Abstract
Oxygen is the sustenance of aerobic life and yet is highly toxic. In early life, antioxidants functioned solely to defend against toxic effects of reactive oxygen species (ROS). Later, as aerobic metabolisms evolved, ROS became essential for signalling. Thus, antioxidants are multifunctional and must detoxify, but also permit ROS signalling for vital cellular processes. Here we conduct metazoan-wide genomic assessments of three enzymatic antioxidant families that target the predominant ROS signaller, hydrogen peroxide: namely, monofunctional catalases (CAT), peroxiredoxins (PRX), and glutathione peroxidases (GPX). We reveal that the two most evolutionary ancient families, CAT and PRX, exhibit metazoan-wide conservation. In the basal animal lineage, sponges (phylum Porifera), we find all three antioxidant families, but with GPX least abundant. Poriferan CATs are distinct from bilaterian CATs, but the evolutionary divergence is small. Amongst PRXs, subfamily PRX6 is the most conserved, whilst subfamily AhpC-PRX1 is the largest; PRX4 is the only core member conserved from sponges to mammals and may represent the ancestral animal AhpC-PRX1. Conversely, for GPX, the most recent family to arise, only the cysteine-dependent subfamily GPX7 is conserved across metazoans, and common across Porifera. Our analyses illustrate that the fundamental functions of antioxidants have resulted in gene conservation throughout the animal kingdom.
Similar content being viewed by others
Introduction
A universal and ancient challenge for all life forms is to deal with the toxifying effects of molecular oxygen1. Reactive derivatives of oxygen, termed reactive oxygen species (ROS), are toxic for non-target molecules2, causing oxidative damage to nucleic acids, proteins and lipids3, reviewed by4,5. To prevent this damage, organisms from all domains of life have antioxidants that function to detoxify and regulate ROS in reduction reactions6. The first antioxidants are estimated to have arisen between 3.5 and 4.1 billion years ago (bya), soon after the origin of life on earth, and far predating aerobic metabolism and the rise of atmospheric oxygen6,7,8,9. For the anaerobic life forms that inhabited the anoxic and sulfidic atmosphere of early Earth, antioxidants provided an evolutionary advantage by protecting against localised or trace oxygen levels1,9,10. By 2.4 bya, in occurrence with the Great Oxidation event, ROS had been recruited into early redox signalling systems1,11,12,13, and thus the roles of antioxidants had expanded to include regulators of redox signalling. The most ancient of these antioxidants still exist in extant organisms across all domains, as evidence of shared evolutionary history and their necessity for the survival of aerobic life6,13,14.
In extant aerobic organisms, redox signalling together with its regulation by antioxidants is critical to a vast array of life-sustaining cellular functions15. These functions include, but are not limited to, innate immunity16, cell cycle transition17,18, neurodevelopment and regeneration19,20, cell differentiation/proliferation2, and the circadian redox clock21. Antioxidants can be either non-enzymatic (e.g., vitamins E, C, A, selenium, transferrin, and lactoferrin, ascorbic acid, glutathione, melatonin, carotenoids, flavonoids, proline), or specialised enzymes that vary in subcellular localisation, predominant substrate, and rate of reactivity7,22,23. The specialist enzymatic antioxidant families include those that target hydrogen peroxide (H2O2), which is the major redox signalling ROS reviewed by5,24,25, and those that target superoxide anion radical (O2•-) for the generation of H2O2 as a product[reviewed by26,27.
The three major families of antioxidants that target H2O2 are monofunctional catalase (CAT), peroxiredoxin (PRX), and glutathione peroxidase (GPX) (Fig. 1A–C)28,29,30,31. Of these, CAT that uses iron as its electron acceptor, and PRX that uses sulfur-based cysteine32, both represent some of the most evolutionary ancient antioxidant enzymes targeting H2O2; both predate the great oxidation event (GOE)7. The third family, GPX that uses glutathione (GSH) as a reductant, has a more recent evolutionary origin, after the GOE7. Both the PRX and GPX enzyme families have been further classified into multiple subfamilies, but because these subfamily classifications have been based largely on mammalian gene complements, their relevance to the rest of the animal kingdom is not clear.
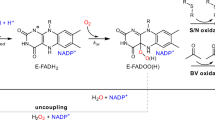
Summary of reaction mechanisms for (A): CAT, (B): PRX, and (C): GPX. In the first step of the reaction mechanism of all PRXs and CysGPXs, and GPX7, H2O2 reacts with the peroxidatic cysteine (CP) to form a sulfenic acid (SOH) intermediate. Whilst in SecGPXs, a catalytic selenocysteine first reacts to form a selenic acid (SeOH). If a second, resolving cysteine (CR) is present (i.e., in AhpC-PRX1, PRX5 & CysGPX), this quickly reacts with the SOH to form either an inter- or intramolecular disulfide bond that is then most commonly reduced by thioredoxin (TRX), reactivating the enzyme. PRX6 instead forms a disulfide with another molecule, commonly GST, and is then recycled by glutathione (GSH), generating oxidised glutathione (GSSG). In SecGPXs the SeOH is similarly reduced by two GSH generating GSSG, whilst GPX7 is described to be reactivated via the ER protein disulfide isomerase (PDI). For PRXs under high concentrations of H2O2, SOH reacts with another molecule of H2O2 to form a sulfenic acid (SO2H), resulting in hyperoxidation. Enzymes within the subfamily, AhpC-PRX1 only may then be slowly re-activated via the enzyme sulfiredoxin (SRX) via the inactivation loop. (D): Generalised domain structure for CAT, PRX, and GPX enzyme families. CAT enzymes comprise a catalase domain and catalase-related immune -responsive (catalase_rel). Stars denote presence of active site, His and Asn respectively. All PRX enzymes comprise the domain, Alkyl hydroperoxide reductase-Thiol specific antioxidant (AphC-TSA). The subfamilies AhpC-PRX1 and PRX6 additionally commonly encode the Peroxiredoxin, C-terminal domain (1-cysPrx_C). CP (red) and CR (blue) conserved active site are displayed, residues in bold denote absolutely conserved, and underlined residues denotes amino acids that deviated from that displayed within more than one metazoan sequence. GPX enzymes comprise a single GSHPx domain. GPX enzymes may encode either CP or a catalytic Sec (S) within the active site.
In this study, we conduct a comparative genomic assessment of these three major antioxidant enzyme families—CAT, PRX, and GPX—in 19 species, with high-quality genomes that span 10 metazoan phyla, from sponges to chordates. In doing so, we provide the first assessment of these enzymes in sponges (phylum Porifera), including four marine and one freshwater species of three different classes. Sponges evolved at least 700 million years ago33 and are widely considered to be the oldest of the extant animal phyletic lineages34,35. As probable sister to all other animal phyla, traits shared by sponges and the rest of animal kingdom can logically be traced back to the last common animal ancestor36. Thus, sponges can provide unique insight into the evolutionary history of these ancient enzymatic antioxidant families that play a critical role throughout the animal kingdom.
Results and discussion
Our metazoan-wide survey has provided the most comprehensive analysis to date of gene number and phylogenetic distribution of three key antioxidant gene families across the animal kingdom. Genes encoding all three families were observed in 18 metazoan species; the exception is the ctenophore Mnemiopsis leidyi that has PRX and GPX, but not CAT (Table 1). Our findings demonstrate that the antioxidants CAT and PRX both are evolutionary ancient and highly conserved enzyme families (Figs. 2 and 4). By comparison, the GPX family is less conserved, with total gene numbers and functional types varying considerably among metazoan species (Table 1). Below we discuss our expanded analysis, detailing substantial gene conservation across evolutionary diverse bilaterian and non-bilaterian phyletic lineages, and for the first time reporting a suite of H2O2-targeting enzymatic antioxidants in the basal metazoan phylum Porifera (the sponges).
