Abstract
Although the Mediterranean Sea is a crucial hotspot in marine biodiversity, it has been threatened by numerous anthropogenic pressures. As flagship species, Cetaceans are exposed to those anthropogenic impacts and global changes. Assessing their conservation status becomes strategic to set effective management plans. The aim of this paper is to understand the habitat requirements of cetaceans, exploiting the advantages of a machine-learning framework. To this end, 28 physical and biogeochemical variables were identified as environmental predictors related to the abundance of three odontocete species in the Northern Ionian Sea (Central-eastern Mediterranean Sea). In fact, habitat models were built using sighting data collected for striped dolphins Stenella coeruleoalba, common bottlenose dolphins Tursiops truncatus, and Risso’s dolphins Grampus griseus between July 2009 and October 2021. Random Forest was a suitable machine learning algorithm for the cetacean abundance estimation. Nitrate, phytoplankton carbon biomass, temperature, and salinity were the most common influential predictors, followed by latitude, 3D-chlorophyll and density. The habitat models proposed here were validated using sighting data acquired during 2022 in the study area, confirming the good performance of the strategy. This study provides valuable information to support management decisions and conservation measures in the EU marine spatial planning context.
Similar content being viewed by others
Introduction
The Marine Strategy Framework Directive (MSFD), Marine Spatial Planning (MSP) and Common Fisheries Policy (CFP) constitute the main policies to maintain the productive, resilient, and good health status (GES) of marine habitats to provide ecosystem services and limit the loss of biodiversity in EU Member States (EEA, 2015). This environmental strategy, although different in terms of achievable objectives, is based on the Ecosystem Based Management approach (EBM), which is assumed to be a holistic and integrated pathway worldwide. In particular, this approach aims to maintain or restore the composition, structure, function, and delivery of services of natural and modified ecosystems to achieve sustainability (Millennium Ecosystem Assessment, 2005). In this light, knowledge of the spatiotemporal distribution and abundance of target species, as well as the extension of their critical habitats and their overlap with highly impacted areas strongly characterized by anthropogenic pressures, is essential, especially in aquatic ecosystems.
Although, on a global scale, the Mediterranean Sea is one of the most important hotspots for its richness in marine biodiversity1,2,3, it has been historically threatened by numerous anthropogenic pressures, such as the presence of commercial maritime and fishing activities, a growing urbanization mostly along coastal zones, and the occurrence of different sources of pollution, from chemical to acoustic4,5. In addition, climate change, the spreading of alien species, and the increasing occurrence of disease outbreaks are considered the most recent verified drivers of impact on the basin6,7,8. The cetaceans of the Mediterranean Sea are among species worthy of conservation distributed in a heavily anthropized basin9. In this context, dolphins and whales can be exposed to several impacts, such as bycatch, competition of resources due to fishing activities10,11,12,13,14, shipping collisions15, chemical pollution from persistent organic pollutants, marine litter, heavy metals16,17,18 and noise pollution19,20.
To date, several studies21,22,23,24,25,26,27,28,29,30,31,32 have been conducted to provide information on the estimated abundance and distribution of regular species occurring in Mediterranean eco-regions. Moreover, over the last three decades, extensive literature describing methodological approaches to assess the abundance of top marine predators has rapidly expanded33,34,35,36, starting from methodologies requiring basic information (e.g., number of individuals, distances, photoidentification data), based on distance sampling37,38,39 and mark-recapture methods40,41,42, to more advanced techniques requiring a greater amount of data36. The latter brings us face to face one of the big challenges in ecology, namely, the identification of environmental predictor variables, which help to forecast bioecological responses based on environmental changes43,44,45. An example of the modeling approaches for an abundance assessment are model-based estimation methods, such as density surface modeling46, species distribution modeling47, and the most powerful machine learning techniques48. Generalized Additive Models49, Neural Networks50, Least Squares Boosting51, Random Forest52 and Support Vector Machines53 are some of the most popular learning models and have already been successfully applied in several application domains43,54,55,56,57,58,59,60,61,62,63. Despite this plethora of information, the question regarding cetacean species abundance in the Mediterranean Sea is far from being closed. In fact, enormous efforts are required in the continuous updating of the collected data, thus covering longer periods of sightings of cetacean populations, as well as larger study areas. Moreover, the evolution of the theoretical and computational improvements of the modeling approaches for the estimation of abundance and for the identification of environmental predictors require a continuous training of the models through the employment of state-of-the-art statistical techniques and strategies.
This study starts with the identification of the main environmental predictors related to the abundance of three cetacean species: the striped dolphin Stenella coeruleoalba, the common bottlenose dolphin Tursiops truncatus, and the Risso’s dolphin Grampus griseus, observed in the Gulf of Taranto (Northern Ionian Sea, Central-eastern Mediterranean Sea). A group of 28 environmental variables, extracted by the Copernicus Marine Service (https://marine.copernicus.eu/it) and EMODnet-bathymetry dataset (https://www.emodnet-bathymetry.eu/data-products), are tested to train three regression models: RF, LSBoost, and NN. Next, estimates of species abundance are provided as approach examples of habitat suitability definitions and baselines for these odontocetes in the Mediterranean Sea. Habitat models were developed using sighting data collected during marine surveys in the study area from July 2009 to October 2021. Finally, the most important variables for building these habitat models were identified and examined, and a validation of the proposed strategy for abundance estimation was provided using sighting data collected in 2022. All the sighting data and environmental variables, used in this study, are freely available.
Results
All data were analyzed using MATLAB (MathWorks, Natick, MA). To build habitat models, dataset D, which collected sighting data in the period 2009–2021, was used (see “Data description” section in the “Materials and methods”). In particular, abundance data from striped dolphin, common bottlenose dolphin and Risso’s dolphin have been analyzed in relation to the following geographical, physical, and biochemical ocean variables: latitude, longitude, distance of the sighting from the coastline (Distance_From_Coast), maximum depth of the water column in the numerical model (Max_Depth), bathymetry (Emodnet_Depth), primary production*, nitrate*, phosphate*, phytoplankton carbon biomass* (PHYC), 3D-chlorophyll* (CHL3D), chlorophyll a (CHLA) at surface (CHLA), temperature*, salinity*, density*, mixed layer depth, thermocline depth (depthOfMaxN2), currents speed* (Currents_Intensity) and direction* (Currents_Direction). The three-dimensional variables have been labeled with * symbols and contain two values: the top value, which is the value computed at the top of the water column. corresponding to the variable mean value in the range [0, 40 m]; the bottom value computed at the bottom of the water column, given by the variable mean value in [50 m, 200 m]. Hence, a total of 28 environmental variables were used to train the models.
