Abstract
Neuroblastoma is a childhood tumour that is responsible for approximately 15% of all childhood cancer deaths. Neuroblastoma tumours with amplification of the oncogene MYCN are aggressive, however, another aggressive subgroup without MYCN amplification also exists; rather, they have a deleted region at chromosome arm 11q. Twenty-six miRNAs are located within the breakpoint region of chromosome 11q and have been checked for a possible involvement in development of neuroblastoma due to the genomic alteration. Target genes of these miRNAs are involved in pathways associated with cancer, including proliferation, apoptosis and DNA repair. We could show that miR-548l found within the 11q region is downregulated in neuroblastoma cell lines with 11q deletion or MYCN amplification. In addition, we showed that the restoration of miR-548l level in a neuroblastoma cell line led to a decreased proliferation of these cells as well as a decrease in the percentage of cells in the S phase. We also found that miR-548l overexpression suppressed cell viability and promoted apoptosis, while miR-548l knockdown promoted cell viability and inhibited apoptosis in neuroblastoma cells. Our results indicate that 11q-deleted neuroblastoma and MYCN amplified neuroblastoma coalesce by downregulating miR-548l.
Similar content being viewed by others
Introduction
Neuroblastoma (NB) is a heterogeneous complex cancer arising from the transient embryonic neural crest1. It is one of the most common extra cranial solid tumours found in children2. NB can also be defined by the international neuroblastoma risk group (INRG)3, suggesting that low and intermediate NB have a good prospect for treatment, whereas, high risk tumors are more difficult to treat4. This difficulty to treat results in a higher mortality. High risk NB tumors have specific genetic alterations and are highly aggressive. Two of the most prevalent forms of high risk NB are; amplification of the MYCN proto-oncogene5 and unbalanced 11q-deleted loss of heterozygosity (LOH) tumors2, which account for approximately 20% and 30% of all cases, respectively. Currently, these two are the most difficult subgroups to treat with current therapies. These two common aggressive NB tumor subtypes rarely occur together and are therefore usually considered to be mutually exclusive6. In the rare cases with both 11q deletion and MYCN amplification the extent of 11q deletion is often reported to be 11q21-11qter and when there is no MYCN amplification the deletion extends to 11q14.1-11qter6. Extensive investigation into 11q has occurred since the identification as a frequent alteration. Currently, no gene or genes in the 11q-region have been proven to be NB tumor suppressors, although there are a number of candidates. Previously identified candidate tumor suppressor genes on 11q include; DLG2 (11q14.1)7, ATM (11q22.3)8, CADM1 (11q23.3)9and H2AFX (11q23.3)6. Common to all of these candidate tumor suppressor genes (TSGs) is the lack of a second hit. Based on the Knudson two hit hypothesis, the lack of a second hit could indicate that gene dosing is involved.
MicroRNAs (miRNAs) are a class of short single-stranded RNA sequences that have a large impact on the regulation of the transcriptome10,11. It has been estimated that as many as 60% of all human genes may be regulated post-transcriptionally by miRNA12. The regulatory effects of miRNAs are exerted by binding to the 3’-untranslated region (UTR) of a target messenger RNA (mRNA) via base pairing. Due to the short base pairs between miRNA and target mRNA one miRNA can bind to several (up to hundreds of) transcripts and a single transcript can be targeted by several miRNAs13,14. MicroRNAs have been demonstrated to play a major role in a broad range of important biological processes such as development, differentiation, growth and metabolism15. It has been shown that aberrant expression of miRNAs correlates with certain features, such as tumorigenesis, differentiation status and clinical outcome16.
Several studies have shown that miRNAs can play a critical role in initiation and progression of cancer by acting as tumour suppressors or oncogenes17. The gain of additional copies of a chromosomal region harbouring miRNAs may lead to overexpression of these miRNAs, which will in turn result in excessive down-regulation of their target genes. In cases where the target gene function as a tumour suppressor, its down-regulation may result in increased tumour growth. Therefore, miRNA expression profiles can potentially be of great prognostic and diagnostic value and should be considered when trying to understand the complexity of neoplasia18,19. miR-21 is located on chromosome 17q21, which is amplified frequently in all subtypes of advanced-stage NB. Wang et al., show that by reducing the expression of miR-21 in NB cell lines, the expression of two tumour suppressor genes (PDCD4 and PTEN) increases, leading to inhibition of cell proliferation and increased apoptosis in the NB cell line20. Conversely, the genetic alterations acquired during cancer development may include the loss of chromosomal regions harbouring miRNAs, and may therefore be one of the underlying reasons for the loss of growth control, since the targets of these miRNAs may include proto-oncogenes. For example, miR-193b has been implicated as causal in NB. Roth et al., have shown that overexpression of miR-193b in NB reduces cell growth via downregulation of the three oncogenes (MCL1, Cyclin D and MYCN). The results support the role of miR-193b as a potential therapeutic biomarker for NB21.
