Abstract
Marine species exhibiting wide distributional ranges are frequently subdivided into discrete genetic units over limited spatial scales. This is often due to specific life-history traits or oceanographic barriers that prevent gene flow. Fine-scale sampling studies revealed distinct phylogeographic patterns in the northeastern Atlantic and the Mediterranean, ranging from panmixia to noticeable population genetic structure. Here, we used mitochondrial sequence data to analyse connectivity in the bogue Boops boops throughout most of its widespread distribution. Our results identified the existence of three clades, one comprising specimens from the Azores and eastern Atlantic/Mediterranean, another with individuals from the Canary Islands, Madeira and Cape Verde archipelagos, and the third with samples from Mauritania only. One of the branches of the northern subtropical gyre (Azores Current) that drifts towards the Gulf of Cádiz promotes a closer connection between the Azores, southern Portugal and the Mediterranean B. boops populations. The Almería-Oran Front, widely recognised as an oceanographic barrier for many organisms to cross the Atlantic-Mediterranean divide, does not seem to affect the dispersal of this benthopelagic species. The southward movement of the Cape Verde Frontal Zone during the winter, combined with the relatively short duration of the pelagic larval stage of B. boops, may be potential factors for preventing the connectivity between the Atlantic oceanic archipelagos and Mauritania shaping the genetic signature of this species.
Similar content being viewed by others
Introduction
The existence of population genetic structure in the marine realm is often considered paradoxical given the apparent absence of physical barriers1. Nonetheless, an increasing number of studies reported significant population diversification over limited spatial scales in species exhibiting large geographic ranges due to various processes2. Among those, life-history traits associated with limited dispersal abilities, such as determined by short pelagic larval durations (PLD)3 or oceanographic barriers that avert gene flow4,5, may play an essential role in shaping the genetic structure of the species. Studies performing meta-analyses and fine-scale sampling revealed the existence of distinct phylogeographic patterns in the northeastern Atlantic and Mediterranean. These patterns range from fish species displaying noticeable population structure6 to others exhibiting panmixia throughout their geographic ranges7. Historical events and oceanographic patterns generate species-specific responses8.
The bogue Boops boops (Linnaeus, 1758) (family Sparidae) is a gregarious, benthopelagic fish species inhabiting depths between 0 and 350 m9. This coastal species is broadly distributed in the Mediterranean and eastern Atlantic (from Norway to Angola), including the oceanic archipelagos at these latitudes10. It represents a vital fishery resource, particularly in the eastern Mediterranean11, inhabiting diverse habitats, including rocky substrates, sandy bare seabeds, seaweeds and seagrass meadows12. Boops boops have a PLD of 16 to 18 days and exhibit a vertical pattern of larval assemblages: smaller larvae occur at the surface while larger individuals are more frequently found at the bottom13. During the summer, groups of adult individuals approach shallow coastal waters, while in winter migrate to waters deeper than 100 m14. Until now, studies on the bogue have focused on biological aspects15,16,17 or on the species' impact on fish community assemblages and aquaculture18,19. Defining the spatial structure of intraspecific genetic diversity is paramount for fisheries stock assessment20 and understanding how oceanographic features affect population connectivity.
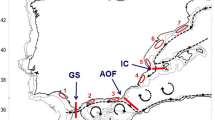
In the present study, we used the mitochondrial control region (D-loop) to infer genetic connectivity in B. boops throughout most of its geographic range in the eastern Atlantic, from the Bay of Biscay to Mauritania, including the archipelagos of Madeira, Azores, Canary Islands and Cape Verde (Fig. 1). The species is distributed across some oceanographic barriers including, e.g. the Almería-Oran Front in the Atlantic-Mediterranean transition zone21. To analyse the putative effect of this oceanographic barrier on the genetic structure of the species, we also included several sampling locations in the Mediterranean (Algeria, Tunisia, Lebanon, Croatia, Spain and Italy).
