Abstract
The ancestors of cyanobacteria generated Earth’s first biogenic molecular oxygen, but how they dealt with oxidative stress remains unconstrained. Here we investigate when superoxide dismutase enzymes (SODs) capable of removing superoxide free radicals evolved and estimate when Cyanobacteria originated. Our Bayesian molecular clocks, calibrated with microfossils, predict that stem Cyanobacteria arose 3300–3600 million years ago. Shortly afterwards, we find phylogenetic evidence that ancestral cyanobacteria used SODs with copper and zinc cofactors (CuZnSOD) during the Archaean. By the Paleoproterozoic, they became genetically capable of using iron, nickel, and manganese as cofactors (FeSOD, NiSOD, and MnSOD respectively). The evolution of NiSOD is particularly intriguing because it corresponds with cyanobacteria’s invasion of the open ocean. Our analyses of metalloenzymes dealing with reactive oxygen species (ROS) now demonstrate that marine geochemical records alone may not predict patterns of metal usage by phototrophs from freshwater and terrestrial habitats.
Similar content being viewed by others
Introduction
Oxygen is essential for complex life forms as it is used during aerobic respiration to create more energy per mol of substrate than other available electron acceptors1. Although today the Earth’s atmosphere contains ~21% oxygen (O2), it was at least 105 times lower in the Archaean 4.0–2.5 billion years ago (Gya)2,3. Just how and when O2 first appeared as a byproduct of biological evolution-oxygenic photosynthesis - remains controversial, with estimates ranging from near the origin of life4 to 3.8 billion years ago (Ga)5 to immediately preceding the Great Oxidation Event (GOE)6, which is estimated to have begun 2.50–2.45 Ga7,8. Before this, O2 could have been produced by physical processes, such as photodissociation of water and carbon dioxide by UV light9,10, but is unlikely to have accumulated at appreciable levels. With the evolution of oxygenic photosynthesis came the potential to produce O2 on a much larger scale. Since O2 is highly reactive, early Cyanobacteria—the first producers of biogenic O2—likely experienced selective pressure, resulting in the evolution of more efficient antioxidants. Such effects have been documented in the photosynthetic machinery, which has been evolving strategies of dealing with reactive oxygen species (ROS) throughout its history11. Therefore, it is reasonable to assume that O2-generating organisms, such as cyanobacteria, would have co-evolved more efficient mechanisms of managing ROS as water oxidation proteins evolved.
Cyanobacteria remove ROS using carotenoids, α-tocopherol, and antioxidant enzymes, including peroxidases, catalases, superoxide reductases (SORs), and superoxide dismutases (SODs)12, and their evolutionary history can be elucidated with phylogenetic methodologies. Although peroxidases and catalases enhance the rate of removal of peroxides (such as H2O2 and R-O-O-H)13, SORs and SODs remove superoxide free radicals (O2.−)14. These O2.− are produced as a byproduct of photosynthetic and respiratory electron transport chains12 as well as extracellular processes on the cell surface15. They can also have beneficial roles in iron acquisition, cell signaling, and growth15,16, but if O2.− are allowed to accumulate inside the cell, they react with solvent-exposed 4Fe-4S clusters in proteins, including those required for amino acid biosynthesis17 and photosynthesis18, generating reactants of the Fenton reaction, which can ultimately lead to extensive DNA damage12. To balance the beneficial effects of O2.− with damage caused by over-exposure, organisms must maintain control over their abundance. Perhaps, it is for this reason that SODs and SORs have been found in all three domains of life—Eukarya, Archaea, and Bacteria14.
Phylogenetic methodologies coupled with protein structural analyses have revealed that three different isoforms of SOD enzymes evolved independently of one another to remove unnecessary O2.− 19. Each has a unique 3D structure, amino acid sequence, and metal cofactor(s); either manganese (MnSOD), nickel (NiSOD), or a combination of copper and zinc (CuZnSOD)19,20. A fourth SOD enzyme shares its evolutionary heritage with MnSOD, but utilizes iron as its cofactor (FeSOD)21. All of these SODs are found within cyanobacteria22,23, but their locations within the cell differ. FeSOD and NiSOD are cytoplasmic22,24,25,26,27, whereas CuZnSODs from Synechococcus and MnSODs from Anabaena and Plectolyngbya are tethered to membranes25,27,28,29,30. The choice of which SOD isoform(s) and associated metal cofactor(s) are used by a given species could reflect its evolutionary heritage, function, and environmental history.
The evolutionary record of cyanobacteria spans at least 1.88 Ga (the oldest undisputed colonial cyanobacterial microfossils;31), and perhaps as many as or >3.22 Ga (based on stromatolite fabric32 and molecular clock analyses of PSII33). During this time, cyanobacteria have diversified into a wide range of marine, freshwater, and terrestrial habitats34,35. Early phylogenetic studies on the evolution of SODs in cyanobacteria were limited by the availability of genomes but found that NiSODs are encoded in the genomes of planktonic marine strains (such as Prochlorococcus spp.), whereas MnSOD and FeSOD are more widespread22,23,36. CuZnSODs, while rare, are not restricted to any particular group22,23. The 3D structures required for metalloproteins to incorporate copper cofactors have previously been predicted to have arisen after those required to incorporate Fe- and Mn-utilizing proteins in the Proterozoic (~2.4–0.5 Ga), following the GOE37. Therefore, it would follow that cyanobacteria used FeSODs and MnSODs before CuZnSODs. Although genes encoding CuZnSODs and FeSODs/MnSODs are widely distributed in bacteria and eukaryotes, NiSODs are restricted to bacteria and may have appeared later in the evolutionary history of life19. Previous approaches aimed at determining metal usage in SODs have not considered the habitats where strains live or evolved. This is problematic because metal availability differs across oceanic and terrestrial environments38,39.
Here, we analyze when cyanobacteria acquired genes encoding NiSOD (sodN), CuZnSOD (sodC), FeSOD (sodB), and MnSOD (sodA) in the context of their habitat and metal cofactor availability through geological time. To do this, we construct phylogenies of all SODs encoded in the genomes of 15,899 bacteria. We also implement a phylogenomic approach and multiple molecular clock analyses to estimate when the crown group of cyanobacteria diverged from their closest non-photosynthetic relatives, the Vampirovibrionia40,41. By studying the timing of the divergence of cyanobacteria from their closest relatives, as well as their acquisition of SODs, we establish points in the Proterozoic and Archaean when these SODs were present in cyanobacteria.
Results
Bacterial SOD diversity
To elucidate which transition metals cyanobacteria first used to protect themselves against the oxidative stress caused by O2.−, the evolutionary history of SOD genes was modeled and mapped onto an updated time-calibrated phylogeny. In order to do this, we began by screening 15,899 bacterial genomes for genes encoding NiSOD, CuZnSOD, and MnSOD. BLASTP was unable to distinguish between genes encoding FeSOD and MnSOD, so they are considered as one hereafter. Results reveal that the sodN gene encoding NiSOD is less common in bacteria than sodC encoding CuZnSOD. The sodN gene was found in 1,464 different species, including gammaproteobacteria, alphaproteobacteria, planctomycetes, bacteroidetes, actinobacteria, cyanobacteria, verrucomicrobia, and chloroflexi (Fig. 1a). By contrast, sodC was absent from the latter two phyla, but present in more genomes (5,723) of beta-, gamma- and alpha–proteobacteria as well as planctomycetes, bacteroidetes, actinobacteria, cyanobacteria, and firmicutes (Fig. 1b). Together, sodA and sodB are more widespread than sodC and sodN combined, being present in 13,748 bacterial genomes across all ten phyla mentioned previously, as well as fibrobacteres, chlorobi, and vampirovibrionia (Fig. 1c and Supplementary Fig. 1).
a NiSOD b CuZnSOD c Fe- and Mn-utilizing SODs. a and b are representative of three independent replicates (Supplementary Figs. 11–12), whereas c is the most likely of three potential topologies (Supplementary Fig. 1). All trees were constructed with ML methodology implemented in IQ-TREE v1.6.146. Branches are colored to represent proteins found in 1 of 13 bacterial phyla: Cyanobacteria (orange), actinobacteria (green), gammaproteobacteria (blue), alphaproteobacteria (yellow), betaproteobacteria (pink), bacteroidetes (cyan), firmicutes (red), chloroflexi (light green), verrucomicrrobia (dark blue), planctomycetes (purple), chlamydia (olive), fibrobacteres (dark green), and chlorobi (gray). Branch lengths represent the number of amino-acid substitutions per site with scale bars representing an average of one substitution per site. Numbered arrows indicate cyanobacterial proteins. Interesting UFBoot values ≥ 95 in all NiSOD replicate trees are indicated with black circles.