Multiple studies have reported that a common breakpoint region (11q14-qtel) exist in 11q-NB6,7,22. In this study, we aimed to identify miRNAs located in this region and their target genes that may be responsible for the development of NB.
Results
Bioinformatics analysis of miRNAs in the 11q14-qtel region and their target genes
To identify miRNAs at the chromosome 11 breakpoint regions we searched miRBase database and found 26 miRNAs within 11q14-qtel region (Fig. 1). We conducted a bioinformatics analysis of the miRNAs to find those with potential target genes that may be involved in development of NB.
Since miRNAs have important roles in various biological processes by post-transcriptional regulation of target mRNAs, we studied the function of the miRNAs by identifying putative target genes. The target genes of the 26 miRNAs were identified in silico, followed by a pathway analysis that was performed with the KEGG database to investigate the possible biological impact of the miRNAs and their target genes in development of NB. We found in total 61 KEGG pathways predicted to be influenced by these 26 miRNAs (Supplementary Table 1). As expected, target genes were enriched in pathways associated with cancer, including proliferation, evading apoptosis and DNA repair. We also noted that three miRNAs (miR-548l, miR-708-5p, and miR-1261) regulated 36% of all genes involved in signaling pathways regulating pluripotency of stem cells (Fig. 2).
Results from KEGG analysis indicated that miR-548l was central, as it alone was predicted to be involved in 26 of 61 pathways and it regulated the central genes in several pathways, among those responsible for DNA repair, the Hippo, PI3K and Foxo signaling pathways (Fig. 3). Thus, miR-548l was chosen for further bioinformatics and experimental analysis in NB cells.
miRNA regulation of genes involved in Hippo, Foxo and PI3K-Akt pathway. The yellow box nodes represent miRNAs; green nodes describe genes located in the Hippo pathway; blue nodes describe genes located in the Foxo pathway; grey nodes describe genes located in the PI3K-Akt pathway; pink nodes describe genes located in both the Hippo and Foxo pathways; orange nodes describe genes located in both the Hippo and PI3K-Akt pathways; purple node describes gene located in the Hippo, PI3K-Akt and Foxo pathways; white nodes describe genes located in both the PI3K-Akt and Foxo pathways.
Gene ontology (GO) analysis of miR-548l target genes showed that target genes corresponding to the biological processes from GO categories were mainly involved in neuron differentiation, nervous system development, generation of neurons and neurogenesis (Fig. 4).
The 209 enriched biological processes controlled by miR-548l from at least 2 of the database lists. (a) the enriched biological processes of genes under the control of miR-548l (b) tabular result of the adjusted p values of 10 selected enriched terms. The data is presented as a Manhattan plot with the y axis representing the -log10 adjusted p value and GO ID number along the x axis, so that similar terms cluster on this plane. The functional enrichment analysis was performed using g:Profiler with g:SCS multiple testing correction method applying significance threshold of 0.05.
Network analysis of protein targets for miR-548l
Additional network analysis and functional enrichment was performed for miR-548l targets to extract hub genes and pathways of potential relevance for disease. Figure 5a shows 20 top-ranked nodes based on Cytoscape plugin cytoHubba23 degree score. The hubs with the largest degree (≥ 10) are CTNNB1, ACTB, HSP90AA1, and MAP2K1 (Fig. 5b). Cytoscape plugins ClueGO and CluePedia24 were used to identify enriched KEGG pathways as well as the interplay between these by showing genes that connect different pathways (Fig. 6, only pathways with p ≤ 0.05 are shown). CTNNB1 was found to overlap between 3 significantly enriched KEGG pathways: adherens junction, focal adhesion and signaling pathways regulating pluripotency of stem cells (Fig. 6a).