Sampling sites across the distributional range of Boops boops in the Northeastern Atlantic and the Mediterranean. Location codes in Table 1. Circles size proportional to samples sizes. Figure generated using the worldHires (http://CRAN.R-project.org/package=mapdata) function implemented in R language24. URL https://www.R-project.org/) (version 3.3.1), which uses publicly available coastline coordinates from the NOAA National Geophysical Data Center (http://www.ngdc.noaa.gov/mgg/shorelines/shorelines.html).
Materials and methods
Sampling collection
Collections of B. boops tissue (n = 335) were obtained from several locations across the distributional range of the species in the Atlantic and Mediterranean: Bouharoun, Algeria (ARG); Azores, Portugal (AZO); Bay of Biscay, Spain (BIS); Cádiz, Spain (CAD); Canary Islands, Spain (CAN); Mindelo, Cape Verde (CPV); Brač Island, Croatia (CRO); Galícia, Spain (GAL); Cesenatico, Italy (ITA); Lampedusa, Italy (LAM); Tyre and Tripoli, Lebanon (LEB); Lisbon and Tróia, Portugal (LIS); Madeira Island, Portugal (MAD); Nouadhbou, Mauritania (MAU); Murcia, Spain (MUR); Ria Formosa, Portugal (ALG); Sicily, Italy (SIC); Bizerte and Kélibia, Tunisia (TUN) (Fig. 1 and Table 1). Specimens were collected by local fishermen or from local markets. After asserting the species identification for each individual, fins were clipped and preserved in 96% ethanol.
DNA extraction, amplification and sequencing
Total genomic DNA was extracted with the REDExtract-N-Amp Kit (Sigma-Aldrich) following the manufacturer's instructions. The mitochondrial control region (D-loop) was amplified in a Bio-Rad Mycycler thermal cycler, using the primers L-pro1 and H-DL122. The PCR protocol was performed in a 20 μl total reaction volume with 10 μl of REDExtract-N-ampl PCR mix (Sigma-Aldrich), 0.8 μl of each primer (10 μM), 4.4 μl of Sigma water and 4 μl of template DNA. PCR conditions were the following: initial denaturation at 94 °C for 2 min., followed by 35 cycles (denaturation at 94 °C for 30 s., annealing at 55 °C for 45 s., and extension at 72 °C for 1 min.) and a final extension at 72 °C for 10 min. The same primers were used for the sequencing reaction, and the PCR products were purified and sequenced in STABVIDA (http://www.stabvida.net/). Chromatograms were edited with Codon Code Aligner (Codon Code Corporation, http://www.codoncode.com/index.htm), and sequences were aligned with Clustal X 2.123. All sequences were deposited in GenBank (Accession numbers ON605268-ON605602).
Data analysis
The R24 packages haplotypes25 and pegas26 were used in RStudio27 to estimate standard descriptive measures of genetic diversity in B. boops, including the number of haplotypes and private haplotypes, haplotype (h,28 and nucleotide (π,28 diversities. The genetic structure among all locations was assessed by AMOVA29 using Arlequin 3.5.2.230 with several site group arrangements. The genetic group constitution was assessed by Φct, while the pairwise differentiation between all locations was assessed by Φst. We used Φst and Φct because, contrary to its analogues Fst and Fct, these parameters consider the genetic differences between haplotypes. The significance of each pairwise comparison was tested with 10,000 random replicates, and multiple comparisons were corrected using the false discovery rate31 to calculate q-values (q-values < 0.05 were set as the significant threshold).
The PopART software32 (https://github.com/jessicawleigh/popart-current.git) was used to build a haplotype network based on the Minimum Spanning algorithm33,34. To further visualize relationships between haplotypes at population level, we used the software Network v. 10.2.0.034 to build a Median Joining network (https://www.fluxus-engineering.com/sharenet.htm). Pairwise nucleotide distances (p-distance, 500 bootstrap replicates) within and between B. boops clusters defined by AMOVA were computed in MEGA735. We used the pheatmap function of the R package with the same name36 (https://cran.r-project.org/web/packages/pheatmap/index.html) to obtain a heatmap, which is a graphical representation of the individuals in a matrix to visualize the similarity and dissimilarity among individuals with colors, and simultaneously generate a dendogram to better visualise the clusters.
