekker, H., Berendsen, H. J. & Fraaije, J. G. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997).
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallog. 26, 283–291 (1993).
Acknowledgements
We are thankful for University Grant Commision (UGC), India for granting research fellowship to Priyanka Sathoria and Bhawna Chuphal (Ref No. F. 44-1/2018 SA-III, Ref. No. 19/06/2016(i)EU-V). We would like to acknowledge Dr. Jogeswar Satchidananda Purohit, Assistant Professor in Biochemistry and Molecular Biology, Cluster Innovation Centre, University of Delhi, India for his support and valuable suggestions. We are thankful to Principal, Maitreyi College, University of Delhi, India for her support and encouragement.
Author information
Authors and Affiliations
Contributions
P.S.: conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing-original draft. B.C.: data curation, formal analysis, molecular modelling, writing and editing. U.R.: conceptualization, resources, supervision, visualization, writing and editing, supervision. B.R.: conceptualization, methodology, validation, resources, writing & editing, supervision.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sathoria, P., Chuphal, B., Rai, U. et al. Molecular cloning, characterization and 3D modelling of spotted snakehead fbn1 C-terminal region encoding asprosin and expression analysis of fbn1. Sci Rep 13, 4470 (2023). https://doi.org/10.1038/s41598-023-31271-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-31271-x
This article is cited by
-
Asprosin: its function as a novel endocrine factor in metabolic-related diseases
Journal of Endocrinological Investigation (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.