Abstract
This study analyzes the genomic findings of the first report of Salmonella isolate carrying the blaCTX-M-55 gene, recovered from a bacteremic patient from Brazil. A bacterial isolate positive for the blaCTX-M-55 gene was submitted to antimicrobial susceptibility testing by disk diffusion and epsilometric test. Whole genome sequencing was performed using Illumina technology. Conjugation assay was performed; plasmid sizes determined by S1-PFGE and plasmid content were investigated by hybrid assembly after MinION long reads sequencing. Isolate 288_18 was identified as sequence type ST13, resistant to ampicillin, cefotaxime, ceftazidime, cefepime, ceftriaxone, and aztreonam. A transferable IncFII plasmid sized approximately 67 kb was found to carry the blaTEM-1 and blaCTX-M-55 in a module consisting of IS26-blaTEM-1B-WbuC-blaCTX-M-55-IS26. In addition, an 117 kb IncI1plasmid was also identified in the 288_18 isolate, but without additional resistance genes. To the best of our knowledge, this is the first report of blaCTX-M-55 in Salmonella isolated from human infection in Brazil. The occurrence of blaCTX-M-55 in the IncFII epidemic plasmid in a relevant clinical human isolate of Salmonella Agona underscores the urgent need for enhanced and effective continuous surveillance for controlling its dissemination.
Similar content being viewed by others
Introduction
Salmonellosis is one of the most common food-borne human diseases worldwide1. Within the last years, Salmonella enterica subsp. enterica serovar Agona has been one of the top 20 most commonly reported serotypes causing human infections2. Although cases of gastroenteritis due to Salmonella enterica usually resolve without medical intervention, antimicrobial therapy is recommended for patients with severe disease3. In these cases, antibiotics are used to treat invasive nontyphoidal Salmonella, like fluoroquinolone, cephalosporin, and azithromycin4. Currently, multidrug-resistant S. Agona poses a significant hazard to human and animal health, given that the emergence of Salmonella isolates resistant to extended-spectrum cephalosporins (ESCs) is a worldwide public health concern5. Infection with extended-spectrum beta-lactamases (ESBL)-producing organisms is particularly concerning since few treatment options exist and worse clinical outcomes can occur.
Plasmid-mediated ESBL plays a vital role in the increasing frequency of multidrug-resistant Enterobacterales, among which CTX-M-type are the most common ESBL type found in recent years6. CTX-M-55 is a variant of CTX-M-15, containing a single amino acid substitution (Val77Ala) that contributes to enhanced cephalosporin-hydrolyzing activity6. This variant has been reported in several countries. In China, CTX-M-55 is the second most common CTX-M ESBL, and identical plasmids have been observed in humans and animals, indicating their spread in different reservoirs7,8. CTX-M-55 was detected to rapidly increase in prevalence, especially in Escherichia coli from animals9.
The first report on CTX-M-55-producing Salmonella in human isolates came from the United States and China in 201110,11. This enzyme has also been reported in different serotypes of Salmonella in Europe, Asia, China, and Thailand12,13,14, but there is no report of CTX-M-55 in Salmonella isolates from Brazil. In this study, we describe the genomic findings of the first report of a Salmonella isolate carrying the blaCTX-M-55 gene recovered from a bacteremic patient from Brazil.
Materials and methods
Bacterial isolate
Adolfo Lutz Institute is a regional reference laboratory for Salmonella isolates from São Paulo, Brazil. Thus, all isolates received are submitted to classical serotyping, antimicrobial susceptibility testing, and molecular-typing by PFGE and next-generation sequencing. Isolate 288_18 was recovered from a blood sample from a patient with clinical symptoms of bacteremia in 2018.