We used ABGD (Automatic Barcode Gap Discovery)37 (https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html) to analyse putative cryptic diversity within B. boops. ABGD is based on identifying the barcode gap, i.e., a larger divergence among individuals of different species than among conspecific individuals. We selected the Jukes-Cantor model, the X-value of 1.5 (minimum relative barcoding gap width), and prior intraspecific divergences ranging from Pmin = 0.001 to Pmax = 0.1 to run the barcode gap analysis.
Ethics
No experiments on live vertebrates were performed in this study.
Results
A 403-fragment of the D-loop region produced 300 haplotypes in 335 B. boops individuals collected from 18 locations throughout the Western Atlantic and Mediterranean distribution. Haplotype diversities were high, ranging between 0.924 (in samples from the Canary Islands) and 1.000 in several locations of the Atlantic and Mediterranean (average = 0.989; all samples combined = 0.998). Nucleotide diversities varied between 0.013 (in samples from the Canary Islands) and 0.042 (in samples from Lisbon) (average = 0.027; all samples combined = 0.049) (see Table 1 for further details). Four indels were observed, and 281 (94%) haplotypes occurred as singletons. The most abundant haplotype was shared among 10 individuals from Algarve (1), Croatia (3), Italy (3), Lampedusa (1) and Lebanon (2), while the second most abundant was shared between Canary Islands (6) and Madeira (2). We also detected five shared haplotypes between the Canary Islands and Madeira.
Several site group arrangements were tried with the AMOVA. The three-group structure revealed (1) the Mediterranean and eastern Atlantic, including the Azores (hereafter designated as ATL/MED), (2) Canary Islands, Madeira and Cape Verde archipelagos (hereafter designated as ILS) and (3) Mauritania (hereafter designated as MAU) is supported by the highest highly significant Φct-AMOVA (0.76, p < 0.001) of the several groups tried (results not shown). The Φst-based values (Table 2) revealed differences compatible with this three-group structure.
Both haplotype network building algorithms, Minimum Spanning (Fig. 2) and Median Joining (Supplementary material S1) identified the three distinct clades (ATL/MED, ISL and MAU). The clade with samples from Mauritania was separated from the ATL/MED by 25 mutational steps. The clade, including individuals from the Atlantic islands (ISL), was also separated by 25 mutational steps from ATL/MED. The hierarchical clustering visualised in the heatmap also shows the existence of those three clusters ATL/MED, ISL—and MAU (Fig. 3). Within-group estimates of average divergence were the following: MAU—2% ± 0.4%; ISL—1.5% ± 0.3% and ATL/MED—2.9% ± 0.4%. Net divergences were the following: between ATL/MED vs. MAU was 7.5% ± 1.2%; between ATL/MED vs. IS L was 7.9% ± 1.2%; between MAU vs. ISL was 9.6% ± 1.4%. ABGD results identified three Evolutionary Significant Units (ESU) within B. boops coinciding with the AMOVA results.
Haplotype network for the mitochondrial control region of Boops boops showing the existence of three main groups. The green shade highlights the group including samples from the Canary Islands, Cape Verde and Madeira Islands. The purple shade represents the clade corresponding to the group including individuals from Mauritania. The third group corresponds to samples from the Atlantic/Mediterranean and the Azores. Colours refer to sampling locations. The area of the circles is proportional to each haplotype frequency. In the case where haplotypes are shared among sampling locations, filling is proportional to the frequency of the haplotype in each sampling location.
Heatmap and dendrogram based on pairwise distances based on Kimura (1980) for the mitochondrial control region of Boops boops. The heatmap and the associated dendrogram were generated using the pheatmap R-package (https://cran.r-project.org/web/packages/pheatmap/index.html).
Discussion
The main finding of the present study is the identification of genetic structure in a benthopelagic short-sized species that disperses over large distances. We found evidence for the existence of three genetic clusters within the bogue B. boops, one comprising specimens from the Azores and eastern Atlantic/Mediterranean, another with individuals from the eastern Atlantic archipelagos (Canary Islands, Madeira and Cape Verde), and the third with samples from Mauritania (Figs. 2 and 3). The genetic divergence between haplogroups (between 7.5% and 9.6%) is compatible with differences at the genus level found in several fish species38. Further molecular and morphological analyses are required to confirm the existence of cryptic lineages within B. boops.