The performances of three regression models, LSBoost, RF and NN, in terms of Root Mean Square Error (RMSE), have been evaluated using a K-fold cross-validation, with K empirically set equal to five. Therefore, each model was trained on 80% of the available examples in the dataset D and tested on the remaining 20% at each run of the cross-validation procedure, and evaluation metrics were obtained by averaging values in the five runs. Parameter tuning for each model was empirically performed (see the “Regression models” section of the “Materials and methods”). The first experiment was conducted on the sighting data of striped dolphin (S), common bottlenose dolphin (T), and Risso’s dolphin (G). The performances of the three models, whose optimized.
hyperparameters are shown in Supplementary material, Table S1, were quite similar (see Table 1).
In particular, RF performances were slightly better than those of NN and LSBoost, with an RMSE of 6 individuals for the T dataset, 8 for the G dataset and 46 for the S dataset. Very important is that RF required a lower training time; hence, in the following, we always refer to this algorithm.
Differences between the results obtained on the T and G datasets with the results on the S dataset are reasonably due to multiple factors. Note that comparisons between datasets or models are out of the scope of this study. A first consideration is that the influence of the 28 variables on the prediction of cetacean abundance, made by RF models, could vary in different ranges of the values of group size, especially when these values vary greatly, thus influencing the performance of the regression model. Moreover, a main concern is the relatively minor dimension of groups of common bottlenose dolphins and Risso’s dolphins with respect to those of striped dolphins. In fact, in the T and G datasets, the maximum number of individuals observed in the groups size during the sightings is equal to 30 and 50 individuals, respectively (see the “Data description” section in the “Materials and methods”), so marine mammal observers can be obviously more accurate when counting. Instead, striped dolphin group size values can reach even hundreds of individuals, making their counting very difficult when group size increases. Obviously, the introduced bias increases with the dimension of the group. Therefore, when the group size counting is inaccurate, the regression model will be trained on wrong data, affecting the value of the RMSE computed. To test this hypothesis, the dataset S has been split into four subsets, according to the size of the observed groups during the sightings: S1 contains 585 sightings whose group size ranges from 1 to 30; S2 counts 362 sightings with group size varying in the interval [31, 60]; S3 contains 135 sightings whose group size ranges from 61 to 90; S4 counts 145 sightings with group size in the interval [91, 150] (see Table 2 and Fig. 1).
Dataset S contains only 37 sightings with group sizes greater than 150, whose values fall into very few bins (see Supplementary material, Fig. S1); hence, these samples were discarded in the following analysis. The four subsets and relative cutoffs were selected considering the max group size values observed in the T and G datasets, which were 30 and 50 individuals, respectively. Moreover, some empirical considerations were made to ensure enough examples in each subset to train the models. Finally, RF models were trained on the four subsets, and their performances in terms of RMSE are shown in Table 2, while their optimized hyperparameters are shown in Supplementary material, Table S2. Experimental results highlight good performances of RF on subsets S1, S2, and S3, showing RMSE values varying from 6 to 9 individuals. Instead, the RMSE of RF on S4 increases up to 19. Figure 1 shows the distribution of the examples in the different group sizes for the S1, S2, S3, S4, G and T datasets. The main difference between S4 and the other datasets is that few examples in a few bins of the S4 histogram are available. It is reasonable that the shape of this histogram affects the RF regression performance, which becomes worse than others. The appearance of the S4 histogram can be imputable to inaccurate observations when individuals are greater than approximately one hundred, while the frequency of sighting striped dolphin groups with this number or greater number of individuals becomes low. The tested hypothesis is confirmed by these results, and the proposed strategy based on RF and the 28 environmental variables for the abundance estimation for these odontocetes is effective, mostly when the group size is lower than 90 individuals. In addition, the identification of the most common influential predictors, among the 28 environmental variables considered here, was assessed by ranking their importance given by the RF models (see Table 3 and Supplementary material, Figs. S2:S7) and analyzing the first ten positions of the ranked lists.
Feature importance measures how variables influence the model when predicting the response. The influence of a predictor variable increases with the value of this measure. The idea underlying the feature importance computed by RF models is that if a variable is influential in prediction, then permuting its values should affect the model error; if a variable is not influential, then permuting its values should have little to no effect on the model error. Overall, nitrate, phytoplankton carbon biomass, temperature, and salinity, at the top of the water column, were included in 83% of the models, followed by latitude, top 3D-chlorophyll and top density in 67% of the models. Note that temperature at the bottom of the water column was also important in half of the models. Globally, the less influential variables (i.e., variables with zero frequency in the first ten positions of the ranked lists) resulted in the maximum depth, chlorophyll a at the surface, thermocline depth and bottom current direction and bottom phosphate.
In particular, the group size of striped dolphin seems to be positively influenced by the concentration of nitrate, especially for datasets S1 and S4, which is contrary to what was observed for T and G, for which this parameter seems negatively affects their group size although in a slight way (Supplementary material, Fig. S9). However, it is currently difficult to hypothesize about the meaning of these relationships, which need to be further investigated, also using larger sighting datasets. Additionally, the salinity and the temperature at the top layer positively affected the group size of this species, especially for dataset S4 and datasets S1 and S2, respectively (Supplementary material, Figs. S10 and S11). Similarly, the temperature at the top layer positively affected the group size of Risso’s dolphin, while the phytoplankton carbon biomass was negatively correlated with its group size (Supplementary material, Figs. S11 and S12). For the common bottlenose dolphin, in addition to the concentration of nitrates, which were found to negatively influence the size of the groups (Supplementary material, Fig. S9), other environmental features that were important for the prediction of abundance were primary production, 3D-chlorophyll and Emodnet_depth (Supplementary material, Fig. S7).
Finally, the proposed strategy for the cetacean abundance estimation was validated using 5 sighting data of striped dolphin and 2 of bottlenose dolphin collected in the study area during 2022, obtaining good performances with an average RMSE equal to 6 individuals.
Discussion
Assessing the abundance of top marine predators and identifying the relationship between their abundance and environmental variables are primary goals in the framework of EU policies aimed to protect and preserve biodiversity and ecosystems9,64,65,66 for the adequate understanding of habitat suitability for different species and the implementation of correct conservation measures.
Here, we propose a modeling strategy that uses RF and a robust statistical methodology to estimate cetacean abundance and to identify the most influential environmental predictors. We tested and validated it using sighting data on three different cetacean species collected in the Gulf of Taranto over a span of over ten years. Although previous studies were conducted in the same area to predict the distribution and abundance of striped and common bottlenose dolphins as a function of environmental and anthropogenic drivers22,24, this study tested a high number of features that go beyond the “classic” physiographic or environmental variables, such as depth, distance from coast, slope, sea surface temperature and chlorophyll a content. This approach allowed us to verify the most powerful statistical method among the most innovative techniques of artificial intelligence to predict the group size of dolphin species according to several predictors and to investigate the importance of other environmental variables less used in the modeling.