Enriched KEGG pathway for miR-548l target genes in ClueGO and CluePedia. (a) Enriched KEGG pathways for miR-548l targets. The node size represents the term enrichment significance (b) Bars represent the number of genes associated with each term/pathway. Percentage of genes per term is shown as bar label.
Two of the significantly enriched pathways containing largest number of proteins that are targeted by miR-548l are focal adhesion (8% genes/term) and signaling pathways regulating pluripotency of stem cells (7% genes/term).
miR-548l expression is repressed in high risk NB cells
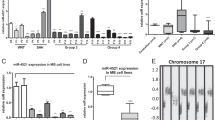
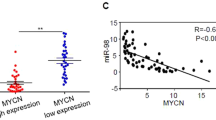
We evaluated the expression of miR-548l in NB cell lines with 11q deletion (del), MYCN amplification (amp.) or neither (no MYCN amp, no 11q del). We could show that there was a significantly lower expression in the MYCN amplified cells (log2 Fold Change (FC) = 4.38, p < 0.001) and in the 11q deleted cells (log2 FC = 4.37, p < 0.001) compared to the NB cell lines lacking either (Fig. 7a). To determine if the effect was as a direct result of MYCN amplification we transfected all three cell lines with miR-34a, a known inhibitor of MYCN. We found that miR-34a mimic transfection resulted in an increase in miR-548l expression in the MYCN amplified cells (log2 FC = 6.69, p < 0.001) when compared to the cells without MYCN amplification or 11q-deletion (Fig. 7b). There was no difference in miR-548l expression in the 11q deleted cells (log2 FC = -0.69, p = 0.66) compared to the cells without MYCN amplification or 11q deletion (Fig. 7b). We verified this correlation in a primary NB dataset, when comparing 33 NB:s without mir-548l expression to 51 NB:s with high mir-548l expression, we found very low expression of mir-34a in the NB:s lacking mir-548l (mean log2 FC = 0.00), while the NB:s with mir-548l expression had a higher mir-34a expression (mean log2 FC = 0.79; p < 0.001)(Fig. 7c). To further test if MYCN was directly responsible we silenced the expression of MYCN using siRNA targeted to MYCN in the MYCN amplified cell lines and detected an increase in miR-548l expression compared to control (log2 FC = 7.52, p < 0.05) (Fig. 7d). We subsequently investigated the mechanism to which MYCN could repress miR-548l expression. We used publicly available ATAC-Seq and CHIP-Seq data (GSE138315)25,26 to investigate the presence of; open chromatin, MYCN and H3K27Ac for 3 MYCN amplified NB cell lines, scaled to 10 KB. We could show that MYCN is bound to the enhancer GH11J094472 which controls miR-548l, with the presence of the H3K27Ac mark consistent across the 3 cell lines (Fig. 7e). The effect of MYCN silencing was determined using publicly available data showing the shutdown of MYCN over three timepoints (0 h, 2 h and 24 h) in SHEP21N cells (GSE80154)27. Here we could visualize the 80 KB upstream region of the miR-548l gene including the MRE11 promoter GH11J094512 which is known to be under direct control of MYCN28. MYCN vacated the GH11J094472 promoter after 2 h indicating weak affinity with the H3K27Ac mark decreasing over time. The GH11J094512 promoter showed gradual decreases in bound MYCN, H3K27Ac and H3K4me3 over time (Fig. 7f).
miR-548l is under the direct control of MYCN. (a) Expression of miR-548l in NB cells with 11q Deletion (Del)., MYCN amplification (amp.) or neither (Normal). (b) Expression of miR-548l expression quantified after miR-34a mimic expression in NB cells with 11q Deletion (Del)., MYCN amplification (amp.) or neither (Normal). (c) Expression of mir-34a in primary NB with no mir-548l expression compared to primary NB with high mir-548l expression (d) Expression of mir-548l after silencing of MYCN (siMYCN) compared to control in MYCN-amplified NB cells. (e) Open chromatin (ATAC-seq) overlay plots of MYCN amplified NB showing open chromatin (blue), MYCN binding (red) and H3K27ac (green) sequences 10 KB upstream of miR-548l. (f) The effect of MYCN silencing in SHEP-N21 cells over time. Chromatin binding (CHIP-seq) plots showing the binding of MYCN (red), H3K27ac (green) and H3K4me3 (pink) 40 KB upstream of miR-548l. The expression data are presented as log2 fold change and plotted as a scatter plot with a line at the mean. The CHIP-Seq and ATAC-Seq are expressed as rpm/bp. Statistical significance was determined by one-way ANOVA with a Holm-Šídák's multiple comparisons test*p < 0.05, **p < 0.01, ***p < 0.001, ns = not significant.