Cyanobacterial SOD diversity
A more specific search among cyanobacteria revealed that most strains with sodN (51 of 55 strains) live in saltwater habitats (Fig. 2). They include representatives from all major clades of marine taxa (Fig. 3). Ten lack a gene (named sodX) encoding NiSOD’s maturation protease (Supplementary Data 1). This maturation protease activates NiSOD42 by cleaving the preprotein43, so these strains may not be able to use NiSOD to remove O2.−. Six of them contain genes encoding other SOD isoforms, while the remaining four picocyanobacteria (i.e., Prochlorococcus and Synechococcus species) do not (Fig. 3). The genome completeness of free-living strains—measured with BUSCO v3.0.2 using lineage data from cyanobacteria44—is estimated at 92–98% so it is possible that the maturation protease gene is present but has not been sequenced (Supplementary Table 1).
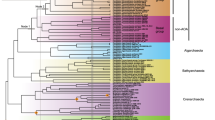
Genes encoding NiSOD (orange circles), NiSOD maturation protease (orange stars), CuZnSOD (red squares), and SODs with Mn- or Fe- cofactors (blue triangles) are highlighted next to the strain names. The earliest node predicted to have sodN, sodC and sodA or sodB are annotated with labels (see Table 2 for posterior age probabilities and Supplementary Fig. 10 for the age distribution of these events). The phylogenetic tree was estimated from SSU and LSU ribosomal RNA and 136 core cyanobacterial proteins from 167 different taxa using the maximum likelihood methodology implemented in IQ-TREE v1.6.146. Node labels represent ultrafast bootstrap approximations <10098. Ages were estimated using a Bayesian relaxed molecular clock with Uncorrelated Gamma Multipliers49 for ribosomal RNA. Black circles represent calibration points (Table 1). The first divergence of cyanobacteria was constrained to occur between 2.7 Ga and 2.32 Ga110,111. The color behind each strain name indicates whether it is from a marine habitat (light blue), freshwater habitat (gray), terrestrial habitat (light brown), or geothermal spring (light pink). GOE Great Oxidation Event, Uni. diaz. unicellular diazotrophs, Phan. Phanerozoic eon, V. Vampirovibrionia.
Many cyanobacterial strains contain multiple SOD isoforms. These include 32 strains with sodN, every strain with sodC, and 45 of 115 with sodA or sodB. The remaining 23 strains which only use NiSOD (including three without sodX) are all marine with small genomes. They include picocyanobacteria and endosymbionts living in larger marine tunicates, algae, and sponges (e.g., Prochloron spp., UCYNA, and two strains of Synechococcus spongiarum). The 70 cyanobacteria which only use FeSOD and MnSOD live in a variety of marine, terrestrial, and freshwater habitats (Fig. 3). Only 3% of cyanobacteria (five of 149 strains discounting plastids) have genes encoding every SOD isoform as well as the NiSOD maturation protease (Fig. 3). Paralogues of sodC were found in three genomes and paralogues of sodA and/or sodB were found in at least 13 genomes (Supplementary Data 1).
The resources needed to make each SOD isoform vary. NiSOD is composed of a mode of 157 amino acids (range 145–166), whereas CuZnSOD is composed of 177 (range 103–236) and FeSOD/MnSOD of 200 (range 197–280) (Supplementary Fig. 2).
Transfer of SODs across phyla
To investigate whether cyanobacteria obtained their SOD genes from other bacterial phyla, maximum likelihood (ML) phylogenies were constructed using the NiSODs, CuZnSODs, and combined Mn- and Fe-SODs from bacteria. If, in each case, all cyanobacterial proteins form a monophyletic group (i.e., they share a recent common ancestor), this would indicate a single origin. Surprisingly, these phylogenetic analyses revealed that cyanobacterial NiSODs, CuZnSODs, MnSODs, and FeSODs are polyphyletic (Fig. 1 and Supplementary Fig. 1), suggesting independent origins perhaps due to several lateral gene transfer events or other modes of reticulated evolution. Furthermore, the SODs with Mn or Fe cofactors found in vampirovibrionia are not related to those in their sister phyla, cyanobacteria40 (Fig. 1c, Supplementary Table 2 and Supplementary Fig. 1).
Molecular phylogenies identified several horizontal gene transfers (HGTs) of each SOD isoform between cyanobacteria and other bacterial phyla: Two of sodN (NiSODs), five of sodC, and three to eight of Mn- and Fe-utilizing SODs (Fig. 1 and Supplementary Fig. 1). A variety of different phyla are involved in these HGTs. For example, most cyanobacterial NiSODs have diversified from a protein resembling that of benthic marine Deltaproteobacteria (namely Geopsychrobacter electrodiphilus, UFBoot 88 and Plesiocystis pacifica, UFBoot 89, Supplementary Fig. 3), whereas other cyanobacterial NiSODs (present in Synechococcus spongiarum) diversified from a protein resembling those of alphaproteobacteria (UFBoot 81, Supplementary Fig. 4).
Genes encoding CuZnSOD may have been present in the shared common ancestor of all extant cyanobacteria. This ancestor gave rise to basal lineages before diverging into macrocyanobacteria and microcyanobacteria (Fig. 3). Although CuZnSODs are rare in macro- and micro–cyanobacteria, they are present in most free-living basal lineages (Fig. 3). Two in particular (Pseudanabaena spp. and Gloeobacter spp.) share sodC genes which are monophyletic (PP 1) and closely related in the same way as the species are to one another (Supplementary Fig. 5). This suggests they have been vertically inherited from the common ancestor of all extant cyanobacteria (otherwise known as the cyanobacteria crown group).
As cyanobacteria diversified to occupy new ecological niches and habitats34, the sodC genes which initially allowed crown cyanobacteria to use copper and zinc to protect against oxidative stress were likely lost. Later, HGTs may have occurred between non-cyanobacterial phyla and picocyanobacteria, diazotrophs, sponge symbionts, Acaryochloris spp. and Microcoleus chthonoplastes (Fig. 1), resulting in the distribution found today.
Bayesian relaxed molecular clock analyses
Divergence times were estimated in Phylobayes 4.145 using SSU and LSU ribosomal RNA from 164 cyanobacteria and eight vampirovibrionia. The molecular clock’s topology was constrained using an ML tree constructed in IQ-TREE v1.6.146 from these same ribosomal RNAs plus 136 core cyanobacterial proteins with similar evolutionary trajectories47. They have a range of functions in metabolism, cellular processes, and information handling34,35,48. All molecular clock analyses implemented six calibration points (Table 1) as previously described48. Divergence times were estimated using uncorrelated gamma multipliers49. All analyses, regardless of calibration points and models, indicate that cyanobacteria diverged from vampirovibrionia between 3.3 and 3.6 Ga in the Archaeaneon (Table 2). Estimates that include 95% credibility intervals allow for a range between 2.8 and 4.3 Ga, suggesting that cyanobacteria diverged from their sister phyla at at least 300 million years before the GOE (Table 2).