Identifying processes generating population differentiation within a species is crucial to understanding its evolutionary dynamics. The interplay between life-history traits (e.g. adult swimming abilities, PLD or larval behaviour) and oceanographic features often plays an important role in the dispersal range of a species39. For instance, the shanny Lipophrys pholis occurs throughout the eastern Atlantic and Mediterranean rocky shores showing no signal of genetic structure along its distribution40. The dispersal of L. pholis occurs exclusively during the 29 days that last its pelagic larval phase, which allows widespread gene flow between distant locations41. The heatmap from Fig. 3 shows that B. boops specimens from the Atlantic and Mediterranean share haplotypes. These results indicate that the Almería-Oran Front, an oceanographic barrier known to prevent gene flow between populations from the Atlantic and Mediterranean in more than 50 marine species42, does not represent a barrier to dispersal in B. boops. The lack of genetic structure between these two basins was also identified in another species of Sparidae (Sarpa salpa), in which the strong swimming abilities of the larvae seem to promote connectivity among populations43. Also, it has been proposed that the Portuguese coast could be considered a sort of "Mediterranean appendix, rather than a biogeographic bridge"44, which is supported by our results.
From June to October, the Cape Verde Frontal Zone migrates eastward from Guinea-Bissau and Cape Verde waters to the Cape Blanc headland (Mauritania), while during the rest of the year it moves southward45. If the dispersal of B. boops in this area depended on the adults' movement, we would expect a closer relationship between Cape Verde and Mauritanian populations. Instead, the genetic analyses revealed that specimens from Cape Verde, Madeira and Canary Islands grouped into a cluster different from the one including the Mauritania samples (Figs. 2 and 3). Boops boops is a demersal coastal species wherein large shoals of adults migrate into deeper waters in the spawning season between January and May, peaking in February,15. The southward direction of the Cape Verde Frontal Zone during the winter and the relatively short PLD of B. boops between 16 and 18 days,13, must prevent the movement of the larvae from Cape Verde to Mauritania. The Mauritanian population persists without admixture of individuals from the eastern Atlantic archipelagos, probably due to the gregarious nature of the adults that, during summer, aggregate in large shoals in shallow waters to feed in one of the most productive marine regions of the world's oceans, the Mauritania upwelling46. Net divergences between groups are larger between the eastern Atlantic archipelagos vs. Mauritania (0.096 ± 0.014) than between Atlantic/Mediterranean vs. Mauritania (0.075 ± 0.012). These results further support the low levels of connectivity between the eastern Atlantic islands and the Mauritanian population.
There are several examples of fish species exhibiting distinct genetic patterns within the same geographic range5,8. For instance, the sand-smelt (Atherina presbyter) populations from the Azores show a closer relationship with the eastern Atlantic archipelagos (Canary Islands and Madeira) and are markedly distinct from specimens of the Mediterranean and southern European Atlantic shores47. This pattern may have resulted from the colonisation of the Azores by the Canary Islands and Madeira acting as stepping stones and glacial refugia for fish coming from West Africa47. Boops boops haplotypes from southern Portugal and the Mediterranean are closely related to samples from the Azores. Yet, it is impossible to quantitatively infer the direction of the dispersal because there are few shared haplotypes and none with the Azores. Nonetheless, a West–East direction is expected due to a branch of the northern subtropical gyre (Azores Current) that drifts towards the Gulf of Cádiz. This current promotes the displacement of larvae and adults from the Azores to southern Portugal and into the Mediterranean, which is consistent with our pattern (Fig. 2).
Although based on a single-marker approach, our results are clear and reflect a comprehensive sampling coverage with a suitable number of individuals. The marker choice, frequently used in traditional phylogeographical approaches, allows comparisons with other species across the same geographical area, preventing biases resulting from different evolutionary rates.