Serotyping and antimicrobial susceptibility testing
The serotype was determined according to the Kauffmann–LeMinor scheme15. Antimicrobial susceptibility testing was performed by the disk-diffusion method according to the guidelines and interpretation criteria of the CLSI16 (nalidixic acid, amoxicillin/clavulanic acid, ampicillin, amikacin, aztreonam, ceftazidime, cefotaxime, ceftriaxone, cefepime, ciprofloxacin, chloramphenicol, streptomycin, gentamicin, imipenem, trimethoprim-sulfamethoxazole, sulfonamide, and tetracycline). Gradient diffusion was performed to nalidixic acid, ciprofloxacin, ceftriaxone, and cefotaxime, and broth microdilution to colistin, and polymyxin B according to CLSI guidelines16.
Conjugation
Conjugation experiments were conducted in Luria–Bertani (LB) broth using sodium azide-resistant E. coli J53 as the recipient strain. Transconjugants were selected on MacConkey agar containing ceftriaxone (2.5 µg/mL) and sodium azide (100 µg/mL). In addition, an antimicrobial susceptibility test was performed to confirm the antimicrobial resistance characteristics of these transconjugants. The donor and the respective transconjugant isolates were submitted to S1-PFGE to identify the size of large plasmids carrying the blaCTX-M-55 gene.
Whole-genome sequencing and analysis
Whole-genome shotgun sequencing was performed using the Illumina MiSeq (Illumina, San Diego, CA, USA). For DNA extraction, strains were grown on Luria Bertani Agar overnight at 37 °C. Subsequently, a single colony was inoculated in 2 mL of Luria–Bertani broth for 12 h at 37 °C. The suspension was used for bacterial cultures to continue extraction and purification by the Wizard Genomic DNA Purification kit (Promega, USA). High-quality DNA (assessed by gel electrophoresis and Qubit quantification) was submitted to library preparation with Illumina DNA Prep Tagmentation and sequenced in a MiSeq (USA) instrument with MiSeq Reagent Kit v3 (150 cycles). Quality control was performed with FastQC and Kraken2 software, within the Galaxy Europe Server (https://usegalaxy.eu).
Long-read sequencing and base-calling of select isolates were performed using Oxford Nanopore technology (Oxford, United Kingdom). Libraries were constructed using a rapid multiplex barcoding kit. Sequencing was performed using an Oxford Nanopore MinION device with R9.4.1 flow cells. Base-calling and read processing was performed using Guppy v2.3.1 using default parameters. The quality of the raw sequence reads was checked by the interactive program FastQC. The genome was assembled using a combination of short and long reads by the Unicycler 0.4.8 hybrid assembler. Genomes were automatically annotated with Prokka v1.14.6. The tools mentioned above were used within the Galaxy Europe Server (https://usegalaxy.eu). Annotations and alignments were visualized in the Artemis Comparison Tool (ACT)17. Online Center for Genomic Epidemiology tools was used to determine in silico serotype and MLST, to detect acquired resistance genes and chromosomal mutations associated with antimicrobial resistance, and for plasmid classification (PlasmidFinder, PBRT, pMLST; https://cge.cbs.dtu.dk/services/). Blast software compared similar plasmids following alignment using BioNumerics v.8.1 (bioMérieux/Belgium). Software BLAST Ring Image Generator (BRIG)18 was used to compare sequence of plasmid 288_18 with other representative plasmids to further generate circular plasmid maps. Core genome MLST (cgMLST) was performed in BioNumerics version 8.1 (bioMérieux/Belgium) using the S.enterica cgMLST scheme of 3002 target loci available on Enterobase (https://enterobase.warwick.ac.uk/). The comparison was performed using the 175 genomic sequences of Salmonella Agona deposited in the Enterobase and PubMLST database (https://pubmlst.org/). We searched for isolates with “ST13” listed as MLST in the strain metadata and “Agona” as the serovar in either the strain metadata or experimental data. The complete nucleotide sequence of strain 288_18 has been deposited into the GenBank database under the accession number: JAODHS000000000.