Overexpression of miR-548l slowed proliferation and promoted apoptosis
WE used publicly available data to investigate miR-548l expression based on INSS stage and age at diagnosis (GSE155945)29. We found that miR-548l expression in NB was low in stage 4 and in patients of older age (Fig. 8a,b).
miR-548l expression in NB primary dataset GSE155945 stratified by (a) INSS stage and (b) age at diagnosis. Violin plots are presented with a slid line at the median and a dashed line showing the quartiles. The effect of miR-548l over expression (mimic) or miR-548l silencing (inhibitor) compared to negative control mimic (NCM) and negative control inhibitor (NCI) was determined in NB69, SKNAS (11q deletion) and SKNBE2 (MYCN amplified) cell lines regarding (c) cell proliferation, (d) cell viability (e) apoptosis initiation measured by caspase 3/7 activity. (f) Cell cycle analysis in response to miR-548I mimic or inhibitor in SKNBE2 cells. (g) miR-548l expression in NB69, SKNAS (11q deletion) and SKNBE2 (MYCN amplified) cell lines after transfection of miR-548l mimic, miR-548l inhibitor, negative control mimic (NCM) and negative control inhibitor (NCI). Boxplots are presented with a line at the median, and ‘ + ’ at the mean. The whiskers represent the 10th to 90th percentiles (Tukeys). The expression and protein data are presented as log2 fold change and plotted as a scatter plot with a line at the mean. Statistical significance was determined by one-way ANOVA with a Holm-Šídák's multiple comparisons test for groups of 3 or more or a paired t-test for exactly 2 groups. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns = not significant.
We next determined the effects on proliferation of miR-548l overexpression or silencing. Overexpression of miR-548l in NB cells (mir-548l mimic) resulted in slower proliferation compared to the negative control mimic (NCM) for NB69, SKNAS and SKNBE cells; 11.85%, 7.92% and 25.78% respectively, p < 0.001 (Fig. 8c). Inhibition of miR-548l increased proliferation in SKNAS and SKNBE by 6.15% and 10.4% respectively compared to negative control inhibitor (NCI), p < 0.001, with no change in NB69 (Fig. 8c). MiR-548l inhibitor promoted the cell viability of SKNBE cells as compared to NCI (19.0% increase, p < 0.01, Fig. 8d), while miR-548l mimic decreased the viability of SKNBE cells compared to NCM (40.2% decrease, p < 0.0001, Fig. 8d). We also found that the activity of caspase 3/7, the effector of apoptotic pathway, was increased by miR-548l mimic (31.4% increase, p < 0.001) and inhibited by miR-548l inhibitor (36.4% decrease, p < 0.001) in comparison with mimic- and inhibitor negative controls (Fig. 8e). We subsequently measured the cell cycle state of SKNBE cells after miR-548l overexpression or silencing. We could show that the cells accumulated in the G1 (12.5%, p < 0.001) and G2/M (8.1%, p < 0.001) phase, with fewer cells in the S Phase after miR-548l overexpression (mimic) (20.59%, p < 0.001) (Fig. 8f). Inhibition of miR-548l expression resulted in a decrease in the number of cells in G1 phase (5.5%, p < 0.05) and an associated increase in S phase cells (5.2%, p < 0.05). No difference in G2/M was detected with inhibition of miR-548l (Fig. 8f). The significant silencing (on average twofold, p < 0.01) or upregulation (on average eightfold, p < 0.001) of miR-548l after inhibitor or mimic transfections compared to NCM and NCI were confirmed by qPCR, for NB69, SKNAS and SKNBE cells (Fig. 8g).