Conclusions
Identifying the processes that generate population differentiation in a species with a broad distribution is pivotal to understand its evolutionary dynamics. Here we show the existence of three clades within the bogue Boops boops that most likely correspond to distinct populations. One comprises samples from the Azores, eastern Atlantic and Mediterranean. Another includes samples from Madeira, Canary Islands and Cape Verde archipelagos. The third cluster is composed of specimens from Mauritania only. The Almería-Oran Front, a known oceanographic barrier for more than 50 species, does not prevent gene flow through the Atlantic-Mediterranean divide. The southward direction of the Cape Verde Frontal Zone during the winter (spawning season of B. boops) combined with the relatively short PLD of this species averts the genetic admixture between populations from Mauritania and the neighbour eastern Atlantic archipelagos. The gregarious nature of the adults that remain in shallow waters during the summer to feed in the highly productive marine system of the Mauritanian upwelling, further prevents connectivity with nearby oceanic islands' populations. The observed genetic discontinuities do not conform to an isolation-by-distance model but instead reflect the role of specific life-history traits and local oceanographic features.
Data availability
All sequences produced in this study were deposited in GenBank under the accession numbers ON605268-ON605602.
References
Palumbi, S. R. Genetic divergence, reproductive isolation, and marine speciation. Annu. Rev. Ecol. Evol. Syst. 25, 547–572 (1994).
Hellberg, M. E. Gene flow and isolation among populations of marine animals. Annu. Rev. Ecol. Evol. Syst. 40, 291–310 (2009).
Bierne, N., Gagnaire, P.-A. & David, P. The geography of introgression in a patchy environment and the thorn in the side of ecological speciation. Curr. Zool. 59, 72–86 (2013).
Cowen, R. K., Lwiza, K. M., Sponaugle, S., Paris, C. B. & Olson, D. B. Connectivity of marine populations: Open or closed?. Science 287, 857–859 (2000).
Reid, K. et al. Secondary contact and asymmetrical gene flow in a cosmopolitan marine fish across the Benguela upwelling zone. Heredity 117, 307–315 (2016).
Woodall, L., Koldewey, H. & Shaw, P. Historical and contemporary population genetic connectivity of the European short-snouted seahorse Hippocampus hippocampus and implications for management. J. Fish Biol. 78, 1738–1756 (2011).
White, T., Fotherby, H., Stephens, P. & Hoelzel, A. Genetic panmixia and demographic dependence across the North Atlantic in the deep-sea fish, blue hake (Antimora rostrata). Heredity 106, 690–699 (2011).
Jenkins, T. L., Castilho, R. & Stevens, J. R. Meta-analysis of northeast Atlantic marine taxa shows contrasting phylogeographic patterns following post-LGM expansions. PeerJ 6, e5684 (2018).
Valle, C., Bayle, J. & Ramos, A. Weight–length relationships for selected fish species of the western Mediterranean Sea. J. Appl. Ichthyol. 19, 261–262 (2003).
Whitehead, P. J. P., Bauchot, M. L., Hureau, J. C., Nielsen, J. & Tortonese, E. v. Fishes of the north-eastern Atlantic and the Mediterranean (1986).
Azab, A. M., El-Far, A. M. & El-Sayed, A. M. Age, growth and population structure of bogue, Boops boops, in the Mediterranean waters front Alexandria Egypt. EJABF 23, 69–81 (2019).
Riera, R., Sanchez-Jerez, P., Rodriguez, M. & Monterroso, O. Artificial marine habitats favour a single fish species on a long-term scale: the dominance of Boops boops around off-shore fish cages. Sci. Mar. 78, 505–510 (2014).
Borges, R., Beldade, R. & Gonçalves, E. J. Vertical structure of very nearshore larval fish assemblages in a temperate rocky coast. Mar. Biol. 151, 1349–1363 (2007).
Ben Abdallah, L., Gaamour, A. & El Abed, A. Mean total length geographical distribution of the bogue (Boops boops, L., 1758) along Tunisian coasts. Biol. Mar. Medit 11, 675–678 (2004).