Maximum likelihood phylogenetic tree of monofunctional CAT enzyme family. (A): Unrooted tree displaying three main CAT clades indicated by branch colour: clade 1 (dark grey), clade 2 (light grey), clade 3 (black). Within clade 3 coloured shapes indicate identified evolutionary groups; Nematoda (orange), Demospongiae (blue), Invertebrate (yellow), inner dashed line indicates bilaterian-invertebrate species only, and Vertebrata (green). (B): Rooted phylogenetic tree using metazoan CAT protein sequences and the choanoflagellate M. brevicollis CAT as an outgroup. (M) denotes mitochondrial localised CAT sequences. Asterisk (*) next to A. queenslandica and T. wilhelma denotes full length CAT gene sequences for these two species. Labels coloured blue denote sequences encoded by phylum Porifera. Black numbers on branches indicate bootstrap support. Circles denote collapse tree nodes. Coloured shapes in B correspond to those displayed in (A). Branch lengths represent evolutionary distances, indicated by tree scale. Constructed based on edited alignment, 1000 bb and the evolutionary models, A: LG + I + G4 and B: LG + G4.
Monofunctional catalase (CAT)
The monofunctional (i.e., “typical”) CAT enzyme family are one of the most evolutionary ancient antioxidants targeting hydrogen peroxide7; the other is the PRX family. The metazoan CATs comprise a relatively small family; most metazoans that we assessed encode just one full length CAT sequence, and the evolutionary divergence among them is relatively small compared with the PRX and GPX enzyme families (Table 1 and Fig. 2).
HMM scans, based on hidden Markov probabilistic models, across coding sequences from 19 metazoan species revealed a total of 56 unique protein sequences encoding at least one CAT-associated domain. Filtering these after sequence alignment and protein structure reduced this number to 44 (Fig. 1D; Supplementary file 2). On this basis, we identified CAT protein sequences in 18 of the 19 metazoans; the exception was the ctenophore M. leidyi, that likely represents evidence of gene loss, given the ancient origins of the CAT family.
CAT enzymes can broadly be assigned to one of three main clades, comprising either small subunit sizes (55–69 kDa) with heme b as the prosthetic group (Clade 3, or clade 1), or large subunit sizes (75–84 kDa) with heme d as the prosthetic group and an additional ‘flavodoxin like’ domain (Clade 2)14,29,38. Of these, Clade 3 enzymes that use NADPH as cofactor are the most widely distributed—all 44 of the animal CAT enzymes we identified belong to Clade 3 and are distinct from the 15 non-metazoan sequences (Fig. 2A). Notably, there are relatively short evolutionary distances among genes within clade 3 compared to genes within the non-metazoan clades 1 and 2. This reflects the relatively recent diversification of metazoan CATs within the much older evolutionary history of this enzyme family (Fig. 2A).
Porifera and cnidaria CAT are phylogenetically distinct from other metazoans
Phylogenetic assessment of 44 animal CATs reveals three well-supported clades. These are Vertebrata (Fig. 2B: 99%), a predominantly bilaterian-invertebrate (Fig. 2B; 90%), and a Poriferan/Cnidarian clade (Fig. 2B; 80%). Our findings are consistent with14,38, but our expanded analysis provides additional evolutionary insight at the base of the metazoan CAT tree. We show that CATs in the basal metazoan phyla Cnidaria and Porifera are evolutionarily distinct from the rest of the metazoan CATs, including those of the phylum Placozoa that sit within an otherwise bilaterian invertebrate clade (Fig. 2B). Moreover, this Poriferan/Cnidarian clade includes two strongly supported subclades of Demospongiae (78%) and Homoscleromorpha/Cnidaria (84%), indicating diversification of these genes before sponges diverged from the metazoan stem. The exception to this is S. ciliatum (class Calcarea) CATs that display greater divergence and low support within Poriferan/Cnidarian clade (Fig. 2b. 26%).
Also consistent with Zámocký et al.14,38, we find that CATs of Caenorhabditis elegans (phylum Nematoda) and Drosophila melanogaster (Arthropoda) are both evolutionarily separated from the rest of the Bilateria, each forming independent monophyletic clades (Fig. 2A,B 100%). Of these, the nematode clade displays the greatest evolutionary divergence, and sits as sister to all other metazoans (Fig. 2B). Notably, observed evolutionary distances within CAT clade 3 are comparatively shorter than within the non-metazoan clades 1 and 2 (Fig. 2A). Considering this and the evolutionary divergence of Nematoda, we hypothesise that diversification of CAT within metazoans is relatively recent compared to the long evolutionarily history of this enzyme.
Consistent with previous descriptions, we found that metazoan CATs from 14 species are predicted to localise to the peroxisome (20 sequences) (Fig. 3; Supplementary file 3)29. However, 17 species have CAT enzymes that localise to multiple subcellular compartments. Overall, we most commonly predicted CATs sequences that localise to the cytoplasm (24 sequences), but also the mitochondria (2 sequences), nucleus, cell membrane and extracellular space (Fig. 3; Supplementary file 3). Moreover, we identified four species, (Xestospongia bergquistia, Nematostella vectensis, Ciona intestinalis, and Branchiostoma floridae) that do not have any peroxisomal CAT but instead encode a cytoplasmic CAT (Fig. 3). That said, 9 of the 24 cytoplasm-localised sequences do encode a peroxisomal targeting signal, whilst 10 sequences encode a nuclear targeting signal (Supplementary file 3). The phylogenetic distribution of cytoplasmic- or peroxisomal-localised CATs has no obvious pattern. However, the two mitochondrial-localised sequences, found in Xenopus tropicalis (Vertebrata) and Sycon ciliatum (Calcarea), are each the most divergent within their respective clades (indicated on Fig. 2b). It has been hypothesised that having multiple CATs localised to various subcellular compartments may confer additional benefits against diseases such as cancer29. However, we cannot assume that all CAT enzymes localised to various subcellular regions are functionally active. For instance, the sponges Amphimedon queenslandica and Tethya wilhelma (class Demospongiae) each encode only one full length CAT sequence localised within the cytoplasm, indicated by asterisks (Fig. 2B), and their additional CAT sequences are of reduced length and thus perhaps non-functional.
Presence of antioxidant enzymes within 8 different subcellular compartments of 19 metazoan species. Shapes denote enzyme family, namely CAT (circle), PRX (triangle), or GPX (square). Subcellular compartments indicated are predictions based on amino acid sequence analysis by DeepLoc-2.0, https://services.healthtech.dtu.dk/service.php?DeepLoc-2.039,40. Colours denote individual PRX and GPX enzyme subfamilies, based on phylogeny corresponding to Figs. 4 and 5. Total number of CAT, PRX, and GPX gene sequences encoded by each species in Table 1.
(A): Maximum likelihood phylogenetic tree of PRX enzyme family. Unrooted tree displaying PRX animal subfamilies; AhpC-PRX1 (green), PRX5 (blue), and PRX6 (pink) that correspond with the three broad classes, typical 2-Cys PRX, atypical 2-Cys PRX, and 1-Cys PRX, respectively. Within AhpC-PRX1 orange shape denotes strongly supported monophyletic clade named PRX4. (B): Depicts zoomed in region of AhpC-PRX1 clade. Monophyletic clades comprising isoforms, PRX1/2 (turquoise) and PRX3 (purple) of Vertebrata are indicated. These isoforms were previously used to classify PRXs across all metazoan species until a revision of PRX system of classification41,42. CNID-PRX denotes recently established subfamily found only within species belonging to phylum Cnidaria43. Orange circle indicates collapsed nodes of PRX4 clade. Labels coloured blue denote sequences encoded by phylum Porifera. Black numbers on branches indicate bootstrap support. Branch lengths represent evolutionary distances, indicated by tree scale. Constructed based on edited alignment, 1000 bb and the WAG + I + G4 evolutionary model. FW: fresh water, SW: sea water.