Discussion
The advent of oxygenic photosynthesis had profound impacts on Earth’s climate, chemistry, and biota. It destroyed a methane greenhouse which kept the world warm50, enhanced the supply of sulfate and redox-active metals to the oceans51,52, marginalized the growth of anoxygenic phototrophs53, and facilitated the chemical oxidation of dissolved Fe(II) in seawater to form banded iron formations (BIF)54. Organisms would have responded by choosing more bioavailable metals where possible and evolving more efficient enzymes, such as SODs, to deal with the expansion of O2 into new habitats34.
Age estimates for the emergence of oxygenic photosynthesis are therefore fundamental to reconciling the biochemistry and use of trace metals. Previous estimates place the divergence of cyanobacteria and their closest relatives, vampirovibrionia40,42, in the neoarchaean between 2.5 and 2.6 Ga55, but our molecular clock analyses point to an earlier origin where the cyanobacterial lineage emerges in the Paleoarchaean between 3.3 and 3.6 Ga based on mean predicted ages (95% confidence intervals range from 2.8 to 4.2 Ga) (Table 2). The difference between these estimates is owing to the choice of molecular clock model, variations in taxa selection, and alternative molecular markers of age divergence. Calibration points are also fundamental for predicting macroevolutionary patterns over long geological timescales. Our clocks are calibrated with cyanobacterial microfossils, whereas Shih, et al.55 are calibrated with non-cyanobacterial fossils. Our finding an earlier origin of cyanobacteria are thus consistent with geochemical studies showing that oxygen was accumulating in shallow coastal habitats ca. 3.0–3.2 Ga32,56,57 and molecular clock analyses of the PSII gene family which predict that photosystems capable of splitting water arose early in the history of life on Earth33,58. We also note that our estimate of 3.3–3.6 Ga is consistent with a recent survey of oxygen utilizing and producing enzymes (e.g., oxygenases) which predicts the rapid proliferation of these enzymes 3.1 Ga59; although differences arise in the dating of FeSOD and MnSOD between this study and that of ref. 59 (see below for discussion).
The action of photosystems (like some other molecules, such as transmembrane NADPH oxidases and flavins) produces O2.− 14 and when production is particularly high (owing to bright light for example), SODs protect the photosynthetic repair mechanisms from damage25,60,61. Key parts of this photosystem repair mechanism (namely membrane-bound FtsH proteases which degrade damaged photosystem components) evolved with the advent of oxygenic photosynthesis in the Archaean62. Our phylogenetic analyses suggest for the first time that CuZnSOD, an antioxidant enzyme capable of protecting photosynthetic repair mechanisms from oxidative damage25, was also present in cyanobacteria in the Archaean, at least 2.9–2.6 Ga (Table 2). This fits with alternative phylogenetic analyses predicting that sodC was present in the last universal common ancestor of life on Earth63.
Our conclusion is based on the close relationship of CuZnSODs from two basal lineages: Pseudanabaena spp. and Gloeobacter spp. (Supplementary Fig. 5). The most recent common ancestor (MRCA) of these two genera appeared ~3.4 Ga (confidence intervals span 4.2–2.8 Ga), and diversified ~2.6 Ga (confidence intervals span 3.0 to 2.5 Ga), giving rise to all extant cyanobacteria (Fig. 3). If the ancestor of crown cyanobacteria had sodC, it could have been inherited by its descendants, resulting in a unique phylogenetic signal whereby the CuZnSODs of Gloeobacter spp. and Pseudanabaena spp. are sisters. Because this phylogenetic pattern exists (Supplementary Fig. 5), it is likely that Archaean cyanobacteria used CuZnSOD. An alternative scenario could result in the same pattern if sodC was transferred laterally from the MRCA of Pseudanabaena spp. to the MRCA of Gloeobacter spp. This would put an upper age constraint on the origin of CuZnSOD ~ 0.95 Ga (confidence intervals span 2.2 to 0.3 Ga) (Fig. 3).
Although sodC has been transferred between bacterial phyla on multiple occasions in the past (Fig. 1b), our phylogenetic analyses provide less evidence of HGT between cyanobacteria of terrestrial and freshwater origin (see Supplementary discussion). As the MRCAs of Pseudanabaena and Gloeobacter spp. were not marine (Supplementary Fig. 6), HGT accompanied by a more recent origin of CuZnSOD ~ 0.95 Ga seems unlikely, if not impossible.
Further insight can be gained by timing the origin of Cu-based metalloenzymes in general. Characteristic protein-folds required to bind Cu are predicted to have evolved during, or following, the GOE37. However, geological evidence suggests that metabolisms which rely on copper metalloenzymes (e.g., nitrification), were present in the Archaean, 2.7 Ga64. If proteins could incorporate Cu cofactors in the Archaean, then CuZnSODs could have been present at the root of cyanobacteria as a mechanism to deal with O2 generated by photosynthesis before it accumulated in the global atmosphere (at the GOE). This would have been particularly important for Mesoarchaean and Neoarchaean individuals living inside benthic mats65,66, where there was a smaller diffusion gradient to pull O2 out of the cells. As a result, higher concentrations of O2 accumulated, raising the potential for superoxide radical generation, particularly in the periplasm67. In the alternative (if unlikely) scenario where basal cyanobacteria acquire sodC via HGT ~ 0.95 Gya, Archaean cyanobacteria might not have used SOD to remove O2.− because we do not find any phylogenetic evidence for NiSOD, MnSOD, or FeSOD having been inherited from an ancestor present before the GOE (Fig. 4). Two scenarios that could reconcile an earlier origin of NiSOD and Fe- or Mn-utilizing SODs with our evolutionary trees (Supplementary Figs. 7–8) are presented in the Supplementary Discussion. All rely on more extensive HGT, so age distributions are presented in Fig. 4, but in paler colors, because most prokaryotic proteins are transferred via vertical inheritance68,69,70.
Coloured bands represent the distribution of age posterior estimates (Table 2) for key nodes in a Bayesian molecular clock parameterized with uncorrelated Gamma Multipliers49 and calibrated with the first radiation of cyanobacteria between 2.32 and 2.7 Ga. Vertical black lines represent the mean in each instance. Dark-colored bands represent the earliest cyanobacteria predicted to have genes encoding the relevant SOD isoform based on a hypothesis of vertical inheritance, whereas paler nodes require horizontal gene transfer or other patterns of reticulated evolution as described in the text. Phan Phanerozoic eon; Meso Mesoproterozoic era; Neo Neoproterozoic era; GOE Great Oxygenation Event, NOE Neoproterozoic Oxygenation Event.
Why are there only a few cyanobacterial strains with the gene encoding CuZnSOD today? Perhaps it is related to physiological considerations resulting in the replacement of CuZnSOD for MnSODs. The thylakoid membranes of Nostoc sp. PCC7120 lack CuZnSOD (Fig. 3) and instead contain MnSOD30. When exposed to intense sunlight these MnSODs protect the cell from photoinhibition60 in a similar manner to CuZnSOD25. Furthermore, the gene encoding MnSOD can be post-translationally processed to produce three proteins of different sizes which vary in concentration between heterocysts and vegetative cells29. Under nitrogen-supplemented conditions, the smallest cytosolic 24 kDa protein predominates the slightly larger 27 kDa protein, but under nitrogen-limiting conditions, both proteins are present in near equal proportions29. No such flexibility has been documented in CuZnSODs. Therefore, the ability to modify the size and localization of MnSOD proteins in response to changes in the environment could explain why the sodA gene encoding MnSOD is present in all major clades of nitrogen-fixing cyanobacteria (defined by71, including all Nostocales, one Trichodesmium spp., seven unicellular diazotrophs, Chroococcidiopsis thermalis, and Leptolyngbya sp. PCC7375, Fig. 3) and many phyla of non-photosynthetic bacteria with diazotrophic representatives (e.g., Firmicutes, Actinobacteria and Proteobacteria72, Fig. 1c and Supplementary Fig. 1). Phylogenetic evidence for the utilization of SODs that incorporate Fe and Mn cofactors appears relatively recently, in the middle of the Proterozoic (Table 2, Fig. 4). Intriguingly, modern cyanobacteria might be less sensitive to oxidative stress caused by O2.− because their proteins with 4Fe-4S clusters have evolved to protect themselves by positioning their Fe atoms in less-solvent-exposed parts of the molecule. It may be with this method that picocyanobacteria are able to thrive with only a cytoplasmic NiSOD (Fig. 3).