The concentration of nitrate, phytoplankton carbon biomass, temperature and salinity, especially in the top layer of the water column, were frequently the most relevant features for the prediction of the group size of the three cetacean species investigated here. This result is somewhat expected because these variables are strongly linked to primary production and to the occurrence of prey (i.e.,36,67,68). The positive influence of the concentration of nitrate and salinity on the group size of striped dolphins is similar to those reported in the eastern tropical Pacific Ocean69, and a positive correlation between the temperature and the group size of striped dolphins was already reported in the Northern Ionian Sea30.
Similarly, the higher abundance of Risso’s dolphin in warmer waters converges with observations reported for the same species off the California coast70,71. Regarding the features that were significant in predicting the group size abundance of common bottlenose dolphins, such as primary production, the concentration of chlorophyll a and depth; these are commonly used features in several studies (i.e.,72,73,74) were positively correlated with the abundance and distribution of species except for depth. In particular, outcomes included in this study converge with results reported by Chavez-Rosales et al. in36, whose scope was to identify the main environmental covariates tied to the abundance of 17 cetacean species in the Western North Atlantic Ocean by using Generalized Additive Models (GAM). Top temperature was highly relevant in the habitat models for Risso's striped dolphins. In addition, primary production was important for bottlenose dolphins, as already found in the Western North Atlantic Ocean. In contrast, distance from the cost is one of the most common covariates in36, while it was influential only for small groups of striped dolphins, of less than 60 individuals, in the present study (see Supplementary material, Figs. S2 and S3). Considering the statistical approaches, to the best of our knowledge, GAM and RF are among the most powerful machine learning algorithms used to predict species abundance. There is an extensive body of literature confirming the predictive ability of GAMs for cetacean abundance estimation75,76,77, as well as an increasing interest in machine-learning techniques, such as RF24,78. A future aim will be to evaluate the most effective method for predicting cetacean species abundance. In machine learning framework it is a common practice to develop studies on the performance comparison of algorithms79,80,81,82, because these can provide meaningful insights into the research topic and can highlight direction to any future studies on that topic. In the modern literature, there are only a few previous studies on this matter83, and the subject is just as intriguing when considering other species; in fact, in84, the authors performed a comparison study between GAM and RF for the density estimation of two different bird species. However, despite this interest, the question needs to be further studied.
Sighting data used in our analysis covered an extended period, from 2009 to 2022. However, a limitation of this paper is that, unfortunately, despite the research effort, the number of available samples remains relatively small; in fact, only 129 sightings for Risso's dolphins and 225 for bottlenose dolphins are available. Instead, 1264 sightings of striped dolphins have been collected; however, in the present study we pointed out that it is convenient to divide this dataset into 4 smaller subsets (S1, S2, S3, and S4), according to the size of the observed groups during the sightings. The number of sightings collected in these four datasets varies from 135 to 585, and also in this case more samples are desirable for the further machine learning analysis. Moreover, the occurrence of group sizes in each dataset varies, with various elements with zero or very low number of available samples.
Another issue raised in this paper concerns the limitations in manually counting the number of individuals encountered by marine mammal observers. Developing innovative strategies based on Unmanned Aerial Vehicle or drones to support them in this task is strongly desirable and should be of great avail85. In addition, a great effort is needed when organizing and labeling data; this task can be time consuming and critical in the present field of application. A standardization of the expert labelling process of complex data, exploiting innovative approaches, is desirable and should be investigated in the future86.
Moreover, the proposed abundance estimation strategy shows good performance on sighting data collected in 2022, never seen before during model training. However, the validation set used here contains only 7 records. A future goal will be the further validation of the proposed strategy using a larger collection of data that will be acquired in the near future.
Last, the proposed strategy is general and could be effectively tested and applied to different geographical areas.
Materials and methods
Study area
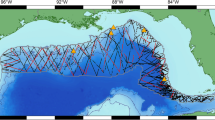
The Gulf of Taranto is in the North-western Ionian Sea (Fig. 2). It is a semienclosed ocean area, covering approximately 14,000 km2,22 and includes the coasts of the Italian regions of Apulia, Basilicata, and Calabria. It is connected to the Northern Ionian Sea and the eastern Mediterranean Sea over an extended section (from Santa Maria di Leuca to Punta Alice), which includes a narrow trench deeper than 2000 m. The Gulf shows a very complex seabed topography characterized by descending terraces on the eastern side and by a narrow continental shelf with a steep slope and several channels on the western side. Centrally, the basin is characterized by the submarine canyon system of Taranto Valley with no clear bathymetric connection to a major river system87,88,89,90 (Fig. 2). The continental shelves—area shallower than 200 m—cover 10% of the total Gulf area. Wider shelves are present on the eastern side (Apulia), and five main rivers (Bradano, Basento, Agri, Sinni, and Crati) discharge from the western coastline with a relatively low annual mean runoff91. The morphology involves a complex distribution of water masses with a mixing of surface and dense bottom waters with the occurrence of high seasonal variability in upwelling currents92,93,94,95. From the oceanographic point of view, the basin-scale circulation of the Gulf is dominated by cyclonic gyres, with reversals in anti-cyclonic patterns occurring only 10–15 times for the period 1993–201891. The ecosystem variabilities and changes (e.g., anomalous chlorophyll a bloom96) could be affected by the formation of downwelling/upwelling in the case of cyclonic/anti-cyclonic patterns. Possible coastal rim currents undergoing instabilities and forming submesoscale structures have been highlighted97. Furthermore, in95 authors described a mixed layer thickness extending down to 30 m during late summer, with an intermediate water salinity maximum—indicative of Modified Levantine Intermediate Waters—in the deep part of the Gulf. Authors in98 and in99 simulated the basin-scale and costal-scale circulation of the Gulf of Taranto using high-resolution models, highlighting the role of the Western Adriatic Coastal Current (WACC) position and strength96,100,101,102 in modulating the circulation patterns in the Gulf.
Data description
Sighting data for striped dolphin, common bottlenose dolphin and Risso’s dolphin were collected from July 2009 to April 2022 during standardized vessel-based surveys carried out onboard a 12 m catamaran investigating an area of 960 km2. The sampling effort was set to approximately 5 h/day along 35 nautical miles (nm). Speed was maintained between 7 and 8 knots, and trips only occurred in favorable weather conditions (Douglas scale ≤ 3 and Beaufort scale ≤ 4). The scientific team onboard included three observers. The first was engaged in searching activity for targets at approximately 180°, while the others supported the activities of the former, searching in a sector from the track line to 90° on the starboard and port sides, respectively.