Peroxiredoxins (PRX)
The PRXs are a large yet highly conserved enzyme family amongst metazoans. Across all 19 metazoan species, we identified a total of 799 unique protein sequences encoding at least one PRX-associated domain. Filtering these by presence of the strictly conserved CP motif (PXXX(T/S)XXC) required for catalytic activity reduced this number to 110, uncovering multiple subfamilies encoded by all 19 metazoan species (Table 1; Supplementary file 2). Our phylogenetic analysis is largely consistent with previous assessments based on smaller numbers of taxa PRX44,43, revealing high support (99–100%) for three monophyletic clades corresponding to the animal subfamilies, AhpC-PRX1, PRX5, and PRX6 (Fig. 4A). However, our taxonomic expansion highlights PRX diversity and supports use of the most recent system of PRX classification based on the peroxidatic cysteine (CP) active site sequence41,42.
AhpC-PRX1 is the largest metazoan PRX subfamily
The PRXs comprise three animal subfamilies, of which AhpC-PRX1 is the largest. For 12 of the 19 metazoan species, we find at least three AhpC-PRX1 genes each, compared to just one or two genes in subfamilies PRX5 and PRX6 (Table 1).
In a previous system of classification that used homology to mammalian PRX isoforms, subfamily AhpC-PRX was subdivided into isoforms PRX1-441,45. However, this system was later deemed insufficient to accurately describe PRXs across diverse animal species45. Indeed, here we show only small subclades of sequences that share similarity to the mammalian isoforms PRX1/2 (turquoise) and PRX3 (purple) (85% & 75%; Fig. 4B). Instead, AhpC-PRX1 comprises multiple independent branches such as the recently described CNID-PRX that is a lineage specific divergence within phylum Cnidaria43 (Fig. 4B). That said, sequences sharing similarity to mammalian isoform PRX4 (orange) do form a strongly supported subclade that is widespread across the animal kingdom, being absent only in C. elegans (99%; Fig. 4A). Indeed, the only AhpC-PRX1 we find in the three marine species of demosponge are these PRX4-like sequences. Additionally, the non-metazoan, M. breviocolis choanoflagellate encodes a single sequence that falls within the PRX4 subclade, indicating that PRX4 may predate the origin of metazoans. Subsequently we propose that PRX4 may be the closest animal orthologue of the ancestral AhpC-PRX1.
Here we use the most recently proposed PRX classification system, that identifies six subfamilies, of which three occur in animals, based on protein sequence similarities at the peroxidatic cysteine (CP) active site41,42 (Table S3). Consistent with this classification, we report highly conserved PXXX(T/S)XXC (CP) active site motifs for each of the three subfamilies across both metazoans and non-metazoans (Fig. 1D). Within metazoans, we find three variable residues within this motif among AhpC-PRX1 sequences, five variable residues among PRX5 sequences and only two variable residues among PRX6 sequences (underlined residues in Fig. 1D). However, we also find that non-metazoan sequences typically display more variability, particularly for subfamilies, PRX5 and PRX6.
We note that PRX classification based on active site profiles has been adopted in recent literature, such as46 and47, although there still are exceptions, such as48,49,50,51. Continuing challenges are the incorrect, vague or ambiguous annotations in online gene databases, in addition to annotations based on older nomenclature thus does not easily correspond with current literature41,45. Antioxidant or peroxiredoxin-specific online databases have been developed in attempts to address these challenges (e.g. PREX: http://csb.wfu.edu/PREX/, or RedoxiBase: http://peroxibase.toulouse.inra.fr/), but are not updated frequently enough to be as useful as larger databases (e.g., NCBI) that capture a greater, and constantly-growing, breadth of PRX sequence diversity. Thus, it is often the less accurate annotations that are most commonly used. Few studies have described PRXs across diverse metazoan phyla, and even less so in an evolutionary context44. Consequently, we suggest that numerous apparently inaccurate online data base annotations may underestimate the true extent of metazoan PRX diversity, and we predict that a greater breadth of PRX research will reveal further lineage-specific PRXs.
PRX5 is the least conserved animal PRX subfamily
The subfamily PRX5 is considered to be the closest animal orthologue to the ancestral, prokaryotic subfamily, PRXQ52; in our study, it also appears to be the least conserved PRX subfamily. PRX5 displays greater sequence diversity at the CR active site than subfamily AhpC-PRX1. Notably, in three sequences from two sponges (phylum Porifera; X. bergquistia and A. queenslandica), the catalytic cysteine of CR is replaced by a Valine (V) residue. Further, the bilaterians B. floridae and Capitella teleta both encode shortened PRX5 sequences in which the CR motif is absent altogether (Supplementary file 1, Fig. S2). Similarly, amongst PRX5 encoded by non-metazoans, we find that the catalytic CR is substituted in all sequences except for that of the choanoflagellate, M. brevicollis. This is consistent with other studies that have noted that CR is not always present within atypical 2-Cys PRXs41. Additionally, PRX5 is absent from five species, making it the only subfamily with evidence of metazoan gene losses (Table 1). However, these five species do encode alternative sequences that are mitochondrially localised, as PRX5 typically is. Indeed, all metazoans except N. vectensis encode at least one mitochondrially-localised PRX (Fig. 3). In mammals, mitochondrially-localised PRX3 is predicted to compensate PRX5 functioning (Table 1)53, and D. melanogaster mutants lacking PRX3 show few effects, supporting a functional redundancy of PRX5 and PRX354.
In phylum Porifera, PRX5 is the only PRX subfamily for which we do not recover a monophyletic demosponge clade, but rather the freshwater (FW) demosponge Ephydatia muelleri branches independently from the three marine (Mar) demosponges (Fig. 4).
In contrast, PRX6 is present in all 19 metazoan species, and is the most consistently localised PRX subfamily; all species have PRX6 genes predicted to localise to the cytoplasm (Fig. 3). Only Oscarella carmela and D. melanogaster that encode multiple PRX6 genes have one of these localised to nuclei as well as to the cytoplasm (Fig. 3; Supplementary file 3). PRX6 is unique amongst the PRXs in that it lacks a resolving cysteine (CR) and is multifunctional, additionally exhibiting both phospholipase, and PLA2 activity55. Of the 28 metazoan PRX6 sequences, we found that 19 encode the full PLA2 catalytic triad, H… S… D, and nine encode the full GXSXG with no substitutions (Purple residues and purple box, respectively; Supplementary file 1, Fig. S3). PRX6 uses the most abundant free radical scavenger, glutathione, and is considered to “moonlight” as a PHGPX55. Its ubiquitous presence and metazoan-wide conservation suggests strong selection for specific PRX6 activity and function.