The relatively late appearance of FeSOD and MnSOD in cyanobacteria is surprising as previous studies have postulated an Archaean origin in bacteria19,59,73,74. What caused this delay? Earth system models suggest that photoferrotrophs outcompeted cyanobacteria for upwelling nutrients in aquatic habitats prior to the GOE53,75. Perhaps, they also limited the soluble Fe2+ available for cyanobacteria, thus facilitating a selective advantage to lineages that used alternative metals for relieving oxidative stress. As the atmosphere became more oxygenated, photoferrotrophs were marginalized to shrinking pools of Fe2+. Our analyses suggest that the Neoproterozoic oxygenation increased cyanobacteria’s requirement for ROS defense mechanisms so much that lineages began using FeSOD or MnSOD regardless of the waning global concentrations of Fe and Mn (Fig. 4). Further study, however, will be needed to assess how effectively FeSODs, MnSODs, and CuZnSODs protect against oxidative stress under different ambient concentrations of oxygen and transition metals.
Phylogenetic evidence also indicates that cyanobacteria started using cytosolic NiSOD isoforms in the MRCA of Leptolyngbya sp. PCC7375 and Nodosilinea nodulosa PCC7104, which diversified in marine Neoproterozoic (Table 2, Fig. 4) habitats (Supplementary Fig. 6). Earlier, during the Paleoproterozoic and Mesoproterozoic, phylogenomic evidence suggests that cyanobacteria were living in terrestrial, coastal brackish, and marine benthic environments34,76. Therefore, NiSOD usage also corresponds with the first appearance of marine planktonic unicellular nitrogen-fixing cyanobacteria and non-nitrogen-fixing picocyanobacteria (including many Synechococcus spp. and all Prochlorococcus spp.) in the open ocean at the end of the Precambrian35,48,77,78. Although only speculative at this time, it is possible that the acquisition of NiSOD and its associated maturation protease assisted the invasion of cyanobacteria into pelagic marine habitats. Pelagic planktonic cyanobacteria are typically limited by P, Fe, and N (in cases of non-nitrogen fixers) owing to their distance from sources of riverine discharge79. As NiSOD is composed of fewer amino acids than other SOD isoforms (Supplementary Fig. 2) and does not require Fe, its utilization by benthic marine cyanobacteria in the Neoproterozoic may have imparted an evolutionary advantage and increased their resilience to nutrient limitation in the open ocean.
By studying only extant taxa, it is inherently difficult to estimate whether extinct lineages of cyanobacteria used NiSOD, MnSOD, FeSOD, or CuZnSOD prior to the estimations provided in Fig. 3, Fig. 4, and Table 2. It is reassuring, however, that our estimates of all SOD isoforms predate the Cretaceous-Paleogene mass extinction of non-avian dinosaurs from terrestrial and aquatic environments 66 Mya80, the Permian-Triassic mass extinction of 56% of all marine animal genera 252 Mya81, and the Snowball Earth glaciations 720–635 Mya, which subjected life to extreme climate fluctuations82. Therefore, our estimations of timing are robust across several past extinction events. Any future discovery of novel lineages of basal cyanobacteria, however, could alter our estimated order of SOD appearance.
It has been proposed that trace metal inventories of ancient marine sediments should reflect trace metal availability and correspond to the emergence of novel metabolisms and metalloenzymes37. In light of this suggestion, the past two decades have seen a number of papers purporting the use of ancient marine chemical sediments (i.e., BIF, shales, pyrite) to track seawater composition through time (see ref. 39 for a review). With regards to the metal cofactors contained in SOD, their temporal trajectories have been reconstructed based on both thermodynamic considerations (e.g.83) and sedimentary records (e.g.,84,85). However, there is a poor record of trace metal inventories outside of marine habitats, so it can be more difficult to establish links between metal availability and diversification of cyanobacteria living in terrestrial and freshwater habitats.
Our ancestral state reconstructions predict that a non-marine cyanobacterium could have acquired CuZnSOD (Supplementary Fig. 6) in the Archaean (Table 2). In the Archaean to Proterozoic ocean, Cu and Zn were supposed to have been present at exceedingly low abundances given the low solubility of sulfide phases and the belief that they would be sequestered into euxinic sediments after the GOE. However, BIF and shale records indicate near static reservoirs for Zn84,85 and Cu86 in the water column. This may hint at either a strong biological control on water column cycling being established very early on or the possible complexation of these trace metals by organic ligands. Although the possible release of Cu through the oxidative weathering of sulfides in the lead up to, and following, the GOE86, may explain the emergence of CuZnSOD in the Neoarchaean, at this point there is a dearth of geochemical evidence to support this hypothesis, at least from a global first-order perspective. Despite this, existing chemical sediment compilations cannot preclude the possibility of local or transient increases in Cu, creating oases of bioavailability that stimulated the emergence of CuZnSODs.
Past reconstructions of Ni in seawater based on BIF and syn-sedimentary to early diagenetic pyrite87,88,89 show a dramatic, and unidirectional, decline that immediately preceded the Great Oxidation Event. This drop has been proposed to reflect a decline in the flux of nickel to the oceans due to waning komatiite weathering and would have marginalized methanogenic microbial communities from the water column to the sediment pile, corresponding to seawater concentrations decreasing from as high as 400 nM down to effectively near modern levels of Ni (~ 9 nM)87,88. In turn, this would have created environmentally favorable conditions for a rise in oxygenic photosynthesis by cyanobacteria. This Neoarchaean to Paleoproterozoic decline in Ni is also reflected in a compilation of shale data, normalized to evolving upper continental crust (Fig. 5). Interestingly, however, there appears to be a slight uptick in the flux of Ni from continental weathering to the oceans in the terminal Neoproterozoic to Phanerozoic as evidenced by elevated Ni/Ti ratios (Fig. 5) that occurs shortly after the appearance of NiSOD in the Tonian period (Table 2), which is not captured in the BIF record87,88. This discrepancy in sedimentary records is likely due to the declining concentration of Fe in seawater through time as Earth’s oceans moved from a dominantly anoxic and ferruginous state in the Archaean to the modern oxic oceans we have today; a trend perhaps best reflected in the disappearance of large superior-type BIF following the Paleoproterozoic (e.g.,90).
Each group represents a time-binned subset of the shale database representing the Archaean (blue), Paleoproterozoic (orange), Mesoproterozoic (yellow), Neoproterozoic (purple), or Phanerozoic (green) with mean values of molar Ni/Ti, normalized to evolving crust116, being bootstrap resampled (n = 10,000 for each bin).
Elevated Ni/Ti ratios during the Neoproterozoic presumably reflects a relatively greater Ni flux to the oceans from either increased oxidative weathering, similar to other sulfide bound metals such as molybdenum or vanadium at this time52,91, or through the weathering of freshly emplaced large igneous provinces as has been proposed for phosphorous92. At present, the exact mechanisms and timing of higher Ni concentrations in Neoproterozoic shales require further investigation as it lags behind the observed increases in molybdenum, vanadium, and phosphorus52,91,92,93.
Given the histories of these trace metals, one may ask whether it is availability driving the assimilation of Cu, Zn, Fe, Mn, and Ni into SOD metalloenzymes, or an inherent competition with other extant lineages at any given time. For instance, metalloenzyme fold superfamilies that incorporate Zn proliferate late and have been proposed to contribute to the delay in eukaryotic evolution37. Alternatively, this has been proposed to reflect an intrinsic biological property of eukaryotic evolution rather than a sudden shift in Zn availability84. In the latter view, the early incorporation of Cu and Zn into SODs would reflect a lack of competition from the yet unevolved eukaryotes in the Archaean. Then in the late Mesoproterozoic, as eukaryotes began to become more abundant and dominate marine and terrestrial environments, they may have outcompeted cyanobacteria for Cu and Zn, providing an impetus for the emergence of other membrane-bound SODs, such as MnSOD—effectively providing a strategy for alleviating limitation imparted by competition with emergent eukaryotes. Although eukaryotes also utilize Mn, protein structures required to bind Mn have not been preferentially retained in their genomes94 and cyanobacteria, having evolved earlier than photosynthetic eukaryotes95, had plenty of time to develop an efficient Mn uptake system.