Once a target had been sighted, the dolphin group was switched to off-effort38, maintaining a minimum distance of approximately 50 m to avoid alteration in its behavioral activity. When the dolphins approached closer, the speed of the research vessel was reduced gradually until the engine was switched off. Sighting date, time of first contact, GPS position, group size and depth (m) were all recorded.
All sighting data are freely available (see Data Availability section) and have been divided into two parts: a dataset, named D, which collected data acquired in the period 2009–2021 and was devoted to training and testing regression models; and a validation set, which collected data from 2022 and was used to validate the models.
Dataset D contains 1618 records of cetacean sightings, with the following attributes: id, date, and position of sighting (latitude and longitude), number of sighted individuals and species (see Table 5). Among the 1618 records, 1264 are sightings of striped dolphin (denoted as S), 225 are sightings of common bottlenose dolphins (denoted ad T), and 129 records are sightings of Risso’s dolphins (denoted as G) (see Table 4).
Figure 3 illustrates the distribution of the number of observations among different group sizes in the S, G and T datasets. The different trends of the data in S compared with that of data collected in T and G are immediately evident; in fact, the group size in S reached higher values, up to 500 individuals, while in G and T, the maximum group size was equal to 50 and 30, respectively. The distribution of observations of S, T and G sightings in the period 2009–2021 is shown in Fig. 4.
Details on the seasonal distribution of these sightings data are shown in Supplementary material, Table S3.
The 1618 entries of the dataset D were enriched by:
-
physical variables: ocean temperature, salinity, density, mixed layer depth, Brunt–Väisälä frequency, currents speed and direction;
-
biogeochemical variables: primary production, nitrate, phosphate, phytoplankton carbon biomass, chlorophyll;
-
auxiliary variables: max depth of the water column of the numerical model, high-resolution bathymetry, and distance of the sighting location from the coastline.
Table 5 shows a detailed description of the features used in this work. The physical features are provided by the Mediterranean Sea Physics reanalysis, produced by CMCC (IT)103,104 and delivered by Copernicus Marine Service. The product is generated by a numerical system composed of a hydrodynamic model, the Nucleus for European Modeling of the Ocean (NEMO,105), and a variational data assimilation scheme (OceanVAR,106). OceanVAR assimilates temperature and salinity vertical profiles and Sea Level Anomaly along satellite track data. The model horizontal grid resolution is 1/24° (ca. 4–5 km), and the unevenly spaced vertical levels are 141. In addition, the water density was computed according to107. The computation of the Brunt–Väisälä frequency (N2) was conducted using Copernicus Marine Service temperature and salinity, according to108.
The simulated biogeochemical features are provided by the Mediterranean Sea biogeochemical reanalysis, produced by OGS (IT) and delivered by Copernicus Marine Service109. The product at 1/24° horizontal resolution (ca. 4–5 km) is produced using the MedBFM3 model system. MedBFM3 includes the transport model OGSTM v4.0 coupled with the biogeochemical flux model BFM v5 and the variational data assimilation module 3DVAR-BIO v2.1 for surface chlorophyll. MedBFM3 is coupled offline with the physical reanalysis103, which provides daily forcing fields (i.e., currents, temperature, salinity, diffusivities, wind, and solar radiation). The ESA-CCI database of surface chlorophyll concentration (CMEMS-OCTAC REP product) is assimilated with a weekly frequency.
The chlorophyll a at the surface (CHL_A) observed from satellite is provided by the product Mediterranean Sea Reprocessed Surface Chlorophyll Concentration from Multi Satellite observations, produced by the Global Ocean Satellite monitoring and marine ecosystem study group (GOS) of the Italian National Research Council (CNR, IT110), within the Copernicus Marine Service. The Level-4 product includes the daily interpolated chlorophyll field with no data voids starting from the multi-sensor (MODIS-Aqua, NOAA-20-VIIRS, NPP-VIIRS, and Sentinel3A-OLCI) and the monthly averaged chlorophyll concentration for the multi-sensor and climatological fields, all at a 1 km resolution. Chlorophyll fields are obtained by means of the Mediterranean regional algorithms: an updated version of the MedOC4 (Case 1 waters,111, with new coefficients) and AD4 (Case 2 waters,112).
Among the auxiliary variables, high-resolution bathymetry was derived from the EMODnet-bathymetry dataset (2020-DTM (https://www.emodnet-bathymetry.eu/); the maximum depth of the water column (Max_Depth) was derived by the Mediterranean Sea Physics reanalysis; and the distance of the sighting location from the coastline (Distance_From_Coast) was computed using the geographical coordinates (lat, lon).
The features described above have been preliminarily processed before feeding the machine learning algorithms. First, the three-dimensional variables were extracted at the surface and at 10 m, 20 m, 30 m, 40 m, 50 m, 100 m, and 200 m. The variables have been limited up to 200 m of depth because of the stability and low variability of the water column below. Then, a sea-overland extrapolation procedure99,113 was used to prevent the presence of missing values interpolating the oceanic fields over each cetacean sightings record. This procedure uses a diffusive boundary layer approach that extrapolates the field values on the areas near the coastline where the Copernicus Marine Service solutions are not defined. The procedure iteratively computes the ocean quantities on the land grid points so that these quantities can be interpolated on the sighting records that are very close to the coast.
From the Brunt–Väisälä frequency N2, only one variable was derived, corresponding to the thermocline depth (i.e., depth of the max value of the N2 vector), entitled “depthOfMaxN2”, for each CMEMS grid point.
Following the temperature climatology in the study area during 2009–2021 (see Supplementary material, Fig. S8), primary production, nitrate, phosphate, phytoplankton carbon biomass, 3D-chlorophyll, temperature, salinity, density, current intensity and direction features were averaged in the intervals of [0–40 m] and [50–200 m]. Therefore, for each variable, two features were considered, named top when mean values are computed in the interval [0, 40 m] and bottom in [50–200 m].
Finally, the dataset, prescribed to the analysis with machine learning algorithms, included 1618 records of sightings, each enriched with the 28 variables (N’) previously described. The labels used to train the models were the number of individuals counted in each sighting.
Last, the validation set contains 7 records of cetacean sightings, of which 5 are of sightings of striped dolphins and 2 are of common bottlenose dolphins. For each record, the same 28 variables, already used for dataset D, were measured and used for models validation.