Discussion
NB is a childhood tumor that is responsible for approximately 15% of all childhood cancer deaths. While survival from various pediatric tumors has increased dramatically, survival of patients with NB continues to be poor. The subgroup of NB with deletion of chromosome arm 11q30 are highly aggressive and one of the most difficult subgroups to treat with current therapies31 with low overall survival30. Multiple studies have reported that a common breakpoint region (11q14-qtel) exist in high stage NB6,7,22. The mutual exclusivity of the 11q loss and MYCN amplification, despite themselves been common alteration in high risk NB indicates that they have a similar mechanism of action.
We identified and selected targets of the twenty-six miRNAs identified with the 11q14-qtel region and established their roles using biological process via KEGG pathway analysis. The biological pathways that were enriched included several cancer related pathways including; Hippo, PI3K and Foxo pathways (Fig. 3). Hippo signaling has been shown to regulate cell proliferation, neurogenesis and apoptosis32. We subsequently selected miR-548l for further investigation as it was expressed in neural tissues, and bioinformatics analysis revealed interesting target genes. We could show that there was a decrease in proliferation of NB cells after the restoration of miR-548l expression as well as a decrease in the percentage of cells in the S phase (Fig. 8). When combined with the results of the target genes using G:profiler we showed an enrichment of the genes associated with the development of the nervous system (Fig. 7) including; neurogenesis and neuron differentiation and neuron development all of which are altered in high risk NB33 and consistent with the function of Hippo signaling. Both miR-548l silencing an upregulation caused significant changes in the miR-548l level, which led to alterations in NB cell viability and apoptosis. Our study shows that six miRNAs from 11q14-qtel region regulate 45% of the genes that are involved in the Hippo signaling pathway (Fig. 3). The Hippo pathway gene, YAP (also known as YAP1) is regulated by miR-548l. Amplification of the YAP gene has been reported in a variety of tumors, including medulloblastomas34, prostate35 and colorectal cancer36 and YAP has been identified as a novel regulator of NB proliferation, tumorigenesis and invasion37.
The PI3K-Akt38 and Foxo39 pathways are also important in maintaining genomic stability in response to DNA damage, with Foxo overexpression in NB predicting adverse clinical outcomes40. The PI3K-Akt pathway was enriched as about 15% of the genes in this pathway were targets of miR-708-3p and miR-34b-3p (p < 0.01, Supplementary Table 1). Our analysis shows that the set of 52 genes implicated in the PI3K-Akt pathway are potential targets for miR-708-5p and miR-34b-3p.
CTNNB1 was found to overlap between 3 significantly enriched KEGG pathways: adherens junction, focal adhesion and signaling pathways regulating pluripotency of stem cells (Fig. 6a). The CTNNB1 encoded protein, β-catenin, has been shown to be highly expressed with aberrant localization in non-MYCN amplified high risk tumors41. Additionally, high expression of β-catenin has been shown to reduce cellular sensitivity to the ALK inhibitor crizotinib42. Among other top ranked hubs, HSP90AA1 has been directly associated with NB in a previous study, showing that Hsp90 inhibition in NB cell lines led to significant growth suppression, a decrease in MYC and MYCN expression, and an increase in protein expression of p5343. Furthermore, it has been shown to be associated with cisplatin resistance in NBs44. In previous work it was also shown that one of our highly ranked hubs, YAP1, promotes the proliferation of NB cells28.
We have shown that miR-548l is predicted to interact with both DLG2 (11q14.1) and ATM (11q22.3), two candidate TSGs in the 11q14.1-ter breakpoint (Fig. 3). As we have previously stated, all of the current candidate TSGs within this region lack a second hit, based on the Knudson two hit hypothesis. The interaction of a miRNA with a transcript often results in translational repression of the target sequence. However, this is not always the case as under specific circumstances a miRNA can promote gene expression45. The presence of two major transcriptional isoforms of DLG2, each with a unique 3´UTR region does provide a mechanism for miRNA induced differential expression. Previously established results, showed differential DLG2 isoform expression in high INSS staged neuroblastoma tumors46, and we could also see a lower expression of mir548l in stage 4 primary neuroblastomas (Fig. 8), similar to the DLG2 expression. ATM has previously been shown to act the DNA damage response pathway leading to intra S phase arrest and activation of the G2/M cell cycle checkpoint47, something that we show is activated in response to the mimic of miR-548l (Fig. 8). We could also see an effect of mir-548l expression on apoptosis signaling, which could be causative to the changes in cell proliferation seen in NB cells after mir-548l silencing or upregulation (Fig. 8).