Phylum porifera encode PRXs sensitive to hyperoxidation, but lack SRX
For each of the 19 metazoan species, including phylum Porifera, we identified at least one AhpC-PRX1 sequence encoding the full motifs (GGLG and YF) that confer sensitivity to hyperoxidation (SO2H) under high concentrations of H2O2 (Fig. 1A; Supplementary file 1 Fig. S1). Sensitivity to hyperoxidation has so far been observed only in animal AhpC-PRX1 (i.e., typical 2-Cys PRXs), and causes its temporary inactivation until reactivated by the ATP-dependent enzyme Sulfiredoxin (SRX) (Fig. 1b)56,57,58. To date, no other mechanisms for reactivation have been described. Thus, it is surprising to find 10 species that encode sensitive PRXs but not the SRX-like reductant; these are the six sponge species, the ctenophore M. leidyi, and the bilaterians C. teleta, C. elegans, and X. tropicalis (Supplementary file 1, Table S4). Specifically, we identified seven species encoding the SRX ParBc domain (PF02195; IPR003115) but that lacked the strictly conserved SRX-N terminal binding motif F(S/G)GCHR required for catalytic activity (Supplementary file 1, Table S4). SRX has not been widely studied, thus SRX sequence structure may exhibit greater diversity than has currently been described. However, for X. bergquistia, O. carmela, and C. elegans, we could not find even the SRX domain ParBc (PF02195) (Supplementary file 1, Table S4). One possible explanation is that, despite encoding the GGLG and YF motifs, the susceptibility to hyperoxidation for each of these 10 species may in fact be sufficiently low that AhpC-PRX1 inactivation does not occur.
Indeed, it is known in mammals that not all AhpC-PRX1 genes are equally sensitive to hyperoxidation; isoforms PRX1, PRX2, and PRX3 are most susceptible59,60,61, whilst PRX4 and PRX5 are more resistant, with PRX4 being protected within the ER53,62,63. In marine demosponges, PRX4 is the only AhpC-PRX1 that we identified, and in two of these species it was predicted to localise extracellularly so would not be protected within the ER (Fig. 3; Supplementary file 3). Recently, Bolduc et al.64 described how substitution of residues within a and b motifs increases susceptibility to hyperoxidation. Our assessment of PRX4 sequences revealed that at least one residue is substituted within these motifs across all species except for C. intestinalis. Most commonly, the missing residue is His from motif a, except for D. melanogaster that instead is missing residue Asn/Gly; five species are also missing additional residues (Supplementary file 1, Fig. S1; Table S5). Furthermore, E. muelleri and C. teleta that lack SRX encode substitutions for two residues (Supplementary file 2, Table S5; C. elegans does not encode PRX4, but PRX1/2 that lacks 3 residues). These substitutions suggest that the PRX4 genes of the species lacking SRX are at least somewhat susceptible to hyperoxidation, even if not to the same degree as PRX1-3.
Alternatively, species may encode PRXs that are sensitive to hyperoxidation but that are not reactivated, given that reactivation may not always confer increased fitness. In SRX-depleted D. melanogaster, McGinnis et al.53 demonstrated that PRX hyperoxidation does not adversely affect resistance to oxidative stress or fly lifespan, but instead results in increased physical fitness and endurance. This result was very surprising given the number of studies that have demonstrated reduced fitness from SRX under expression in cell cultures, plants, and mammals65,66,67. One possible explanation is that hyperoxidized PRXs in the SRX mutant could either signal as damage associated molecular patterns (DAMPs) themselves or alter post-translation modifications of other proteins that in turn signal as DAMPs, to induce beneficial response pathways53. DAMPs serve as alarm signals within the innate immune system, alerting cells to any damage or to the presence of non-native microbes, which in turn activates host immune responses68. Thus, perhaps species that encode sensitive PRX, but not SRX, use hyperoxidized PRXs for other diverse important signalling functions.
The glutathione peroxidase family
GPX, the most evolutionary recent antioxidant family to emerge, is considerably less conserved than CAT or PRX. Here we expand on previous assessments69,37 by surveying an additional four species of Porifera, as well as the annelid C. teleta and urochordate C. intestinalis, not included by Trenz et al.37. In these six species, we identified 19 unique protein sequences encoding the GSHPx domain. Filtering by domain structure characteristic of a GPX reduced this number to 15 (Fig. 1D; Supplementary file 2). Aside from subfamily GPX7/8, selenocysteine GPXs are widespread across the other six GPX subfamilies. The only exceptions to this include cysteine-dependent GPXs in D. melanogaster GPX4, C. elegans GPX4 and GPX3/5/6, S trongylocentrotus purpuratus GPX3, B. floridae GPX1/2, and Homo sapiens GPX5 (starred sequences Fig. 5). We find the total number and functional subfamilies of GPX genes encoded by each species is variable, with multiple cases of gene loss. Typically, we find fewer GPX genes in non-bilaterian species, and indeed GPX represents the smallest of the three antioxidant families within phylum Porifera (Table 1). Specifically, we find that GPX7 is most common within phylum Porifera, encoded by four species of classes Homoscleromorpha, Calcarea, and two marine species of Demospongiae, but absent from X. bergquistia and the freshwater demosponge E. muelleri (Fig. 5). Aside from this, we reveal class Demospongiae encode GPX4-like sequences, and class Homoscleromorpha encodes a putative GPX1/2 but for which only partial sequences were obtained.
Maximum likelihood phylogenetic tree of GPX enzyme family. Unrooted tree displaying GPX subfamilies, GPX1/2 (peach), GPX3/GPX5/GPX6 (orange), GPX7/GPX8 (turquoise), and PHGPX/GPX4 (grey). Labels in blue denote sequences encoded by phylum Porifera. Labels with a star indicate cysteine GPXs, where cysteine is the catalytic residue. Subfamily GPX7 is exclusively cysteine dependant for all species. Black numbers on branches indicate bootstrap support. Branch lengths represent evolutionary distances, indicated by tree scale. Constructed based on edited alignment, 1000 bb and the LG + G5 evolutionary model.
Phylogenetic analysis revealed support for four main evolutionary groups, namely GPX1/GPX2 (100%), GPX3/GPX5/GPX6 (84%), GPX7/GPX8 (99%), and PHGPX (i.e., Vertebrata GPX4) (77%; Fig. 5). All metazoan GPX sequences fell into one of these four clades, whilst non-metazoan sequences were paraphyletic and phylogenetically distinct. Only one sequence encoded by the choanoflagellate M. brevicollis clustered together with animal GPXs from basal metazoans (phyla Porifera, Ctenophora and Cnidaria) within the subfamily PHGPX/GPX4 (Fig. 5). We found the subfamilies GPX4 and GPX7 are the most abundant across the metazoans.
Subfamily GPX7 is most conserved across the animal kingdom
Subfamily GPX7, which is exclusively cysteine dependent, is the most commonly encoded GPX in metazoans (Fig. 5). It also shows highly conserved subcellular localisation, being predicted to localise to the ER in 13 of the 14 metazoans that encode it (Fig. 3); this finding is consistent with previous observations70.
GPX7 is an animal-specific subfamily that has a key role in facilitating ER protein folding71 and has been described as the novel GPX72. GPX7 is similar to typical CysGPXs in more efficiently using thiols as its reductant rather than GSH, but different in lacking the second resolving cysteine within the canonical site (Fig. 1C)73,74. Instead, GPX7 uses the endoplasmic reticulum (ER) protein disulfide isomerase (PDI) as its reductant, thus helping to recycle it73, reviewed by75. Within the ER, newly synthesised proteins are oxidised by PDI, which in turn are again re-oxidised by ER oxidoreductase 1 (ERO1α) in a reaction that generates H2O272, reviewed by76. GPX7 can increase PDI-oxidising activity of ERO1α70,76, which promotes the refolding of misfolded proteins, and prevents ER oxidative stress response through H2O2 scavenging76. This unique function may explain the strong conservation of GPX7 gene number and localisation across the Metazoa.