Regression models
Random Forest52 is an ensemble method that uses multiple decorrelated decision trees that are merged to perform regression or classification tasks: each tree is built using a random subset of features and examples, while the results on the test set are obtained by computing the average of the results of each tree. LSBoost51 a variant of the Adaboost algorithm58,114, was used for the regression. Through a weighted combination of the outputs produced by a set of weak classifiers, LSBoost defines a function able to estimate the abundance of the dolphin groups. More specifically, at each step, the algorithm fits a new classifier to the difference between the observed response and the aggregated prediction of all classifiers grown previously. The aim is to minimize the mean-squared error. All new classifiers are fitted to \({y}_{n}-\upeta \mathcal{F}\left({x}_{n}\right)\), where \({y}_{n}\) is the observed response, \(\mathcal{F}\left({x}_{n}\right)\) is the aggregated prediction from all weak classifiers grown thus far for observation \({x}_{n}\), and \(\upeta\) is the learning rate.
Finally, to compare the traditional regression models with modern deep learning techniques, a feedforward fully connected NN was developed (see Fig. 5). This class of networks consists of multiple layers of computational units, usually interconnected in a feed-forward way50. Each neuron in one layer has directed connections to the neurons of the subsequent layer. RF, LSBoost and NN models have been trained with automatic parameter tuning using Bayesian optimization115. This means that the model settings are initialized as default; after a first full cross validation on the available dataset, these settings are updated according to the just obtained performance and the current learning rate; this process is iterated for a certain number of times or until the model converges. Of course, in the end, the settings that provided the best performance were considered. Bayesian optimization uses a surrogate for the objective function, which is much easier to optimize than the objective function. It works by finding the next set of hyperparameters to evaluate the actual objective function by selecting hyperparameters that perform best on the surrogate function. In our case, the optimizable parameters of the RF models are the maximum number of splits, minimum leaf size, numbers of predictors to sample and number of ensembles learning cycles116, while the optimizable parameters of the LSBoost models are the minimum leaf size, number of ensembles learning cycles, maximum number of splits and learning rate. Last, the optimizable parameters for the neural network models are the number of hidden layers, the size of each hidden layer, the activation function, and the regularization term strength. In our application, the number of hidden layers of the networks varied from 1 to 5, and for each layer, the number of neurons was in the range [1, 100]. The activation functions used in our analysis were ReLu, tanh, sigmoid and the identity function. The regularization term strength is optimized over continuous values in the range [1e−5,1e5]/(number of observations), where the value is chosen uniformly in the log transformed range.
Root mean square error (RMSE)
The performance of a regression model is evaluated in terms of Root Mean Square Error (RMSE), a measure of the residuals between values predicted by a model and the values observed. It is defined as follows:
where N is the number of samples, \({y}_{i}\) is the real estimation of the pod abundance and \(\widehat{{y}_{i}}\) is the pod abundance predicted by the regression model.
Data availability
The datasets generated and analyzed during the current study are available on GitHub datarepository1/Environmental-variables-and-machine-learning-models-to-predict-cetacean-abundance repository at https://github.com/datarepository1/Environmental-variables-and-machine-learning-models-to-predict-cetacean-abundance. Further inquiries can be directed to the corresponding author.
References
Myers, N., Mittermeier, R. A., Mittermeier, C. G., Fonseca, G. & Kent, J. M. Biodiversity hotspots for conservation priorities. Nature 403, 853–858 (2000).
Cuttelod, A., García, V., Abdul Malak, D., Temple, H. & Katariya, V. The Mediterranean: A biodiversity hotspot under threat. In Wildl. a Chang. World an Anal. 2008 IUCN Red List Threat. Species 89–101 (2008).
Coll, M. et al. The biodiversity of the Mediterranean Sea: Estimates, patterns, and threats. PLoS ONE 5, e11842–e11842 (2010).
Coll, M. et al. The Mediterranean Sea under siege: Spatial overlap between marine biodiversity, cumulative threats and marine reserves. Glob. Ecol. Biogeogr. 21, 465–480 (2012).
Micheli, F. et al. Cumulative human impacts on mediterranean and black sea marine ecosystems: Assessing current pressures and opportunities. PLoS ONE 8, e79889 (2013).
Lejeusne, C., Chevaldonné, P., Pergent-Martini, C., Boudouresque, C. F. & Pérez, T. Climate change effects on a miniature ocean: The highly diverse, highly impacted Mediterranean Sea. Trends Ecol. Evol. 25, 250–260 (2010).
Tsirintanis, K. et al. Bioinvasion impacts on biodiversity, ecosystem services, and human health in the Mediterranean Sea. Aquatic Invasions, 17(3), 308–352 (2022).
Sanderson, C. E. & Alexander, K. A. Unchartered waters: Climate change likely to intensify infectious disease outbreaks causing mass mortality events in marine mammals. Glob. Chang. Biol. 26, 4284–4301 (2020).
EEC, 1992. European Commission. In EU Council Directive 92/43/EEC on the Conservationof Natural Habitats and of Wild Fauna and Flora. Orkesterjournalen L 7–50 206 (1992).
Bearzi, G. Interactions between cetacean and fisheries in the Mediterranean Sea. In: G. Notarbartolo di Sciara (Ed.), Cetaceans of the Mediterranean and Black Seas: state of knowledge and conservation strategies. A report to the ACCOBAMS Secretariat, Monaco, 9, 20 (2002).
Reeves, R. R., Smith, B. D., Crespo, E. A. & Notarbartolo di Sciara, G. Dolphins, Whales and Porpoises : 2002–2010 Conservation Action Plan for the world’s Cetaceans (2003).
Dolman, S., Evans, P., Ritter, F., Simmonds, M. & Swabe, J. Implications of new technical measures regulation for cetacean bycatch in European waters. Mar. Policy 124, 1043 (2020).
Carlucci, R. et al. Managing multiple pressures for cetaceans’ conservation with an Ecosystem-Based Marine Spatial Planning approach. J. Environ. Manage. 287, 112240 (2021).
Carlucci, R. et al. Assessment of cetacean–fishery interactions in the marine food web of the Gulf of Taranto (Northern Ionian Sea, Central Mediterranean Sea). Rev. Fish Biol. Fish. 31, 135–156 (2020).
Fossi, C. & Lauriano, G. Impacts of shipping on the biodiversity of large marine vertebrates: Persistent organic pollutants, sewage and debris. Marit. Traffic Eff. Biodivers. Mediterr. Sea Rev Impacts Prior. Areas Mitig. Meas. 3, 65–73 (2008).
Cardellicchio, N. Persistent contaminants in dolphins: An indication of chemical pollution in the mediterranean sea. Water Sci. Technol. 32, 331–340 (1995).