Later, in Neoproterozoic marine habitats, cyanobacteria began supplementing their ROS defense mechanisms with a new and smaller SOD isoform (NiSOD) that requires less phosphorus and nitrogen to manufacture. This timing of sodN incorporation into the cyanobacterial genomic repertoire coincides with Earth’s second planetary oxygenation3 and the emergence of planktonic marine cyanobacteria35. Therefore, we suggest that cyanobacteria began assimilating nickel into SODs to invade Neoproterozoic planktonic communities as Neoproterozoic oxygen levels rose.
Methods
Acquisition of SOD sequences
Bacterial SOD diversity was assessed by downloading 15,899 bacterial genomes from the NCBI RefSeq database (https://www.ncbi.nlm.nih.gov/refseq/) in 2018. They were searched for genes encoding NiSOD, CuZnSOD, FeSOD, and MnSOD using the basic local alignment search tool (BLAST) for proteins (https://blast.ncbi.nlm.nih.gov/Blast.cgi) with a word size of six, gap opening penalty of 11, gap extension penalty of 1, and e value cut off of <1 × 10−5. Query sequences are detailed in Supplementary Table 3. They were aligned to relevant subject sequences using the BLOSUM62 substitution matrix. Each genome was treated as a separate subject (functionally equivalent to a unique database) and if multiple homologs of a single SOD isoform were found in a given genome, only one with the highest bitscore was retained, except for the cyanobacteria and vampirovibrionia where all hits were included (Supplementary Data 2). The resulting hit’s functions were verified by choosing a representative subsample of hits with distant evolutionary relationships and noting functional domains predicted by the NCBI conserved domain search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Supplementary Data 3 and Supplementary Fig. 9). If possible, query sequences with experimentally defined functions were utilized.
Cyanobacterial SOD diversity was assessed by sampling 153 additional genomes (strains are listed in Supplementary Data 1 and their habitat in Supplementary Table 4), including representatives from all major clades of salt-tolerant and freshwater cyanobacteria35. These were supplemented with five eukaryotic plastid genomes95 and eight vampirovibrionial genomes representing all four orders (Gastranaerophilales, Vampirovibrionales, Caenarcaniphilales, and Obscuribacterales)40.
SOD gene phylogenies
Amino-acid sequences for all SOD isoforms (including paralogs found in cyanobacteria) were aligned using MAFFT96 and gaps present in ≥80% of sequences removed. The evolutionary history of each isoform was estimated by constructing nine ML phylogenies with IQ-TREE version 1.6.146. Substitution models were identified by ModelFinder97 and node supports measured using ultrafast bootstrap two approximations98.
Species phylogeny of cyanobacteria
An evolutionary tree of cyanobacteria was generated from a data set including SSU rRNA (1687 nucleotides), LSU rRNA (3387 nucleotides), and 136 core proteins (52,227 aa). Each protein is encoded by an orthologous gene, which is conserved among cyanobacteria and involved in a key cellular function, such as information processing, metabolism, or cellular processes. For a full description of these genes, see previous papers34,35,77. Each protein and ribosomal RNA were aligned using MAFFT96 and concatenated using an alignment viewer (http://sdsssdfd.altervista.org/arklumpus/AlignmentViewer/AlignmentViewer.html). Gaps were removed if present at the same position in ≥80% sequences and ML methodology was implemented in IQ-TREE v1.6.146 to estimate the cyanobacterial phylogeny. Each protein and rRNA was characterized under a unique partition with -SP to account for heterotachy and appropriate substitution models identified by ModelFinder97. Ultrafast bootstrap values were calculated to measure support for branching relationships by resampling sites within partitions 1000 times98.
Divergence time estimation
Bayesian relaxed molecular clocks were implemented using the topology described above. This topology was fixed, and ages estimated based on predicted mutation rates for ribosomal RNA (SSU and LSU, 5074 aligned positions). Substitutions were modeled using a flexible general time-reversible model inferred from the alignment and a Dirichlet process prior99 (CAT-GTR) to account for different rates of evolution between distant sites of the molecule. Divergence times were estimated in phylobayes 4.145 using uncorrelated gamma multipliers,49, a birth-death prior on divergence times, and root prior chosen from a Gamma probability distribution with mean 3060 and standard deviation 404. As a result, 97% of the prior distribution specified that the ancestor of all vampirovibrionia and cyanobacteria originated after the end of the late heavy bombardment 3.9 Ga100,101.
We also implemented microfossil calibrations as follows; filamentous cyanobacteria >1.9 Ga102,103, akinete-forming cyanobacteria 1.6–1.888 Ga102,104,105, and apical cells of cyanobacteria 1.7–1.888 Ga102,104,106. These fossil constraints were supplemented with evidence dating the appearance of eukaryotic hosts of endosymbiotic cyanobacteria. For example, Richelia species diversify before their diatom host, named Hemiaulus107, appeared 110 Mya108, and UCYNA species diversify before their prymnesiophyte host, named Braarudosphaera bigelowii, appeared 91 Mya109. Geochemical evidence was used to constrain cyanobacteria to first diversify before the Great Oxygenation Event of 2.32 Ga110, but after either; (a) 3 Ga when molybdenum oxides document the first “whiffs” of atmospheric oxygen56; or (b) 2.7 Ga based on the earliest fossilized evidence of cyanobacterial stromatolites111. Soft bounds were applied throughout to allow 5% of the prior probability density to fall outside of the specified minimum and maximum ages.
Models were considered complete when four replicate independent chains converged. This was tested by estimating effective sample sizes and relative differences using Tracecomp (in Phylobayes with effsize > 50, reldiff < 0.3). Chronograms and divergence times were calculated from a single representative chain using Readdiv (in Phylobayes) with the same burn-in (25% of mean chain length) and sampling frequency (1 in every 10 points) as Tracecomp.
Chronology of SOD isoforms
The order in which SOD isoforms appeared in cyanobacteria, was estimated using a method described as “topological comparison”112. First, a Bayesian protein phylogeny was created for each SOD isoform using only cyanobacterial sequences (Supplementary Figs. 5 and 7). Bayesian phylogenetic reconstructions of MnSODs and FeSODs had not converged after 2 weeks, so the sequences were separated into seven groups based on their position in the ML bacterial phylogenies (Supplementary Fig. 1) to conduct alignments and phylogenetic reconstructions on more similar sequences (Supplementary Fig. 8). The resulting protein phylogenies were then compared with the species phylogeny of cyanobacteria (Fig. 3) to identify monophyletic groups whose NiSOD, CuZnSOD, or MnSOD/FeSOD had evolved as expected by vertical inheritance (without HGT). The last common ancestor of each of these clades was assumed to have been capable of using the corresponding SOD.
Ancestral state reconstruction of habitat preference
To find out which habitats ancestral cyanobacteria lived in when SOD isoforms appeared, Bayesian stochastic character mapping113 was implemented with SIMMAP v1.5114 in the phytools package115 of R using our time-calibrated trees. Prior distribution on the root node of the tree was estimated based on the data, 1000 simulations were performed and the all rates different model was utilized to allow transition rates between marine and non-marine habitats to vary based on the data (as implemented in95). Character states were coded as either “marine” or “non-marine” (Supplementary Data 1).