In contrast, subfamilies GPX1/2, GPX3/5/6, and GPX4, all of which are predominantly Sec-dependent, are comparatively less conserved. One explanation for this may be their functional redundancy shared with certain PRXs. The subfamily PRX6 that is so well conserved across metazoans is known to “moonlight” as a PHGPX with its similar dependence on GSH54. Moreover, typical CysGPXs share a similar catalytic cycle to 2-Cys PRXs and are hypothesised to function in the same way54,73 (Fig. 1b,c). Interestingly, GPXs1-6 show positive selection at residues located at or close to active sites, or at the dimer interface77. Notably, the catalytic residue within the active site, Sec (U), is encoded by the nucleotide sequence UGA that also encodes the STOP codon78,79. It thus requires additional, energetically costly machinery to be encoded80,81,82. However, selenocysteine GPXs do exhibit significantly greater efficiency than Cys because of their higher nucleophilic activity, and capacity of Sec to efficiently catalyse both one-electron, as well as two-electron reactions83,84,85. Thus, we hypothesise that selection on GPX1-6 may favour seleno-dependent GPXs (i.e., extreme phenotype) that is harder to encode but exhibits greater efficiency. However, without supporting Sec machinery, Sec may not be maintained in the protein, leading to a loss of function. Indeed, in selenocysteine-dependent subfamilies, GPX gene duplications and partial sequences are notably common, particularly within larger genomes of species such as H. sapiens (Supplementary file 3) 77,86, likely reflecting a more rapid rate of evolution.
Conclusions
In our survey of 19 species spanning 10 animal phyla, we find that gene number and distribution are highly conserved in the antioxidant families CAT and PRX, but much less so in the GPX family. We reveal for the first time that all three families—CAT, PRX, and GPX—are encoded by the six species of the basal metazoan phylum Porifera, considered sister to all other animal phyletic lineages. From this we can infer the distribution of these three ancient antioxidant families in the last common animal ancestor (LCAA).
Monofunctional CAT comprises a comparatively small and conserved family in animals; its diversification since the LCAA is recent compared to the very long evolutionary history of this enzyme family. We find both peroxisomal and cytoplasmic forms are common among metazoans; the exceptions are that we did not find any of the peroxisomal form in the marine demosponges or cnidarians surveyed in our study. This suggests that the peroxisomal form may have arisen after the cnidarian-bilaterian split, with the addition of signal peptides.
In contrast, the PRXs comprise a large enzyme family. Subfamilies AhpC-PRX1 and PRX6 are the most widely distributed and conserved, whilst PRX5 exhibits notable gene losses. Interestingly, PRX5, the closest animal orthologue to ancestral PRXQ, appears to have been lost in several species that exhibit gene expansion of subfamily AhpC-PRX1. We show that phylum Porifera encode all three animal PRX subfamilies. However, marine demosponges encode just a single AhpC-PRX1, belonging to PRX4, which is the only subclade conserved across the animal kingdom. This indicates that PRX4, which is also found within non-metazoan choanoflagellate, may be the ancestral AhpC-PRX1.
GPX is the most evolutionary recent origin of all the antioxidant enzyme families, is the least conserved among metazoans, and is the least abundant in phylum Porifera. The subfamilies GPX4 and cysteine-dependent GPX7 are the most common in poriferans, with GPX7 present in all three classes, and GPX4 in Demospongiae only. We find strong conservation across the animal kingdom of ER-localised GPX7, which may reflect its unique role of preventing oxidative damage during protein folding within the ER.
That the enzyme families CAT and PRX have been so widely conserved since their ancient origins predating the evolution of aerobic life suggest a core role that is conserved across the animal kingdom. Thus, our comparative genomic analyses illustrate that the fundamental functions of antioxidants have resulted in gene conservation throughout the animal kingdom, paving the way for functional analyses on these enzyme families in diverse animal phyla.
Methods
Enzyme identification and subfamily classification
We searched for gene sequences encoding candidate members of the CAT, PRX, and GPX families in high quality genomes of 19 metazoan species representing 10 phyla (Supplementary file 1, Table S1). Specifically, predicted coding sequences were scanned against the Pfam A database using hmmscan in HMMER v3.1b2 (hmmer.org) for sequences encoding domains specific to each enzyme family, and their respective subfamilies (Fig. 1D)39. Specifically, HMMER allows us to identify protein sequences encoding functional domains through implementing probabilistic Hidden Markov Models (HMM) to search for protein sequence homologs against a profile database such as Pfam. The number and position of all identified domains was determined. For all identified candidate gene sequences, we predicted protein subcellular localisation regions using DeepLoc-2.0, https://services.healthtech.dtu.dk/service.php?DeepLoc-2.040,87, which uses protein sequences as input for Neural Networks algorithm trained on Uniprot proteins with experimental evidence. The algorithm incorporates the importance (“attention”) of particular amino acids and those within adjacent positions of the region. Positions in the sequence with high “attention” give more weight to the final prediction of the model. DeepLoc-2.0 is able to predict proteins that are located in more than one compartment. The methodology for enzyme identification was cross-validated by comparing the number and type of CAT, PRX, and GPX genes identified through our analysis with those that have previously been described.
Candidate CAT amino acid sequences were identified based on the presence of both of the CAT associated domains PF00199 (catalase) and PF06628 (catalase_rel) (Fig. 1D). No further criteria were applied.
Candidate PRX amino acid sequences were identified based on the presence of at least the PRX domain PF00578, in addition to one or both of PF08534 (AhpC-TSA) and PF10417 (1-cysPrx_C) (Fig. 1D). Candidates were then scanned and filtered based on the presence of the strictly conserved CP active site motif, PXXX(T/S)XXC (where X may be any amino acid Wood et al. 2003). The CP motif is required for PRX catalytic activity on H2O2, so sequences that did not contain this motif were excluded from further analysis. Additionally, we scanned for the presence of subfamily-specific motifs. For subfamily PRX6, this included the catalytic tetrad His, Ser, Asp encoding phospholipase A2 (PLA2) and GXSXG motif encoding phospholipase/ esterase (lipase) activity55. For subfamily AhpC-PRX1, we searched for the motifs GGLG and YF that encode sensitivity to hyperoxidation, as well as a and b motifs that contribute to determining the degree of PRX sensitivity to hyperoxidation56,57,58,64. Accordingly, we also searched for the presence of enzymatic reductant sulfiredoxin (SRX) that can reactivate hyperoxidized PRXs (Fig. 1B). To do this, we first scanned predicted coding sequences for the presence of the ParB-like nuclease domain (PF02195; IPR003115) and then filtered based on the presence of the strictly conserved SRX-N terminal binding motif F(S/G)GCHR that is required for SRX catalytic activity88.
Candidate GPX amino acid sequences considered in our study include 61 sequences obtained previously by37 from 13 metazoan species indicated in Table S1 (Supplementary file 1). In addition to these, we assessed protein coding sequences of six species, that includes four sponges, that were not assessed by37. For these six species, we retained candidate GPX enzyme sequences encoding the domain GSHPx (PF00255; Fig. 1D) and scanned these for the presence of the conserved GPX catalytic tetrad, Sec/Cys (U/C), Gln (Q), Trp (W), and Asn (N) required for GPX activity89,90. Sequences encoding Sec at the first residue of the catalytic tetrad were classified as selenium-dependent and those encoding Cys at the first residue of the catalytic tetrad were classified as cysteine dependant GPXs90.
Multiple sequence alignment and phylogenetic assessment
For each enzyme family, candidate sequences were aligned using MAFFT91,92 (https://mafft.cbrc.jp/alignment/software/) with default parameters and visualised in the multiple sequence alignment editor AliView v1.2793 (https://ormbunkar.se/aliview). To provide evolutionary context to the metazoan phylogenetic relationships, we also incorporated non-metazoan sequences; these included the phylum Choanoflagellata that is closest extant animal relative, as well as other non-metazoan eukaryotics representing Amoebozoa, Red algae, and fungi. Sequences were obtained from UniProt (https://www.uniprot.org/) with specific details provided in Table S2 (Supplementary file 1).