Results
Strain 288_18 was isolated from the blood of a 73-year-old patient and identified as Salmonella enterica subsp. enterica serovar Agona, sequence type ST13. The phenotypic test (disk diffusion) revealed resistance to ampicillin, cefotaxime, ceftazidime, cefepime, ceftriaxone, and aztreonam. The isolate was susceptible to the remaining antimicrobials tested. The double disk synergism test was positive for ESBL production. Minimal inhibitory concentration (MIC) showed that isolate 288_18 was sensitive to nalidixic acid (6 µg/mL), and ciprofloxacin (0.015 µg/mL). Still, it confirmed resistance in high levels to ceftriaxone (32 µg/mL), cefotaxime (256 µg/mL), cefepime (16 µg/mL), and aztreonam (64 µg/mL).
A transferable IncFII plasmid, pMLST F33:A-:B-, sized approximately 67 kb, was found to carry the blaTEM-1B and blaCTX-M-55, in a module consisting of IS26- blaTEM-1B-WbuC-blaCTX-M-55-IS26, associated with phenotypic resistances detected to ampicillin, amoxicillin/clavulanate, cefotaxime, ceftazidime, ceftriaxone, and cefepime. Further comparative analysis of the complete plasmid indicated that blaCTX-M-55 was inserted in an IncFII plasmid in a nearly identical manner to E.coli strain SCEC020023 plasmid pEC020023-FII (GenBank Accession nº CP025949.1), K. pneumoniae strain KP32558 plasmid pKP32558-4-FII (GenBank Accession nº CP076034.1) and S. Enteritidis strain SE104 plasmid pSE104-FII (GenBank accession nº CP050713.1) (Fig. 1). These three plasmids were identified in Chinese isolates recovered from urine, bronchoalveolar lavage fluid, and stool, respectively, all of them also carrying the blaCTX-M-55 and blaTEM-1B genes.
BRIG (Blast Ring Image generator) comparison of the p288_18 of IncFII- type bla CTX-M-55-carrying plasmid against the two IncFII plasmids (accession numbers: CP025949.1, CP076034.1, CP050713.1). P288_18 containing the blaCTX-M-55 gene was used as a reference. Each ring corresponds to a plasmid (indicated at the right of the figure together with the color code). Circle 1 (innermost) displayed the scale in kilobase pairs. Circle 2 and 3 displayed the GC content and GC skew, respectively. Regions of the plasmid are labelled on the outer circle.
Conjugative transfer of blaCTX-M-55 was successfully achieved to E. coli J53 as the recipient. The ceftriaxone and cefotaxime MIC values of the transconjugant strain remained the same observed for the wild strain, ceftriaxone (32 µg/mL) and cefotaxime (256 µg/mL). S1-PFGE identified two plasmids compatible with hybrid assemblies generated with short and long reads.
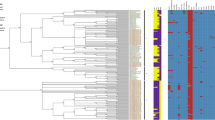
In addition, an 117 kb IncI1 plasmid (ST 231) was also identified in the 288_18 isolate, but without additional resistance genes. The chromosome fosA7 and aac(6′)-Iaa genes were found, which can confer resistance to fosfomycin and aminoglycosides, respectively. Still, the phenotypic resistance test was sensitive to these drug classes. The minimum spanning tree showed that isolate 288_18 presented the highest similarity (27 alleles distance) with an epidemiologically unrelated isolate recovered from a human source in the United Kingdom, dated 1972 (Fig. 2; Supplementary Table 1).
A Minimum-spanning tree based on cgMLST analysis of 175 Salmonella Agona strains (Supplementary Table 1). The figure was generated using BioNumerics version 8.1 (bioMérieux/Belgium). The entire set of genomes and all its metadata and genotyping results are available to the registered users of PubMLST at (https://pubmlst.org/) and Enterobase (https://enterobase.warwick.ac.uk/). (A) Isolates are color-coded by their source of origin as shown in the legend. (B) Clinical isolate (288_18) was grouped within a clade formed with ST13 S. Agona isolates related to strains of human, animal and food origin from different countries and it was linked with an isolate from the human from United Kingdom with 27 alleles distance. (C) Isolates are color-coded by their Inc type plasmids as shown in the legend. (D) Isolates are color-coded by the presence and ∕or absence of ESBL genes as shown in the legend. ND (not detected).