Compilation of Ni data
From a database of >4000 literature shale analyses spanning the Archaean to modern, 1584 had both Ni and Ti data available and are used here to reconstruct the trajectory of Ni in seawater (Supplementary Data 4). Molar Ni/Ti ratios were normalized to evolving continental crust116 then time-binned based on age (Archaean, Paleoproterozoic, Mesoproterozoic, Neoproterozoic, Phanerozoic). This normalization accounts for secular changes in the composition of Earth’s continental crust that reflect the emergence, growth, and subsequent differentiation of continental crust from the Archaean through the Phanerozoic. The evolution of continental crust influences the relative availability of trace and major elements weathered from land to the oceans, and thus the normalization removes this temporal variability, facilitating a comparison of Ni/Ti ratios in shales through time. Mean values for each time bin were bootstrap resampled (n = 10,000; Fig. 5) in Matlab® 2019b using the bootstrap function.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The sequence data analyzed in this study and accession numbers of genomes from the NCBI RefSeq database (https://www.ncbi.nlm.nih.gov/refseq/) are available in the open science framework repository, https://osf.io/yj7qb/?view_only=cc861929817e4913a7795cde10ae64fc.
References
Wheelis, M. Principles of Modern Microbiology. 140–167 (Jones and Bartlett Publishers, London, 2008).
Kump, L. R. The rise of atmospheric oxygen. Nature 451, 277–278 (2008).
Lyons, T. W., Reinhard, C. T. & Planavsky, N. J. The rise of oxygen in Earth’s early ocean and atmosphere. Nature 506, 307–315 (2014).
Oliver, T., Sanchez-Baracaldo, P., Larkum, A. W., Rutherford, A. W. & Cardona, T. Time-resolved comparative molecular evolution of oxygenic photosynthesis. Biochim. Biophys. Acta Bioenerg. 1862, 148400 (2021).
Rosing, M. T. & Frei, R. U-rich Archaean sea-floor sediments from Greenland - indications of >3700 Ma oxygenic photosynthesis. Earth Planet. Sci. Lett. 217, 237–244 (2004).
Fischer, W. W., Hemp, J. & Johnson, J. E. Evolution of oxygenic photosynthesis. Annu. Rev. Earth Planet. Sci. 44, 647–683 (2016).
Konhauser, K. O. et al. Aerobic bacterial pyrite oxidation and acid rock drainage during the Great Oxidation Event. Nature 478, 369–374 (2011).
Warke, M. R. et al. The Great Oxidation Event preceded a Paleoproterozoic “snowball Earth”. Proc. Natl Acad. Sci. USA 117, 13314–13320 (2020).
Lu, Z., Chang, Y. C., Yin, Q. & Jackson, W. M. Evidence for direct molecular oxygen production in CO2 photodissociation. Science 346, 61–64 (2014).
Meadows, V. S. Reflections on O2 as a biosignature in exoplanetary atmospheres. Astrobiology 17, 1022–1052 (2017).
Orf, G. S., Gisriel, C. & Redding, K. E. Evolution of photosynthetic reaction centers: insights from the structure of the heliobacterial reaction center. Photosynth. Res. 138, 11–37 (2018).
Latifi, A., Ruiz, M. & Zhang, C. C. Oxidative stress in cyanobacteria. FEMS Microbiol. Rev. 33, 258–278 (2009).
Bernroitner, M., Zamocky, M., Furtmuller, P. G., Peschek, G. A. & Obinger, C. Occurrence, phylogeny, structure, and function of catalases and peroxidases in cyanobacteria. J. Exp. Bot. 60, 423–440 (2009).
Sheng, Y. et al. Superoxide dismutases and superoxide reductases. Chem. Rev. 114, 3854–3918 (2014).
Diaz, J. M. & Plummer, S. Production of extracellular reactive oxygen species by phytoplankton: past and future directions. J. Plankton Res. 40, 655–666 (2018).
Rose, A. L. The influence of extracellular superoxide on iron redox chemistry and bioavailability to aquatic microorganisms. Front. Microbiol. 3, 124 (2012).
Wallace, M. A. et al. Superoxide inhibits 4Fe-4S cluster enzymes involved in amino acid biosynthesis. Cross-compartment protection by CuZn-superoxide dismutase. J. Biol. Chem. 279, 32055–32062 (2004).
Vassiliev, I. R., Ronan, M. T., Hauska, G. & Golbeck, J. H. The bound electron acceptors in green sulfur bacteria: Resolution of the g-tensor for the F(X) iron-sulfur cluster in Chlorobium tepidum. Biophys. J. 78, 3160–3169 (2000).
Miller, A. F. Superoxide dismutases: ancient enzymes and new insights. FEBS Lett. 586, 585–595 (2012).
Perry, J. J., Shin, D. S., Getzoff, E. D. & Tainer, J. A. The structural biochemistry of the superoxide dismutases. BBA Proteins Proteom. 1804, 245–262 (2010).
Wolfe-Simon, F., Grzebyk, D., Schofield, O. & Falkowski, P. G. The role and evolution of superoxide dismutases in algae. J. Phycol. 41, 453–465 (2005).
Priya, B. et al. Comparative analysis of cyanobacterial superoxide dismutases to discriminate canonical forms. BMC Genomics 8, 435 (2007).
Dupont, C. L., Neupane, K., Shearer, J. & Palenik, B. Diversity, function and evolution of genes coding for putative Ni-containing superoxide dismutases. Environ. Microbiol. 10, 1831–1843 (2008).
Caiola, M. G., Canini, A. & Ocampo-Friedmann, R. Iron superoxide dismutase (Fe-SOD) localization in Chroococcidiopsis sp (Chroococcales). Phycologia 35, 90–94 (1996).
Ke, W. T., Dai, G. Z., Jiang, H. B., Zhang, R. & Qiu, B. S. Essential roles of iron superoxide dismutase in photoautotrophic growth of Synechocystis sp. PCC 6803 and heterogeneous expression of marine Synechococcus sp. CC9311 copper/zinc superoxide dismutase within its sodB knockdown mutant. Microbiol 160, 228–241 (2014).
Regelsberger, G. et al. The iron superoxide dismutase from the filamentous cyanobacterium Nostoc PCC 7120. Localization, overexpression, and biochemical characterization. J. Biol. Chem. 279, 44384–44393 (2004).
Okada, S., Kanematsu, S. & Asada, K. Intracellular distribution of manganese and ferric superoxide dismutases in blue-green algae. FEBS Lett. 103, 106–110 (1979).
Regelsberger, G. et al. Biochemical characterization of a membrane-bound manganese-containing superoxide dismutase from the cyanobacterium Anabaena PCC 7120. J. Biol. Chem. 277, 43615–43622 (2002).
Raghavan, P. S., Rajaram, H. & Apte, S. K. Membrane targeting of MnSOD is essential for oxidative stress tolerance of nitrogen-fixing cultures of Anabaena sp. strain PCC7120. Plant Mol. Biol. 88, 503–514 (2015).
Li, T. et al. Differential expression and localization of Mn and Fe superoxide dismutases in the heterocystous cyanobacterium Anabaena sp strain PCC 7120. J. Bacteriol. 184, 5096–5103 (2002).
Javaux, E. J. Challenges in evidencing the earliest traces of life. Nature 572, 451–460 (2019).
Homann, M. Earliest life on earth: evidence from the Barberton Greenstone Belt, South Africa. Earth-Sci. Rev. 196, 102888 (2019).
Cardona, T., Sanchez-Baracaldo, P., Rutherford, A. W. & Larkum, A. W. Early Archean origin of Photosystem II. Geobiology 17, 127–150 (2019).
Blank, C. E. & Sanchez-Baracaldo, P. Timing of morphological and ecological innovations in the cyanobacteria - a key to understanding the rise in atmospheric oxygen. Geobiology 8, 1–23 (2010).
Sanchez-Baracaldo, P. Origin of marine planktonic cyanobacteria. Sci. Rep. 5, 17418 (2015).
Schmidt, A., Gube, M., Schmidt, A. & Kothe, E. In silico analysis of nickel containing superoxide dismutase evolution and regulation. J. Basic Microbiol. 49, 109–118 (2009).