To assess phylogenetic relationships, alignments were manually edited in AliView v1.2793 (https://ormbunkar.se/aliview) to remove regions containing more than 50% gaps. Edited alignments were then imported to IQ-TREE94 to construct maximum likelihood trees using ultrafast bootstrap95, based on 1000 bb and the most appropriate evolutionary model as identified by ModelFinder96. Models identified and used to construct each enzyme family tree were as follows: CAT, LG + I + G4, and CAT metazoan LG + G4; PRX, WAG + I + G4; GPX, LG + G5. Resultant phylogenetic trees were first visualised in iTOL v.6.2.197 before annotating in Adobe Illustrator. Classification of PRX and GPX gene subfamilies were inferred from the relative placing of putative sequences within known subfamily clades of phylogenetic trees.
Data availability
All data generated or analysed during this study are included in this published article, its supplementary information files, and publicly available repositories.
References
Ślesak, I., Ślesak, H. & Kruk, J. Evolution of life on earth: In silico comparative analysis of biochemical pathways. Astrobiology 12, 775–784. https://doi.org/10.1089/ast.2011.0704 (2012).
Gough, D. & Cotter, T. Hydrogen peroxide: a Jekyll and Hyde signalling molecule. Cell Death Dis 2, e213 (2011). https://doi.org/10.1038/cddis.2011.96
Stadtman, E. R. & Berlett, B. S. Reactive oxygen-mediated protein oxidation in aging and disease. Chem. Res. Toxicol. 10(5), 485–494 (1997).
Halliwell, B. & Gutteridge, J.M.C. Free Radicals in Biology and Medicine. Oxford: Oxford Univ. Press. 5th ed. (2015)
Sies, H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: Oxidative eustress. Redox Biol. 11, 613–619 (2017).
Case, A. J. On the origin of superoxide dismutase: An evolutionary perspective of superoxide-mediated redox signaling. Antioxidants (Basel). 6(4), 82. https://doi.org/10.3390/antiox6040082 (2017).
Inupakutika, M. A., Sengupta, S., Devireddy, A. R., Azad, R. K. & Mittler, R. The evolution of reactive oxygen species metabolism. J. Exp. Bot. 67(21), 5933–5943 (2016).
Neubeck, A. & Freund, F. Sulfur Chemistry May Have Paved the Way for Evolution of Antioxidants. Astrobiology 20, 670–675. https://doi.org/10.1089/ast.2019.2156 (2020).
Khademian, M. & Imlay, J. A. How microbes evolved to tolerate oxygen. Trends Microbiol. 29(5), 428–440 (2021).
Jabłońska, J. & Tawfik, D. S. The evolution of oxygen-utilizing enzymes suggests early biosphere oxygenation. Nat. Ecol. Evol. 5, 442–448. https://doi.org/10.1038/s41559-020-01386-9 (2021).
Mittler, R. et al. ROS signaling: the new wave?. Trends Plant Sci. 16(6), 300–309 (2011).
Anbar, A. D. Elements and evolution. Science 322, 1481–1482 (2008).
Halliwell, B. Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol. 141:312–322 (2006). https://doi.org/10.1104/pp.106.077073
Zámocký, M., Gasselhuber, B., Furtmüller, P. G. & Obinger, C. Molecular evolution of hydrogen peroxide degrading enzymes. Arch. Biochem. Biophys. 525, 131–144. https://doi.org/10.1016/j.abb.2012.01.017 (2012).
Sies, H. & Jones, D. P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 21(7), 363–383 (2020).
Nathan, C. & Cunningham-Bussel, A. Beyond oxidative stress: an immunologist’s guide to reactive oxygen species. Nat. Rev. Immunol. 13, 349–361. https://doi.org/10.1038/nri3423 (2013).
Sarsour, E. H., Kumar, M. G., Chaudhuri, L., Kalen, A. L. & Goswami, P. C. Redox control of the cell cycle in health and disease. Antioxid. Redox Signal. 11, 2985–3011 (2009).
Rhee, S. G., Woo, H. A. & Kang, D. The role of peroxiredoxins in the transduction of H2O2 signals Antioxid. Redox Signal. 28, 537–557 (2018).
Weaver, C. J., Leung, Y. F. & Suter, D. M. Expression dynamics of NADPH oxidases during early zebrafish development. J. Comp. Neurol. 524, 2130–2141 (2016).
Weaver, C. J. et al. nox2/cybb deficiency affects zebrafish retinotectal connectivity. J. Neurosci. 38, 5854–5871 (2018).
Edgar, R. S. et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature 485, 459–464. https://doi.org/10.1038/nature11088 (2012).
Nimse, S. & Pal, D. Free radicals, natural antioxidants, and their reaction mechanisms. RSC Adv. 5, 27986–28006 (2015).
Das, K. & Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2, 53. https://doi.org/10.3389/fenvs.2014.00053 (2014).
Rhee, S. G. Redox signaling: Hydrogen peroxide as intracellular messenger. Exp. Mol. Med. 31, 53–59 (1999).
D’Autreaux, B. & Toledano, M. B. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 8, 813–824 (2007).
Parascandolo, A. & Laukkanen, M. O. Carcinogenesis and reactive oxygen species signaling: Interaction of the NADPH oxidase NOX1-5 and superoxide dismutase 1–3 signal transduction pathways. Antioxid. Redox Signal. 30, 443–486. https://doi.org/10.1089/ars.2017.7268 (2019).
Hewitt, O. H. & Degnan, S. M. Distribution and diversity of ROS-generating enzymes across the animal kingdom, with a focus on sponges (Porifera). BMC Biol. 20, 212 (2022).
Mittler, R., Vanderauwera, S., Gollery, M. & Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498 (2004).
Glorieux, C. & Calderon, P. B. Catalase, a remarkable enzyme: targeting the oldest antioxidant enzyme to find a new cancer treatment approach. Biol. Chem. 398(10), 1095–1108. https://doi.org/10.1515/hsz-2017-0131 (2017).
Rhee, S. G. & Kil, S. Mitochondrial H2O2 signaling is controlled by the concerted action of peroxiredoxin III and sulfiredoxin: Linking mitochondrial function to circadian rhythm. Free Rad. Biol. Med. 100, 73–80 (2016).
Brigelius-Flohé, R. & Flohé, L. Regulatory phenomena in the glutathione peroxidase superfamily. Antioxid Redox Signal. 33(7), 498–516. https://doi.org/10.1089/ars.2019.7905 (2020).
Rhee, S.G. Overview on Peroxiredoxin. Mol. Cells. 39(1):1–5 (2016). https://doi.org/10.14348/molcells.2016.2368.
Erwin, D.H., Laflamme, M., Tweedt, S.M., Sperling, E.A., Pisani, D. & Peterson, K.J. The Cambrian conundrum: early divergence and later ecological success in the early history of animals. Science. 25, 334(6059), 1091–7 (2011). https://doi.org/10.1126/science.1206375.
Simion, P. et al. A large and consistent phylogenomic dataset supports sponges as the sister group to all other animals. Curr. Biol. 27, 1–10. https://doi.org/10.1016/j.cub.2017.02.031 (2017).
Nielsen, C. Early animal evolution: A morphologist's view. R. Soc. Open Sci. 6(7), 190638, (2019). https://doi.org/10.1098/rsos.190638
Srivastava, M. et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726. https://doi.org/10.1038/nature09201 (2010).