Discussion
To the best of our knowledge, this is the first report of blaCTX-M-55 in Salmonella isolated from human infection in Brazil. Salmonella Agona is a serovar is increasingly recognized as a cause of foodborne disease outbreaks and has the potential to both be multidrug resistant and cause disease2. The blaCTX-M-55 gene has already been found in S. Agona in Australia isolated in silver gull3, but it was not localized in an epidemic plasmid like IncF. Wild birds, implicated as vectors of antimicrobial resistance internationally, are colonized by MDR and virulent lineages of Enterobacteriaceae that threatent to human health3.
The IncFII plasmid identified in this study was found to be disseminated in other Salmonella serotypes, indicating a potential well-adapted plasmid spreading several antimicrobial resistance genes in different reservoirs19,20. IncF-like plasmids represent the main mechanisms by which epidemic clones can arise3. In Brazil, the blaCTX-M-55 gene is circulating into a lineage of IncFII plasmids especially in E. coli from animal origin21. In Salmonella, the blaCTM-55 gene is mainly found in IncHI222, IncI2, and IncF replicon-type plasmids19,23. Occurrence of blaCTX-M-55 was also reported to exist in the bacterial chromosome24. The identification of the blaCTX-M-55 gene in a genetic environment surrounded by IS26 in an IncFII plasmid represents potential clinical, animal and environment issues and need to be monitored.
The phylogenetic analysis showed that isolate 288_18 presented the highest similarity with a strain from the human isolated in the United Kingdom. The occurrence of blaCTX-M-55 in the IncFII epidemic plasmid in a relevant clinical human isolate of Salmonella Agona underscores the urgent need for enhanced and effective continuous surveillance for controlling its dissemination.
Conclusions
The antimicrobial surveillance of Salmonella strains using whole genome sequencing allowed the first identification of blaCTX-M-55, which confers resistance to third-generation cephalosporin in Brazil. Salmonella Agona is a serovar is increasingly recognized as a cause of foodborne disease outbreaks and has the potential to be both multidrug-resistant and disease-causing. The occurrence of the ESBL gene in an epidemic plasmid reinforces the need for enhanced continuous surveillance of antimicrobial resistance in isolates of both human and non-human sources and alerts to prevent possible failures in the antimicrobial treatment of severe invasive salmonellosis.
Code availability
Accession numbers: This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAODHS000000000. The version described in this paper is version JAODHS010000000.
References
Dangel, A. et al. Genetic diversity and delineation of Salmonella agona outbreak strains by next generation sequencing, Bavaria, Germany, 1993 to 2018. Eurosurveillance 24, 1800303 (2019).
Hoffmann, M. et al. Temporal dynamics of Salmonella enterica subsp. enterica Serovar Agona isolates from a recurrent multistate outbreak. Front. Microbiol. 11, 478 (2020).
Cummins, M. L. et al. Whole-genome sequence analysis of an extensively drug-resistant Salmonella enterica Serovar agona isolate from an australian silver gull (Chroicocephalus novaehollandiae) reveals the acquisition of multidrug resistance plasmids. mSphere 5, (2020).
Crump, J. A., Sjölund-Karlsson, M., Gordon, M. A. & Parry, C. M. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive salmonella infections. (2015). https://doi.org/10.1128/CMR.00002-15
Arlet, G. et al. Salmonella resistant to extended-spectrum cephalosporins: Prevalence and epidemiology. Microbes Infect. 8, 1945–1954 (2006).
Castanheira, M., Simner, P. J. & Bradford, P. A. Extended-spectrum β-lactamases: an update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 3, (2021).
Lv, L. et al. Genetic characterization of IncI2 plasmids carrying blaCTX-M-55 spreading in both pets and food animals in China. Antimicrob. Agents Chemother. 57, 2824 (2013).
Zhang, J. et al. Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect. Dis. 14, (2014).
Norizuki, C. et al. Detection of Escherichia coli producing CTX-M-1-group extended-spectrum β-lactamases from pigs in Aichi Prefecture, Japan, between 2015 and 2016. Jpn. J. Infect. Dis. 71, 33–38 (2018).