Dobroslavić, T., Mozara, R., Glamuzina, B. & Bartulović, V. Reproductive patterns of bogue, Boops boops (Sparidae), in the southeastern Adriatic Sea. Acta Adriat. 58, 117–125 (2017).
Mahé, K. et al. Directional bilateral asymmetry in otolith morphology may affect fish stock discrimination based on otolith shape analysis. ICES J. Mar. Sci. 76, 232–243 (2019).
Labidi, M. B. et al. Morphometric and meristic character variability and relationships among populations of (L.) from four marine stations along the Tunisian coast. Fish. Aquat. Life 29, 13–28 (2021).
Mladineo, I., Šegvić, T. & Grubišić, L. Molecular evidence for the lack of transmission of the monogenean Sparicotyle chrysophrii (Monogenea, Polyopisthocotylea) and isopod Ceratothoa oestroides (Crustacea, Cymothoidae) between wild bogue (Boops boops) and cage-reared sea bream (Sparus aurata) and sea bass (Dicentrarchus labrax). Aquaculture 295, 160–167 (2009).
Aglieri, G. et al. Environmental DNA effectively captures functional diversity of coastal fish communities. Mol. Ecol. 30, 3127–3139. https://doi.org/10.1111/mec.15661 (2021).
Valenzuela-Quiñonez, F. How fisheries management can benefit from genomics?. Brief Funct. Genomics 15, 352–357. https://doi.org/10.1093/bfgp/elw006 (2016).
Patarnello, T., Volckaert, F. A. M. J. & Castilho, R. Pillars of Hercules: Is the Atlantic-Mediterranean transition a phylogeographical break?. Mol. Ecol. 16, 4426–4444. https://doi.org/10.1111/j.1365-294X.2007.03477.x (2007).
Ostellari, L., Bargelloni, L., Penzo, E., Patarnello, P. & Patarnello, T. Optimization of single-strand conformation polymorphism and sequence analysis of the mitochondrial control region in Pagellus bogaraveo (Sparidae, Teleostei): rationalized tools in fish population biology. Anim. Genet. 27, 423–427 (1996).
Larkin, M. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
R: a language and environment for statistical computing. R Foundation for Statistical Computing v. 4.1.3 (2021).
Aktas, C. Haplotypes: an R package v. 1.1.2 for manipulating DNA sequences and estimating unambiguous haplotype network with statistical parsimony. https://CRAN.R-project.org/package=haplotypes; https://CRAN.R-project.org/package=haplotypes (2020).
Paradis, E. pegas: An R package for population genetics with an integrated–modular approach. Bioinformatics 26, 419–420 (2010).
RStudio: Integrated Development for R. RStudio (2020).
Nei, M. Molecular Evolutionary Genetics. (1987).
Excoffier, L., Smouse, P. E. & Quattro, J. M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491 (1992).
Excoffier, L. & Lischer, H. E. L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat. Soc. B 57, 289–300 (1995).
Leigh, J. W. & Bryant, D. POPART: full-feature software for haplotype network construction. MEE 6, 1110–1116 (2015).
Excoffier, L. & Smouse, P. E. Using allele frequencies and geographic subdivision to reconstruct gene trees within a species: molecular variance parsimony. Genetics 136, 343–359 (1994).
Bandelt, H. J., Forster, P. & Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. https://doi.org/10.1093/molbev/msw054 (2016).
Kolde, R. pheatmap: Pretty Heatmaps. R package version 1.0. 12. R Packag. version 1.0 8 (2019).
Puillandre, N., Lambert, A., Brouillet, S. & Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 21, 1864–1877 (2012).
Ward, R. D., Zemlak, T. S., Innes, B. H., Last, P. R. & Hebert, P. D. DNA barcoding Australia's fish species. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 360, 1847–1857 (2005).
Galarza, J. A. et al. The influence of oceanographic fronts and early-life-history traits on connectivity among littoral fish species. Proc. Natl. Acad. Sci. U.S.A. 106, 1473–1478 (2009).