Fossi, M. C., Panti, C., Baini, M. & Lavers, J. L. A review of plastic-associated pressures: Cetaceans of the Mediterranean Sea and Eastern Australian Shearwaters as case studies. Front. Mar. Sci. 5, 125 (2018).
Marsili, L., Jiménez, B. & Borrell, A. Persistent Organic Pollutants in Cetaceans Living in a Hotspot Area (Elsevier, 2018).
Dolman, S. J., Evans, P. G. H., Notarbartolo-di-Sciara, G. & Frisch, H. Active sonar, beaked whales and European regional policy. Mar. Pollut. Bull. 63, 27–34 (2011).
di Sciara, G. N. et al. Place-based approaches to marine mammal conservation. Aquat. Conserv. Mar. Freshw. Ecosyst. 26, 85–100 (2016).
Holcer, D., Fortuna, C. M., Mackelworth, P., Cebrian, D. & Requena Moreno, S. Adriatic Sea: Important Areas for Conservation of Cetaceans, Sea Turtles and Giant Devil Rays (2015).
Carlucci, R. et al. Modeling the spatial distribution of the striped dolphin (Stenella coeruleoalba) and common bottlenose dolphin (Tursiops truncatus) in the Gulf of Taranto (Northern Ionian Sea, Central-eastern Mediterranean Sea). Ecol. Indic. 69, 707–721 (2016).
Carlucci, R., Ricci, P., Cipriano, G. & Fanizza, C. Abundance, activity and critical habitat of the striped dolphin Stenella coeruleoalba in the Gulf of Taranto (northern Ionian Sea, central Mediterranean Sea). Aquat. Conserv. Freshw. Ecosyst. 28, 324–336 (2018).
Carlucci, R. et al. Random Forest population modelling of striped and common-bottlenose dolphins in the Gulf of Taranto (Northern Ionian Sea, Central-eastern Mediterranean Sea). Estuar. Coast. Shelf Sci. 204, 177–192 (2018).
Arcangeli, A., Campana, I. & Bologna, M. A. Influence of seasonality on cetacean diversity, abundance, distribution and habitat use in the western Mediterranean Sea: Implications for conservation. Aquat. Conserv. Mar. Freshw. Ecosyst. 27, 995–1010 (2017).
Panigada, S. et al. Estimating cetacean density and abundance in the Central and Western Mediterranean Sea through aerial surveys: Implications for Management. Deep. Res. Part II-Top. Stud. Oceanogr. 141, 41–58 (2017).
Mannocci, L. et al. Assessing cetacean surveys throughout the Mediterranean Sea: A gap analysis in environmental space. Sci. Rep. 8, 1 (2018).
Panigada, S. et al. Estimates of Abundance and Distribution of Cetaceans, Marine Mega-Fauna and Marine Litter in the Mediterranean Sea from 2018–2019 surveys. ACCOBAMS vol. ACCOBAMS S (2021).
Paiu, R.-M. et al. Estimates of abundance and distribution of cetaceans in the Black Sea from 2019 surveys. ACCOBAMS 54, 45 (2021).
Azzolin, M. et al. Spatial distribution modelling of striped dolphin (Stenella coeruleoalba) at different geographical scales within the EU Adriatic and Ionian Sea Region, central-eastern Mediterranean Sea. Aquat. Conserv. Freshw. Ecosyst. 30, 1194–1207 (2020).
Renò, V. et al. A SIFT-based software system for the photo-identification of the Risso’s dolphin. Ecol. Inform. 50, 95–101 (2019).
Maglietta, R. et al. DolFin: an innovative digital platform for studying Risso’s dolphins in the Northern Ionian Sea (North-eastern Central Mediterranean). Sci. Rep. 8, 17185 (2018).
Hammond, P. S. et al. Estimating the abundance of marine mammal populations. Front. Mar. Sci. 8, 96 (2021).
Fontaine, M. C. et al. History of expansion and anthropogenic collapse in a top marine predator of the Black Sea estimated from genetic data. Proc. Natl. Acad. Sci. 109, E2569–E2576 (2012).
Alter, S. E., Rynes, E. & Palumbi, S. R. DNA evidence for historic population size and past ecosystem impacts of gray whales. Proc. Natl. Acad. Sci. 104, 15162–15167 (2007).
Chavez-Rosales, S., Palka, D. L., Garrison, L. P. & Josephson, E. A. Environmental predictors of habitat suitability and occurrence of cetaceans in the western North Atlantic Ocean. Sci. Rep. 9, 5833 (2019).
Buckland, S. et al. Introduction to Distance Sampling: Estimating Abundance of Biological Populations (Oxford University Press, 2001).
Buckland, S. T. et al. Advanced Distance Sampling: Estimating Abundance of Biological Populations (OUP Oxford, 2004).
Laake, J. S. T., Buckland, E. A., Rexstad, T. A., Marques, C. S. & Oedekoven, F. Distance sampling: Methods and applications. Biometrics 72, 1389–1390 (2016).
Hammond, P. S., Mizroch, S. A. & Donovan, G. P. Individual recognition of cetaceans: Use of photo-identification and other techniques to estimate population parameters. In Incorporating the Proceedings of the Symposium and Workshop on Individual Recognition and the Estimation of Cetacean Population Parameters (1990).
Sandercock, B. K. Handbook of capture-recapture analysis. Biometrics 62, 1276–1277 (2006).
Hammond, P. S. Mark-Recapture. In Encyclopedia of Marine Mammals (Third Edition) (eds Würsig, B. et al.) 580–584 (Academic Press, 2018).
Pless, E., Saarman, N. P., Powell, J. R., Caccone, A. & Amatulli, G. A machine-learning approach to map landscape connectivity in Aedes aegypti with genetic and environmental data. Proc. Natl. Acad. Sci. 118, 9 (2021).
Belanger, C. L. et al. Global environmental predictors of benthic marine biogeographic structure. Proc. Natl. Acad. Sci. 109, 14046–14051 (2012).
Frainer, A. et al. Climate-driven changes in functional biogeography of Arctic marine fish communities. Proc. Natl. Acad. Sci. USA 114, 12202–12207 (2017).
Miller, D. L., Burt, M. L., Rexstad, E. A. & Thomas, L. Spatial models for distance sampling data: Recent developments and future directions. Methods Ecol. Evol. 4, 1001–1010 (2013).
Zurell, D. et al. A standard protocol for reporting species distribution models. Ecography (Cop.) 43, 1261–1277 (2020).
Redfern, J. V. et al. Techniques for cetacean-habitat modeling. Mar. Ecol. Prog. Ser. 310, 271–295 (2006).