Sjölund-Karlsson, M. et al. CTX-M-producing non-Typhi Salmonella spp. isolated from humans, United States. Emerg. Infect. Dis. 17, 97–99 (2011).
Yu, F. et al. High prevalence of extended-spectrum beta lactamases among Salmonella enterica Typhimurium isolates from pediatric patients with diarrhea in China. PLoS One 6, (2011).
Torpdahl, M. et al. Detection of mcr-1-encoding plasmid-mediated colistin-resistant Salmonella isolates from human infection in Denmark. Int. J. Antimicrob. Agents 49, 261–262 (2017).
Wong, M. H. Y., Liu, L., Yan, M., Chan, E. W. C. & Chen, S. Dissemination of IncI2 plasmids that harbor the blaCTX-M element among clinical Salmonella isolates. Antimicrob. Agents Chemother. 59, 5026 (2015).
Luk-in, S. et al. High prevalence of ceftriaxone resistance among invasive Salmonella enterica serotype Choleraesuis isolates in Thailand: The emergence and increase of CTX-M-55 in ciprofloxacin-resistant S. Choleraesuis isolates. Int. J. Med. Microbiol. 308, 447–453 (2018).
Grimont, P. A. D. & Weill, F.-X. WHO collaborating centre for reference and research on Salmonella. In Antigenic Formulae of the Salmonella Serovars 2007, 9th ed.
M100 Performance Standards for Antimicrobial. (2021).
Carver, T. J. et al. ACT: The artemis comparison tool. Bioinformatics 21, 3422–3423 (2005).
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L. & Beatson, S. A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 12, (2011).
Zhang, Z. et al. Dissemination of IncFII plasmids carrying fosA3 and bla CTX-M-55 in clinical isolates of Salmonella enteritidis. Zoonoses Public Health 68, 760–768 (2021).
Guo, L. & Zhao, Y. Global spread and molecular characterization of CTX-M-producing Salmonella Typhimurium isolates. Antibiot. (Basel, Switzerland) 10, (2021).
dos Anjos Adur, M. et al. Escherichia coli ST224 and IncF/bla CTX-M-55 plasmids drive resistance to extended-spectrum cephalosporins in poultry flocks in Parana, Brazil. Int. J. Food Microbiol. 380, 109885 (2022).
Li, L. et al. Genetic context of bla CTX-M-55 and qnrS1 genes in a foodborne Salmonella enterica serotype Saintpaul isolate from China. Front. Microbiol. 13, (2022).
Octavia, S., Chew, K. L., Lin, R. T. P. & Teo, J. W. P. Whole genome sequencing of Salmonella enterica serovar Saintpaul for elucidating the mechanisms of resistance to third generation cephalosporins. Pathology 53, 768–772 (2021).
Zhang, C. Z. et al. The emergence of chromosomally located bla CTX-M-55 in Salmonella from foodborne animals in China. Front. Microbiol. 10, (2019).
Acknowledgements
We thank to the Strategic Laboratory at Adolfo Lutz Institute for library preparation and whole-genome sequencing.
Funding
This study was supported by grant nº 2017/50333–7 and nº 2021/12219–3 São Paulo Research Foundation (FAPESP), São Paulo, Brazil. This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq project 402563/2021–2). This study was carried out as part of our routine work. MRTC has received Productivity Research Fellows from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Author information
Authors and Affiliations
Contributions
All authors contributed to conception and design of the study. A.M.J.B and M.R.T.C wrote the first draft of the manuscript. A.M.J.B and C.H.C organized the database and reviewed the manuscript. A.M.J.B, T.V, C.A.S, E.A.A performed the phenotypic assays, A.D.R performed whole genome sequencing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
de Jesus Bertani, A.M., Vieira, T., Reis, A.D. et al. Whole genome sequence analysis of the first reported isolate of Salmonella Agona carrying blaCTX-M-55 gene in Brazil. Sci Rep 13, 2299 (2023). https://doi.org/10.1038/s41598-023-29599-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-29599-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.