Trenz, T. S. et al. Going forward and back: The complex evolutionary history of the GPx. Biology (Basel). 10(11), 1165. https://doi.org/10.3390/biology10111165 (2021).
Zámocký, M., Furtmüller, P. G. & Obinger, C. Evolution of catalases from bacteria to humans. Antioxid. Redox Signal. 10(9), 1527–1546 (2008).
Eddy, S. R. Accelerated profile HMM searches. PLOS Comp. Biol. 7, e1002195 (2011).
Almagro Armenteros, J. J., Sønderby, C. K., Sønderby, S. K., Nielsen, H. & Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics. 33, 3387–3395. https://doi.org/10.1093/bioinformatics/btx431 (2017).
Nelson, K. J. et al. Analysis of the peroxiredoxin family: Using active-site structure and sequence information for global classification and residue analysis. Proteins 79(3), 947–964. https://doi.org/10.1002/prot.22936 (2011).
Harper, A.F., Leuthaeuser, J.B., Babbitt, P.C., Morris, J.H., Ferrin, T.E., Poole, L.B. & Fetrow, J.S. An atlas of peroxiredoxins created using an active site profile-based approach to functionally relevant clustering of proteins. PLoS Comput. Biol. 13(2), e1005284 (2017). https://doi.org/10.1371/journal.pcbi.1005284.
Helm, R. R., Martín-Díaz, M. L. & Tarrant, A. M. Phylogenetic analysis of cnidarian peroxiredoxins and stress-responsive expression in the estuarine sea anemone Nematostella vectensis. Comp. Biochem. Physiol. A. 221, 32–43. https://doi.org/10.1016/j.cbpa.2018.03.009 (2018).
Bakiu, R. & Santovito, G. New Insights into the molecular evolution of metazoan peroxiredoxins. Acta Zool. Bulg. 67(2), 305–317 (2015).
Poole, L.B. & Nelson, K.J. Distribution and features of the six classes of peroxiredoxins. Mol. Cells. 39(1), 53–9 (2016). https://doi.org/10.14348/molcells.2016.2330.
de Oliveira, M. A. et al. Relevance of peroxiredoxins in pathogenic microorganisms. Appl. Microbiol. Biotechnol. 105, 5701–5717. https://doi.org/10.1007/s00253-021-11360-5 (2021).
Abbas, M. N., Kausar, S. & Cui, H. The biological role of peroxiredoxins in innate immune responses of aquatic invertebrates. Fish Shellfish Immunol. 89, 91–97. https://doi.org/10.1016/j.fsi.2019.03.062 (2019).
Drago, L., Ferro, D., Bakiu, R., Ballarin, L. & Santovito, G. Typical 2-Cys peroxiredoxins as a defense mechanism against metal-induced oxidative stress in the solitary ascidian Ciona robusta. Antioxidants 11, 93. https://doi.org/10.3390/antiox11010093 (2022).
Shrivastava, A.K., Singh, P.K., Sitther, V., Singh, S. & Srivastava, S. Chapter 17 - Cyanobacterial peroxiredoxins and their role in cyanobacterial stress biology in Advances in Cyanobacterial Biology (ed. Singh, P.K., Singh, V.K., Kumar, A. & Shrivasta, A.K.) 249–268 (Elsevier, 2020). https://doi.org/10.1016/C2018-0-05196-8
Xie, D., Wang, S., Zhao, C., Wang, P. & Qiu, L. Molecular characterization and functional analysis of peroxiredoxin3 cDNA from black tiger shrimp (Penaeus monodon). Aquac. Res. 50(7), 1862–1875. https://doi.org/10.1111/are.14069 (2019).
Tolomeo, A.M., Carraro, A., Bakiu, R., Toppo, S., Garofalo, F., Pellegrino, D., Gerdol, M., Ferro, D., Place, S.P. & Santovito, G. Molecular characterization of novel mitochondrial peroxiredoxins from the Antarctic emerald rockcod and their gene expression in response to environmental warming. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 225, 108580 (2019). https://doi.org/10.1016/j.cbpc.2019.108580.
Knoops, B., Loumaye, E. & Van Der Eecken, V. Evolution of the Peroxiredoxins in Peroxiredoxin Systems. Subcellular Biochemistry, vol 44. (ed. Flohé, L. & Harris, J.R.) (Springer, Dordrecht, 2007). https://doi.org/10.1007/978-1-4020-6051-9_2
McGinnis, A., Klichko, V. I., Orr, W. C. & Radyuk, S. V. Hyperoxidation of peroxiredoxins and effects on physiology of drosophila. Antioxidants 10, 606. https://doi.org/10.3390/antiox10040606 (2021).
Radyuk, S. N. et al. Mitochondrial peroxiredoxins are critical for the maintenance of redox state and the survival of adult Drosophila. Free Radic. Biol. Med. 49, 1892–1902 (2010).
Fisher, A. B. The phospholipase A2 activity of peroxiredoxin 6. J. Lipid Res. 59, 1132–1147 (2018).
Biteau, B., Labarre, J. & Toledano, M.B. ATP-dependent reduction of cysteine–sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 425, 980–984 (2003). https://doi.org/10.1038/nature02075
Jonsson, T. J., Murray, M. S., Johnson, L. C. & Lowther, W. Reduction of cysteine sulfinic acid in peroxiredoxin by sulfiredoxin proceeds directly through a sulfinic phosphoryl ester intermediate. J. Biol. Chem. 283, 23846–23851. https://doi.org/10.1074/jbc.M803244200 (2008).
Jonsson, T. J., Tsang, A. W., Lowther, W. T. & Furdui, C. M. Identification of intact protein thiosulfinate intermediate in the reduction of cysteine sulfinic acid in peroxiredoxin by human sulfiredoxin. J. Biol. Chem. 283, 22890–22894. https://doi.org/10.1074/jbc.C800124200 (2008).
Yang, K. S. et al. Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J. Biol. Chem. 277, 38029–38036 (2002).
Cox, A. G. et al. Mitochondrial peroxiredoxin 3 is more resilient to hyperoxidation than cytoplasmic peroxiredoxins. Biochem. J. 421, 51–58 (2009).
Winterbourn, C. C. & Peskin, A. V. Kinetic approaches to measuring peroxiredoxin reactivity. Mol. Cells. 39, 26–30 (2016).
Knoops, B., Goemaere, J., Van der Eecken, V. & Declercq, J. P. Peroxiredoxin 5: Structure, mechanism, and function of the mammalian atypical 2-Cys peroxiredoxin. Antioxid. Redox Signal. 15, 817–829 (2011).
Cao, Z., Subramaniam, S. & Bulleid, N. J. Lack of an efficient endoplasmic reticulum-localized recycling system protects peroxiredoxin IV from hyperoxidation. J. Biol. Chem. 289, 5490–5498 (2014).
Bolduc, J., Koruza, K., Luo, T., Pueyo, J.M., Vo, T.N., Ezerina, D. & Messens, J. Peroxiredoxins wear many hats: Factors that fashion their peroxide sensing personalities. Redox Biol. 42, 101959 (2021). https://doi.org/10.1016/j.redox.2021.101959
Liu, X. P. et al. Molecular and functional characterization of sulfiredoxin homologs from higher plants. Cell Res. 16, 287–296 (2006).
Baek, J. Y. et al. Sulfiredoxin protein is critical for redox balance and survival of cells exposed to low steady-state levels of H2O2. J. Biol. Chem. 287, 81–89 (2012).
Wu, L. et al. Tumor promoter-induced sulfiredoxin is required for mouse skin tumorigenesis. Carcinogenesis 35, 1177–1184 (2014).