Hastie, T. J. & Tibshirani, R. J. Generalized Additive Models (Taylor & Francis, 1990).
Goodfellow, I., Bengio, Y. & Courville, A. Deep Learning (MIT Press, 2016).
Friedman, J. H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 29, 1189–1232 (2001).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Vapnik, N.V. Statistical Learning Theory (1998).
Culley, C., Vijayakumar, S., Zampieri, G. & Angione, C. A mechanism-aware and multiomic machine-learning pipeline characterizes yeast cell growth. Proc. Natl. Acad. Sci. 117, 18869–18879 (2020).
Moore, B. M. et al. Robust predictions of specialized metabolism genes through machine learning. Proc. Natl. Acad. Sci. 116, 2344–2353 (2019).
Renò, V. et al. Combined color semantics and deep learning for the automatic detection of dolphin dorsal fins. Electronics 9, 75 (2020).
Maglietta, R., Milella, A., Caccia, M. & Bruzzone, G. A vision-based system for robotic inspection of marine vessels. Signal Image Video Process. 12, 471–478 (2018).
Maglietta, R. et al. Automated hippocampal segmentation in 3D MRI using random undersampling with boosting algorithm. Pattern Anal. Appl. 19, 579–591 (2016).
Ancona, N., Maglietta, R. & Stella, E. Data representations and generalization error in kernel based learning machines. Pattern Recognit. 39, 1588–1603 (2006).
Martín, B., González-Arias, J. & Vicente-Virseda, J. A. Machine learning as a successful approach for predicting complex spatial temporal patterns in animal species abundance. Anim. Biodivers. Conserv. 2021, 25 (2021).
Dimauro, G. et al. A novel approach for biofilm detection based on a convolutional neural network. Electronics 9, 88 (2020).
Inglese, P. et al. Multiple RF classifier for the hippocampus segmentation: Method and validation on EADC-ADNI Harmonized Hippocampal Protocol. Phys. Med. 31(8), 1085–1091 (2015).
Maglietta, R. et al. Convolutional neural networks for Risso’s Dolphins identification. IEEE Access 8, 80195–80206 (2020).
Conference on Biological Diversity—Nagoya 2010 European Parliament resolution of 7 October 2010 on the EU strategic objectives for the 10th Meeting of the Conference of the Parties to the Convention on Biological Diversity (CBD), to be held in Nagoya (2010).
EU. In Commission Decision (EU) 2017/848 of 17 May 2017 Laying Down Criteria and Methodological Standards on Good Environmental Status of Marine Waters and Specifications and Standardised Methods for Monitoring and Assessment, and Repealing Decision 2 (2017).
European Commission. Directive 2014/89/EU of the European Parliament and of the Council of 23 July 2014 establishing a framework for maritime spatial planning. In Off. J. Eur. Union 2014, L 257, 135; MSFD (2008/56/EC) (2014).
Muckenhirn, A., Baş, A. A. & Richard, F.-J. Assessing the influence of environmental and physiographic parameters on common bottlenose dolphin (Tusiops truncatus) distribution in the southern Adriatic Sea. In Proc. 1st Int. Electron. Conf. Biol. Divers. Ecol. Evol. (2021).
Correia, A. et al. Predicting Cetacean Distributions in the Eastern North Atlantic to Support Marine Management. Front. Mar. Sci. 8, 256 (2021).
Redfern, J. V., Barlow, J., Ballance, L. T., Gerrodette, T. & Becker, E. A. Absence of scale dependence in dolphin-habitat models for the eastern tropical Pacific Ocean. Mar. Ecol. Prog. Ser. 363, 1–14 (2008).
Kruse, S. L. Aspects of the Biology, Ecology, and Behavior of Risso’s dolphins (Grampus griseus) off the California Coast (University of California, Santa Cruz, 1989).
Kruse, S., Caldwell, D. K., Caldwell, M. C., Ridgway, S. H. & Harrison, R. Risso’s dolphin Grampus griseus (G. Cuvier, 1812). Handb. Mar. Mamm. Sec. B Dolphins Porpoises 6, 12 (1999).
Gómez-de-Segura, A., Hammond, P. S. & Raga, J. A. Influence of environmental factors on small cetacean distribution in the Spanish Mediterranean. J. Mar. Biol. Assoc. UK 88, 1185–1192 (2008).
Pitchford, J. et al. Predictive spatial modelling of seasonal bottlenose dolphin (Tursiops truncatus) distributions in the Mississippi Sound: Seasonal spatial distributions of bottlenose dolphins. Aquat. Conserv. Mar. Freshw. Ecosyst. 26, 289–306 (2015).
La Manna, G., Ronchetti, F. & Sarà, G. Predicting common bottlenose dolphin habitat preference to dynamically adapt management measures from a Marine Spatial Planning perspective. Ocean Coast. Manag. 130, 317–327 (2016).
Becker, E. A. et al. Predicting cetacean abundance and distribution in a changing climate. Divers. Distrib. 25, 626–643 (2019).
Cañadas, A. & Hammond, P. S. Abundance and habitat preferences of the short-beaked common dolphin Delphinus delphis in the southwestern Mediterranean: Implications for conservation. Endanger. Spec. Res. 4, 309–331 (2008).
Mannocci, L. et al. Predicting cetacean and seabird habitats across a productivity gradient in the South Pacific gyre. Prog. Oceanogr. 120, 383–398 (2014).
Carretta, J. V. Estimates of Marine Mammal, Sea Turtle, and Seabird Bycatch in the California Large-Mesh Drift Gillnet Fishery: 1990–2019 U.S. Department of Commerce, NOAA Technical Memorandum NMFS-SWFSC-654. https://doi.org/10.25923/7emj-za90 (2021).
Rustam, F. et al. A performance comparison of supervised machine learning models for Covid-19 tweets sentiment analysis. PLoS ONE 16, e0245909 (2021).
D’Addabbo, A. & Maglietta, R. Parallel selective sampling method for imbalanced and large data classification. Pattern Recognit. Lett. 62, 61–67 (2015).
Dimauro, G. et al. An intelligent non-invasive system for automated diagnosis of anemia exploiting a novel dataset. Artif. Intell. Med. 136, 102477 (2023).
Spooner, A. et al. A comparison of machine learning methods for survival analysis of high-dimensional clinical data for dementia prediction. Sci. Rep. 10, 20410 (2020).
Becker, E. A. et al. Performance evaluation of cetacean species distribution models developed using generalized additive models and boosted regression trees. Ecol. Evol. 10, 5759–5784 (2020).
Kosicki, J. Z. Generalised additive models and random forest approach as effective methods for predictive species density and functional species richness. Environ. Ecol. Stat. 27, 273–292 (2020).