Dupont, C. L., Butcher, A., Valas, R. E., Bourne, P. E. & Caetano-Anolles, G. History of biological metal utilization inferred through phylogenomic analysis of protein structures. Proc. Natl Acad. Sci. USA 107, 10567–10572 (2010).
Sigel, H. & Sigel, R. Metal Ions in Biological Systems. 1st edn, Vol. 43 (Taylor & Francis Group, 2005).
Robbins, L. J. et al. Trace elements at the intersection of marine biological and geochemical evolution. Earth-Sci. Rev. 163, 323–348 (2016).
Soo, R. M. et al. An expanded genomic representation of the phylum. Genome Biol. Evol. 6, 1031–1045 (2014).
Di Rienzi, S. C. et al. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to cyanobacteria. Elife 2, e01102 (2013).
Eitinger, T. In vivo production of active nickel superoxide dismutase from Prochlorococcus marinus MIT9313 is dependent on its cognate peptidase. J. Bacteriol. 186, 7821–7825 (2004).
Barondeau, D. P., Kassmann, C. J., Bruns, C. K., Tainer, J. A. & Getzoff, E. D. Nickel superoxide dismutase structure and mechanism. Biochemistry 43, 8038–8047 (2004).
Simao, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Lartillot, N., Lepage, T. & Blanquart, S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25, 2286–2288 (2009).
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A. & Minh, B. Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235 (2016).
Shi, T. & Falkowski, P. G. Genome evolution in cyanobacteria: the stable core and the variable shell. Proc. Natl Acad. Sci. USA 105, 2510–2515 (2008).
Sanchez-Baracaldo, P., Bianchini, G., Di Cesare, A., Callieri, C. & Chrismas, N. A. M. Insights into the evolution of picocyanobacteria and phycoerythrin genes (mpeBA and cpeBA). Front. Microbiol. 10, 45 (2019).
Drummond, A. J., Ho, S. Y. W., Phillips, M. J. & Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, 699–710 (2006).
Kopp, R. E., Kirschvink, J. L., Hilburn, I. A. & Nash, C. Z. The Paleoproterozoic snowball earth: a climate disaster triggered by the evolution of oxygenic photosynthesis. Proc. Natl Acad. Sci. USA 102, 11131–11136 (2005).
Reinhard, C. T., Raiswell, R., Scott, C., Anbar, A. D. & Lyons, T. W. A late Archean sulfidic sea stimulated by early oxidative weathering of the continents. Science 326, 713–716 (2009).
Scott, C. et al. Tracing the stepwise oxygenation of the Proterozoic ocean. Nature 452, 456–459 (2008).
Ozaki, K., Thompson, K. J., Simister, R. L., Crowe, S. A. & Reinhard, C. T. Anoxygenic photosynthesis and the delayed oxygenation of Earth’s atmosphere. Nat. Commun. 10, 3026 (2019).
Schad, M., Konhauser, K. O., Sanchez-Baracaldo, P., Kappler, A. & Bryce, C. How did the evolution of oxygenic photosynthesis influence the temporal and spatial development of the microbial iron cycle on ancient earth? Free Radic. Biol. Med. 140, 154–166 (2019).
Shih, P. M., Hemp, J., Ward, L. M., Matzke, N. J. & Fischer, W. W. Crown group oxyphotobacteria postdate the rise of oxygen. Geobiology 15, 19–29 (2017).
Planavsky, N. J. et al. Evidence for oxygenic photosynthesis half a billiion years before the Great Oxidation Event. Nat. Geosci. 7, 283–286 (2014).
Satkoski, A. M., Beukes, N. J., Weiqiang, L., Beard, B. L. & Johnson, C. M. A redox-stratified ocean 3.2 billion years ago. Earth Planet. Sci. Lett. 430, 43–53 (2015).
Louca, S. et al. Bacterial diversification through geological time. Nat. Ecol. Evol. 2, 1458–1467 (2018).
Jabłońska, J. & Tawfik, D. S. The evolution of oxygen-utilizing enzymes suggests early biosphere oxygenation. Nat. Ecol. Evol. 5, 442–448 (2021).
Zhao, W. X., Guo, Q. X. & Zhao, J. D. A membrane-associated Mn-superoxide dismutase protects the photosynthetic apparatus and nitrogenase from oxidative damage in the cyanobacterium Anabaena sp. PCC 7120. Plant Cell Physiol. 48, 563–572 (2007).
Sae-Tang, P. et al. Overexpressed superoxide dismutase and catalase act synergistically to protect the repair of PSII during photoinhibition in Synechococcus elongatus PCC 7942. Plant Cell Physiol. 57, 1899–1907 (2016).
Shao, S., Cardona, T. & Nixon, P. J. Early emergence of the FtsH proteases involved in photosystem II repair. Photosynthetica 56, 163–177 (2018).
Weiss, M. C. et al. The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 1, 16116 (2016).
Moore, E. K., Jelen, B. I., Giovannelli, D., Raanan, H. & Falkowski, P. G. Metal availability and the expanding network of microbial metabolisms in the Archaean eon. Nat. Geosci. 10, 629–636 (2017).
Lalonde, S. V. & Konhauser, K. O. Benthic perspective on Earth’s oldest evidence for oxygenic photosynthesis. Proc. Natl Acad. Sci. USA 112, 995–1000 (2015).
Wilmeth, D. T. et al. Neoarchean (2.7 Ga) lacustrine stromatolite deposits in the Hartbeesfontein Basin, Ventersdorp Supergroup, South Africa: Implications for oxygen oases. Precambrian Res. 320, 291–302 (2019).
Korshunov, S. & Imlay, J. A. Detection and quantification of superoxide formed within the periplasm of Escherichia coli. J. Bacteriol. 188, 6326–6334 (2006).
Kunin, V., Goldovsky, L., Darzentas, N. & Ouzounis, C. A. The net of life: reconstructing the microbial phylogenetic network. Genome Res 15, 954–959 (2005).
Beiko, R. G., Harlow, T. J. & Ragan, M. A. Highways of gene sharing in prokaryotes. Proc. Natl Acad. Sci. USA 102, 14332–14337 (2005).
Novichkov, P. S. et al. Genome-wide molecular clock and horizontal gene transfer in bacterial evolution. J. Bacteriol. 186, 6575–6585 (2004).
Sanchez-Baracaldo, P., Hayes, P. K. & Blank, C. E. Morphological and habitat evolution in the cyanobacteria using a compartmentalization approach. Geobiology 3, 145–165 (2005).
Hartmann, L. S. & Barnum, S. R. Inferring the evolutionary history of Mo-dependent nitrogen fixation from phylogenetic studies of nifK and nifDK. J. Mol. Evol. 71, 70–85 (2010).
Inupakutika, M. A., Sengupta, S., Devireddy, A. R., Azad, R. K. & Mittler, R. The evolution of reactive oxygen species metabolism. J. Exp. Bot. 67, 5933–5943 (2016).
Kirschvink, J. L. et al. Paleoproterozoic snowball earth: Extreme climatic and geochemical global change and its biological consequences. Proc. Natl Acad. Sci. USA 97, 1400–1405 (2000).
Jones, A. C., Nomosatryo, S., Crowe, S. A., Bjerrum, C. & Canfield, D. Iron oxides, divalent cations, silica and the early earth phosphorus crisis. Geology 43, 135–138 (2015).
Raven, J. A., Beardall, J., Larkum, A. W. D. & Sanchez-Baracaldo, P. Interactions of photosynthesis with genome size and function. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 368, 20120264 (2013).
Sanchez-Baracaldo, P. & Cardona, T. On the origin of oxygenic photosynthesis and cyanobacteria. N. Phytol. 225, 1440–1446 (2020).
Sanchez-Baracaldo, P., Ridgwell, A. & Raven, J. A. A Neoproterozoic transition in the marine nitrogen cycle. Curr. Biol. 24, 652–657 (2014).