Francisco, S. et al. Phylogeography of the shanny Lipophrys pholis (Pisces: Blenniidae) in the NE Atlantic records signs of major expansion event older than the last glaciation. J. Exp. Mar. Biol. Ecol. 403, 14–20 (2011).
Faria, C., Borges, R., Gil, F., Almada, V. C. & Gonçalves, E. J. Embryonic and larval development of Lipophrys pholis (Pisces: Blenniidae). Sci. Mar. 66, 21–26 (2002).
El Ayari, T., Trigui El Menif, N., Hamer, B., Cahill, A. E. & Bierne, N. The hidden side of a major marine biogeographic boundary: a wide mosaic hybrid zone at the Atlantic–Mediterranean divide reveals the complex interaction between natural and genetic barriers in mussels. Heredity 122, 770–784 (2019).
Paiva, R. B. Salema, Sarpa salpa (Linnaeus 1758): stock structure in the eastern Atlantic and biological characterization off the Portuguese coast PhD thesis, University of Lisbon (2015).
Plicanti, A., Iaciofano, D., Bertocci, I. & Brutto, S. L. The amphipod assemblages of Sabellaria alveolata reefs from the NW coast of Portugal: An account of the present knowledge, new records, and some biogeographic considerations. Mar. Biodivers. 47, 521–534 (2017).
Fernández‐Peralta, L. & Sidibé, A. in Oceanographic and biological features in the Canary Current Large Marine Ecosystem (ed Intergovernmental Oceanographic Commission) 215–229 (2015).
Romero, O. E., Ramondenc, S. & Fischer, G. A 2-decade (1988–2009) record of diatom fluxes in the Mauritanian coastal upwelling: impact of low-frequency forcing and a two-step shift in the species composition. Biogeosciences 18, 1873–1891 (2021).
Francisco, S. et al. Phylogeography and demographic history of Atherina presbyter (Pisces: Atherinidae) in the North-eastern Atlantic based on mitochondrial DNA. Mar. Biol. 156, 1421–1432 (2009).
Acknowledgements
The manuscript was improved by useful comments from two anonymous reviewers. This study received Portuguese national funds from FCT—Foundation for Science and Technology through projects MARE/UIDB/MAR/04292/2020, MARE/UIDP/04292/2020 and LA/P/0069/2020 granted to MARE/ARNET and UIDB/04326/2020, UIDP/04326/2020 and LA/P/0101/2020 granted to CCMAR. Regina L. Cunha was funded by the transitional norm—DL 57/2016/CP1361/CT0013. The authors acknowledge several researchers and institutions for supplying specimens (Azores: Gisela Dionisio—Naturalist, contact@naturalist.pt; Madeira: João Canning-Clode—Marine and Environmental Sciences Centre—MARE and Regional Agency for the Development of Research Technology and Innovation, ARDITI; Cape Verde: Evandro Lopes-Atlantic Technical University, Cape Verde; Mediterranean: Marga L. Rivas—University of Almería, Spain; Lebanon: Michel Bariche—American University of Beirut. We thank Cristina Lima and Karen Avellaneda for the help with molecular procedures at the lab. We also thank the IT Services of the University of Algarve for hosting and maintaining the R2C2 computational cluster facility (http://rcastilho.pt/R2C2/R2C2_cluster.html).
Author information
Authors and Affiliations
Contributions
J.R. and R.C.: conceptualisation, investigation, methodology, results interpretation, revising and editing the draft, funding acquisition; R.L.C.: investigation, methodology and data analysis, results interpretation, writing draft and editing final manuscript, corresponding author; R.C.: prepared the figures; S.F.: data analysis and draft revision; L.C.: laboratory procedures and draft revision; A.B.F., M.D.; I.G. and A.K.: sampling and revising draft; J.V. and R.S.: conceptualisation, sampling and revising the draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cunha, R.L., Faleh, A.B., Francisco, S. et al. Three mitochondrial lineages and no Atlantic-Mediterranean barrier for the bogue Boops boops across its widespread distribution. Sci Rep 12, 22124 (2022). https://doi.org/10.1038/s41598-022-26651-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-26651-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.