Barreto, J. et al. Drone-monitoring: Improving the detectability of threatened marine megafauna. Drones 5, 14 (2021).
Sarr, J.-M.A. et al. Complex data labeling with deep learning methods: Lessons from fisheries acoustics. ISA Trans. 109, 113–125 (2021).
Capezzuto, F. et al. The bathyal benthopelagic fauna in the north-western Ionian Sea: Structure, patterns and interactions. Chem. Ecol. 26, 199–217 (2010).
Harris, P. & Whiteway, T. Global distribution of large submarine canyons: Geomorphic differences between active and passive continental margins. Mar. Geol. 285, 69–86 (2011).
Pescatore, T. & Senatore, M. R. A comparison between a present.day (Taranto Gulf) and a Miocene (Irpinian Basin) foredeep of the Southern Apennine (Italy). Spec. Publ. 1986, 169–182 (1986).
Rossi, S. & Gabbianelli, G. Geomorfologia del Golfo di Taranto. Ital. J. Geosci. 97, 423–437 (1978).
Federico, I. et al. Observational evidence of the basin-wide gyre reversal in the Gulf of Taranto. Geophys. Res. Lett. 47, 1030 (2020).
Carlucci, R., Battista-Capezzuto, F., Serena, F. & Sion, L. Occurrence of the basking shark Cetorhinus maximus (Gunnerus, 1765) (Lamniformes: Cetorhinidae) in the central-eastern Mediterranean Sea. Ital. J. Zool. 81, 280–286 (2014).
Matarrese, R., Chiaradia, M. T., Tijani, K., Morea, A. & Carlucci, R. Chlorophyll A multi-temporal analysis in coastal waters with MODIS data. Eur. J. Remote Sens. 2011, 39–48 (2011).
Civitarese, G., Gačić, M., Lipizer, M. & Eusebi-Borzelli, G. L. On the impact of the Bimodal Oscillating System (BiOS) on the biogeochemistry and biology of the Adriatic and Ionian Seas (Eastern Mediterranean). Biogeosciences 7, 3987–3997 (2010).
Pinardi, N. et al. Marine rapid environmental assessment in the \hack{\newline} Gulf of Taranto: A multiscale approach. Nat. Hazards Earth Syst. Sci. 16, 2623–2639 (2016).
Ciancia, E. et al. Investigating the chlorophyll-a variability in the Gulf of Taranto (North-western Ionian Sea) by a multi-temporal analysis of MODIS-Aqua Level 3/Level 2 data. Cont. Shelf Res. 155, 34–44 (2018).
Trotta, F., Pinardi, N., Fenu, E., Grandi, A. & Lyubartsev, V. Multi-nest high-resolution model of submesoscale circulation features in the Gulf of Taranto. Ocean Dyn. 67, 1609–1625 (2017).
Federico, I. et al. Coastal ocean forecasting with an unstructured grid model in the southern Adriatic and northern Ionian seas. Nat. Hazards Earth Syst. Sci. 17, 45–59 (2017).
Trotta, F. et al. A relocatable ocean modeling platform for downscaling to shelf-coastal areas to support disaster risk reduction. Front. Mar. Sci. 8, 103 (2021).
Artegiani, A. et al. The Adriatic Sea general circulation. Part I: Air-sea interactions and water mass structure. J. Phys. Oceanogr. 27, 1492–1514 (1997).
Artegiani, A. et al. The Adriatic Sea general circulation. Part II: Baroclinic circulation structure. J. Phys. Oceanogr. 27, 1515–1532 (1997).
Cushman-Roisin, B., Gacić, M., Poulain, P. M. & Artegiani, A. Physical Oceanography of the Adriatic Sea (2001).
Escudier, R. et al. Mediterranean sea production centre MEDSEA_MULTIYEAR_PHY_006_004 (2021).
Clementi, E. et al. Mediterranean sea physical analysis and forecast (CMEMS MED-Currents, EAS6 system) (Version 1) set. In Copernicus Monitoring Environment Marine Service (CMEMS) (2021).
Madec, G. NEMO Ocean Engine (2008).
Dobricic, S. & Nadia, P. An oceanographic three-dimensional variational data assimilation scheme. Ocean Model 22, 89–105 (2008).
Roquet, F., Madec, G., McDougall, T. J. & Barker, P. M. Accurate polynomial expressions for the density and specific volume of seawater using the TEOS-10 standard. Ocean Model 90, 29–43 (2015).
IOC, SCOR & IAPSO. In The International Thermodynamic Equation of Seawater—2010: Calculation and Use of Thermodynamic Properties 196 (2010).
MEDSEA_MULTIYEAR_BGC_006_008 (2020).
Mediterranean Sea Monthly and Daily Reprocessed Surface Chlorophyll Concentration from Multi Satellite observations + SeaWiFS daily climatology (2020).
Volpe, G. et al. Mediterranean ocean colour Level 3 operational multi-sensor processing. Ocean Sci. 25, 1527–1532 (2019).
Berthon, J.-F. & Zibordi, G. Bio-optical relationships for the northern Adriatic Sea. Int. J. Remote Sens. 25, 1527–1532 (2004).
De Dominicis, M. et al. A relocatable ocean model in support of environmental emergencies. Ocean Dyn. 64, 667–688 (2014).
Freund, Y. & Schapire, R. E. A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 55, 119–139 (1997).
Wu, J. et al. Hyperparameter optimization for machine learning models based on bayesian optimizationb. J. Electron. Sci. Technol. 17, 26–40 (2019).
Yang, L. & Shami, A. On hyperparameter optimization of machine learning algorithms: Theory and practice. Neurocomputing 415, 295–316 (2020).
Acknowledgements
We thank Mr. Fabio Garziano, whose valuable support was essential during the data acquisition campaigns and for the editing to Elena Montrasio and John Ockey.
Author information
Authors and Affiliations
Contributions
R.M., R.C., and G.Co. conceptualized the project. R.M., L.S., R.L., and S.C. designed and performed the experiments. C.F., G.Co., R.L., S.C., and I.F. provided materials. Paper written by R.M., L.S., R.C., G.Co., R.L., S.C., I.F., G.Ci. Critical improvement of the paper by V.T., G.D. and G.Ci. Manuscript reviewed by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Maglietta, R., Saccotelli, L., Fanizza, C. et al. Environmental variables and machine learning models to predict cetacean abundance in the Central-eastern Mediterranean Sea. Sci Rep 13, 2600 (2023). https://doi.org/10.1038/s41598-023-29681-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-29681-y
This article is cited by
-
Distribution and co-occurrence patterns of charophytes and angiosperms in the northern Baltic Sea
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.