Nyholm, S. V. & Graf, J. Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat. Rev. Microbiol. 10(12), 815–827. https://doi.org/10.1038/nrmicro2894 (2012).
Margis, R., Dunand, C., Teixeira, F. K. & Margis-Pinheiro, M. Glutathione peroxidase family—an evolutionary overview. FEBS J. 275, 3959–3970. https://doi.org/10.1111/j.1742-4658.2008.06542.x (2008).
Nguyen, V. D. et al. Two endoplasmic reticulum PDI peroxidases increase the efficiency of the use of peroxide during disulfide bond formation. J. Mol. Biol. 406, 503–515 (2011).
Wang, L., Zhang, L., Niu, Y., Sitia, R. & Wang, C. C. Glutathione peroxidase 7 utilizes hydrogen peroxide generated by Ero1α to promote oxidative protein folding. Antioxid. Redox Signal. 20(4), 545–556. https://doi.org/10.1089/ars.2013.5236 (2014).
Utomo, A. et al. Identification of a novel putative non-selenocysteine containing phospholipid hydroperoxide glutathione peroxidase (NPGPx) essential for alleviating oxidative stress generated from polyunsaturated fatty acids in breast cancer cells. J. Biol. Chem. 279, 43522–43529 (2004).
Brigelius-Flohé, R. & Maiorino, M. Glutathione peroxidases. Biochim. Biophys. Acta. 1830, 3289–3303 (2013).
Flohé, L., Toppo, S., Cozza, G. & Ursini, F. A comparison of thiol peroxidase mechanisms. Antioxid. Redox Signal. 15, 763–780 (2011).
Ramming, T., Kanemura, S., Okumura, M., Inaba, K. & AppenzellerHerzog, C. Cysteines 208 and 241 in Ero1α are required for maximal catalytic turnover. Redox Biol. 7, 14–20 (2016).
Buday, K. & Conrad, M. Emerging roles for non-selenium containing ER-resident glutathione peroxidases in cell signaling and disease. Biol. Chem. 402(3), 271–287. https://doi.org/10.1515/hsz-2020-028 (2020).
Tian, R., Geng, Y., Yang, Y., Seim, I. & Yang, G. Oxidative stress drives divergent evolution of the glutathione peroxidase (GPX) gene family in mammals. Integr. Zool. 16(5), 696–711. https://doi.org/10.1111/1749-4877.12521 (2021).
Zinoni, F., Birkmann, A., Stadtman, T. C. & Böck, A. Nucleotide sequence and expression of the selenocysteine containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc. Natl. Acad. Sci. USA 83, 4650–4654 (1986).
Hüttenhofer, A. & Böck, A. RNA structures involved in selenoprotein synthesis. Cold Spring Harb. Monogr. Arch. 35, (1998).
Lobanov, A. V., Hatfield, D. L. & Gladyshev, V. N. Eukaryotic selenoproteins and selenoproteomes. Biochim. Biophys. Acta. 1790, 1424–1428 (2009).
Chapple, C. E., Guigó, R. & Krol, A. SECISaln, a web-based tool for the creation of structure-based alignments of eukaryotic SECIS elements. Bioinformatics 25, 674–675 (2009).
Lu, J. & Holmgren, A. Selenoproteins. J. Biol. Chem. 284, 723–727 (2009).
Nauser, T., Dockheer, S., Kissner, R. W. H. & Koppenol, W. H. Catalysis of electron transfer by selenocysteine. Biochemistry 45, 6038–6043 (2006).
Steinmann, D. et al. Kinetics of tyrosyl radical reduction by selenocysteine. Biochemistry 47, 9602–9607 (2008).
Arnér, E. S. J. Selenoproteins—What unique properties can arise with selenocysteine in place of cysteine?. Exp. Cell. Res. 316, 1296–1303. https://doi.org/10.1016/j.yexcr.2010.02.032 (2010).
Cubas-Gaona, L. L., Francisco, P., Martin-González, A. & Gutiérrez, J. C. Tetrahymena glutathione peroxidase family: A comparative analysis of these antioxidant enzymes and differential gene expression to metals and oxidizing agents. Microorganisms 8(7), 1008. https://doi.org/10.3390/microorganisms8071008 (2020).
Thumuluri, V., Almagro Armenteros, J.J., Johansen, A.R., Nielsen, H. & Winther, O. DeepLoc 2.0: multi-label subcellular localization prediction using protein language models. Nucleic Acid Res. 50(W1), W228–W234 (2022). https://doi.org/10.1093/nar/gkac278
Zhang, J., Ye, Z., Bowers, R.R., Townsend, D.M. & Tew, K.D. Sulfiredoxin in Encyclopedia of Signaling Molecules. (ed. Choi, S.) 5221–5232 (Springer, Cham, 2018) https://doi.org/10.1007/978-3-319-67199-4_258
Maiorino, M. et al. Probing the presumed catalytic triad of selenium-containing peroxidases by mutational analysis of phospholipid hydroperoxide glutathione peroxidase (PHGPx). Biol. Chem. Hoppe Seyler. 376, 651–660 (1995).
Tosatto, S. C. E. et al. The catalytic site of glutathione peroxidases. Antioxid. Redox Signal. 10, 1515–1526. https://doi.org/10.1089/ars.2008.2055 (2008).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14), 3059–3066. https://doi.org/10.1093/nar/gkf436 (2002).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30(4), 772–780. https://doi.org/10.1093/molbev/mst010 (2013).
Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 30(22), 3276–3278. https://doi.org/10.1093/bioinformatics/btu531 (2014).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. https://doi.org/10.1093/molbev/msu300 (2015).
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. https://doi.org/10.1093/molbev/msx281 (2018).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 14, 587–589. https://doi.org/10.1038/nmeth.4285 (2017).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49(W1), W293–W296. https://doi.org/10.1093/nar/gkab301 (2021).
Acknowledgements
The authors thank Nick Rhodes from the Queensland Cyber Infrastructure Foundation (QCIF), Haojing Shao, and Enya Hermsen for their advice on bioinformatical analyses and computational assistance, Alastair McEwan and Mathias Jönsson for their editorial assistance on early drafts of the manuscript, and all members of the Degnan lab for their valuable discussions.
Funding
This research was supported by an Australian Research Council grant awarded to SM Degnan (DP190102521) and by a University of Queensland Research Training Program scholarship awarded to OH Hewitt.
Author information
Authors and Affiliations
Contributions
O.H.H. and S.M.D. conceptualized this project and the methodological strategies. O.H.H. conducted all bioinformatical analyses. O.H.H. prepared the original draft of text and figures, and SMD made significant contributions to revising the text and figures. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- Bya
-
Billion years ago
- CAT
-
Catalase
- CP
-
Peroxidatic cysteine
- CR
-
Resolving cysteine
- DAMPs
-
Damage associated molecular patterns
- DASP
-
Deacon active site profiler
- ER
-
Endoplasmic reticulum
- ERO1α
-
ER oxidoreductase 1
- GOE
-
Great oxidation event
- GPX
-
Glutathione peroxidase
- GSH
-
Glutathione
- H2O2
-
Hydrogen peroxide
- LCAA
-
Last common animal ancestor
- O2 •-
-
Superoxide anion radical
- PDI
-
Protein disulfide isomerase
- PRX
-
Peroxiredoxin
- ROS
-
Reactive oxygen species
- SRX
-
Sulfiredoxin
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hewitt, O.H., Degnan, S.M. Antioxidant enzymes that target hydrogen peroxide are conserved across the animal kingdom, from sponges to mammals. Sci Rep 13, 2510 (2023). https://doi.org/10.1038/s41598-023-29304-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-29304-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.