Bristow, L. A., Mohr, W., Ahmerkamp, S. & Kuypers, M. M. M. Nutrients that limit growth in the ocean. Curr. Biol. 27, R474–R478 (2017).
Brusatte, S. L. et al. The extinction of the dinosaurs. Biol. Rev. 90, 628–642 (2015).
Bambach, R. K. Phanerozoic biodiversity mass extinctions. Annu. Rev. Earth Planet. Sci. 34, 127–155 (2006).
Hoffman, P. F. et al. Snowball earth climate dynamics and cryogenian geology-geobiology. Sci. Adv. 3, e1600983 (2017).
Saito, M. A., Sigman, D. M. & Morel, F. M. M. The bioinorganic chemistry of the ancient ocean: the co-evolution of cyanobacterial metal requirements and biogeochemical cycles at the archean-proterozoic boundary? Inorg. Chim. Acta 356, 308–318 (2003).
Robbins, L. J. et al. Authigenic iron oxide proxies for marine zinc over geological time and implications for eukaryotic metallome evolution. Geobiology 11, 295–306 (2013).
Scott, C. et al. Bioavailability of zinc in marine systems through time. Nat. Geosci. 6, 125–128 (2013).
Fru, E. C. et al. Cu isotopes in marine black shales record the Great Oxidation Event. Proc. Natl Acad. Sci. USA 113, 4941–4946 (2016).
Konhauser, K. O. et al. Oceanic nickel depletion and a methanogen famine before the Great Oxidation Event. Nature 458, U750–U785 (2009).
Konhauser, K. O. et al. The Archean nickel famine revisited. Astrobiology 15, 804–815 (2015).
Large, R. R. et al. Trace element content of sedimentary pyrite as a new proxy for deep-time ocean–atmosphere evolution. Earth Planet. Sci. Lett. 389, 209–220 (2014).
Konhauser, K. O. et al. Iron formations: a global record of Neoarchaean to Palaeoproterozoic environmental history. Earth-Sci. Rev. 172, 140–177 (2017).
Sahoo, S. K. et al. Ocean oxygenation in the wake of the Marinoan glaciation. Nature 489, 546–549 (2012).
Horton, F. Did phosphorus derived from the weathering of large igneous provinces fertilize the Neoproterozoic ocean? Geochem. Geophy. Geosy. 16, 1723–1738 (2015).
Reinhard, C. T. et al. Evolution of the global phosphorus cycle. Nature 541, 386–389 (2017).
Dupont, C. L., Yang, S., Palenik, B. & Bourne, P. E. Modern proteomes contain putative imprints of ancient shifts in trace metal geochemistry. Proc. Natl Acad. Sci. USA 103, 17822–17827 (2006).
Sanchez-Baracaldo, P., Raven, J. A., Pisani, D. & Knoll, A. H. Early photosynthetic eukaryotes inhabited low-salinity habitats. Proc. Natl Acad. Sci. USA 114, E7737–E7745 (2017).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–591 (2017).
Hoang, D. T. C., O., Haeseler, A.V. Minh, B.Q. Vinh,L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2017).
Lartillot, N. & Philippe, H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21, 1095–1109 (2004).
Sleep, N. H., Zahnle, K. J., Kasting, J. F. & Morowitz, H. J. Annihilation of ecosystems by large asteroid impacts on the early Earth. Nature 342, 139–142 (1989).
Zahnle, K. et al. Emergence of a habitable planet. Space Sci. Rev. 129, 35–78 (2007).
Golubic, S. & Lee, S. J. Early cyanobacterial fossil record: preservation, palaeoenvironments and identification. Eur. J. Phycol. 34, 339–348 (1999).
Hofman, H. J. Precambrian microflora, belcher islands, canada: significance and systematics. J. Paleontol. 50, 1040–1073 (1976).
Sergeev, V. N., Gerasimenko, L. M. & Zavarzin, G. A. The Proterozoic history and present state of cyanobacteria. Microbiology 71, 623–637 (2002).
Golubic, S., Sergeev, V. N. & Knoll, A. H. Mesoproterozoic archaeoellipsoides: akinetes of heterocystous cyanobacteria. Lethaia 28, 285–298 (1995).
Zhang, Y. & Golubic, S. Endolithic microfossils (cyanophyta) from early proterozoic stromatolites, Hebei, China. Acta Micropaleontol. Sin. 4, 1–12 (1987).
Foster, R. A. et al. Nitrogen fixation and transfer in open ocean diatom-cyanobacterial symbioses. ISME J. 5, 1484–1493 (2011).
Sims, A. P., Mann, D. G. & Medlin, L. K. Evolution of the diatoms: insights from fossil, biological and molecular data. Phycologia 45, 361–402 (2006).
Cornejo-Castillo, F. M. et al. Cyanobacterial symbionts diverged in the late Cretaceous towards lineage-specific nitrogen fixation factories in single-celled phytoplankton. Nat. Commun. 7, 11071 (2016).
Bekker, A. et al. Dating the rise of atmospheric oxygen. Geochim. Cosmochim. Acta 68, A780–A780 (2004).
Bosak, T., Liang, B., Sim, M. S. & Petroff, A. P. Morphological record of oxygenic photosynthesis in conical stromatolites. Proc. Natl Acad. Sci. USA 106, 10939–10943 (2009).
Chan, C. X., Beiko, R. G. & Ragan, M. A. Scaling up the phylogenetic detection of lateral gene transfer events. Methods Mol. Biol. 1525, 421–432 (2017).
Huelsenbeck, J. P., Nielsen, R. & Bollback, J. P. Stochastic mapping of morphological characters. Syst. Biol. 52, 131–158 (2003).
Bollback, J. P. SIMMAP: Stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics 7, 88 (2006).
Revell, L. J. Phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012).
Condie, K. C. Chemical-composition and evolution of the upper continental-crust - contrasting results from surface samples and shales. Chem. Geol. 104, 1–37 (1993).
Acknowledgements
We thank Andrew H. Knoll, Paul J. Valdes, John Raven, and Paul Falkowski for helpful comments on the manuscript. We also thank Giorgio Bianchini for technical help in designing scripts for automated BLASTP and leaf coloring as well as developing a trimming tool for alignments. All phylogenetic analyses were performed at high-performance computing facilities (BlueCrystal 3 and 4) at the University of Bristol. Funding support for this work came from a Royal Society University Research Fellowship to P.S.-B. and a University of Bristol Graduate Teaching Scholarship to J.S.B. L.J.R. gratefully acknowledges the support of a Donnelley Environmental Postdoctoral Fellowship from the Yale Institute for Biospheric Studies.
Author information
Authors and Affiliations
Contributions
J.S.B. and P.S-B. conceived the project; J.S.B. and P.S-B. designed the phylogenetic and molecular clocks analyses, J.S.B. performed analyses, J.S.B. and P.S-B. interpreted evolutionary results; K.O.K and L.J.R. interpreted the Ni shale records; J.S.B., K.O.K, L.J.R., and P.S-B. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Communications thanks Luisa Falcón, Kevin Sutherland, and the other, anonymous, reviewer for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Boden, J.S., Konhauser, K.O., Robbins, L.J. et al. Timing the evolution of antioxidant enzymes in cyanobacteria. Nat Commun 12, 4742 (2021). https://doi.org/10.1038/s41467-021-24396-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-021-24396-y
This article is cited by
-
Replicated life-history patterns and subsurface origins of the bacterial sister phyla Nitrospirota and Nitrospinota
The ISME Journal (2023)
-
Globally distributed Myxococcota with photosynthesis gene clusters illuminate the origin and evolution of a potentially chimeric lifestyle
Nature Communications (2023)
-
Extreme summers impact cropland and grassland soil microbiomes
The ISME Journal (2023)
-
Pb toxicity at initial level is managed by Desmonostoc muscorum PUPCCC 405.10 by activating antioxidant defense system
Biologia (2023)
-
Distribution and diversity of ROS-generating enzymes across the animal kingdom, with a focus on sponges (Porifera)
BMC Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.