Taken together, the heterozygous loss of 11q14.1-ter has been shown to result in alterations to miR-548l (Fig. 7), DLG27 and ATM48 expression. The potential for interactions between miR-548l, DLG2 and ATM is an interesting foundation for a cluster of genes on 11q coalescing within similar pathways. Further investigation into the additional effects of miR-548l on these genes is certainly warranted.
In this study we analyzed miRNAs from the 11q14.1-ter breakpoint, aiming to discover potential biomarkers for NB. We could show that 11q-deleted and NB MYCN amplified NB coalesce by downregulating miR-548l.
It is important to stress that bioinformatics approaches for miRNA target prediction and enrichment that were conducted in this study, although highly beneficial for explorative research, suffer from well-known drawbacks, such as lack of sensitivity and specificity. Future work needs to include more functional validation to future strengthen our findings and provide evidence for miR-548l potential role in disease pathogenesis and therapy, but it out of scope for this study.
Methods
Bioinformatics analysis of miRNAs in the 11q14-qtel region and their target genes
In order to identify miRNAs located at the chromosome 11 breakpoints, we used miRBase (release 22.1). MicroRNA target genes were collected from two sources; experimentally validated targets and computational predicted targets. Validated miRNA target genes were generated from TarBase (Version 8)49, which provides curated experimentally validated miRNA target genes from published literature. TargetScan (release 7.2)50 and microT (Version 5)51 algorithms were used to receive the predicted target genes. Both algorithms use miRNA sequence and 3’UTR of potential target mRNA as input files in FASTA format and determine their binding ability by calculating the minimum free energy for hybridization. Pathway analysis was also performed on miRNA target genes based on the KEGG database (http://www.genome.jp/keg/)52. The count number larger than 4 and FDR less than 0.01 were chosen as cut-off criterion. Determination and visualization of the enriched biological processes was performed using g:Profiler using the previously established gene lists 53. Analysis of hubs in network of proteins targets for miR-548l was performed with Cytoscape plugin cytoHubba23 (version 0.1). Plugins ClueGO (version 2.5.8) and CluePedia24 (version 1.5.8) were used to identify enriched KEGG pathways among miR-548l targets.
Publicly available data
We used GSE138315 and GSE80154 from Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/). ChIP-seq for MYCN, as well as ATAC-seq was performed in various neuroblastoma cell lines (GSE138315 and GSE80154). Data for analyses and comparison of miR-548l expression between the different patient subgroups for the miRNA dataset GSE155945 was imported from the R2 platform (http://r2.amc.nl).
Cell lines and tissue culture
Human NB cell lines SHSY-5y (no MYCN, no 11q deletion), IMR32 (MYCN amplification), SKNAS (11q deletion), SKNBE (2) (MYCN amplification), and NB69 (no MYCN, no 11q deletion) were obtained from the ATCC Cell Line Collection (ATCC). SHSY-5y, IMR32, SKNAS and SKNBE (2) were maintained in RPMI 1640 culture medium supplemented with 10% FBS, 1% L-Glutamine, 1% HEPES solution, and 1% sodium pyruvate. NB69 was cultured in RPMI 1640 medium supplemented with 15% FBS, 1% L-Glutamine, 1% HEPES solution, and 1% sodium pyruvate. The cells were cultured in a humidified incubator at 37 °C with 5% CO2.
Transfections
Cell lines were grown to 80% confluence and subsequently transfected with 5 pmol mirVana® miRNA corresponding to; hsa-miR-548l Mimic (4,464,066, Thermo Fischer Scientific), hsa-miR-548l Inhibitor (4,464,084, Thermo Fischer Scientific), hsa-miR-34a-5p Mimic (4,464,066, Thermo Fischer Scientific), mirVana™ miRNA Mimic, Negative Control #1 (4,464,058, Thermo Fischer Scientific) or mirVana™ miRNA Inhibitor, Negative Control #1 (4,464,079, Thermo Fischer Scientific) using Lipofectamine™ RNAiMAX Transfection Reagent (13,778,030, Invitrogen) and subsequently harvested 48 h post transfection. Cell lines with MYCN amplification were grown to 80% confluence and transfected with either siMYCN or scrambled control and harvested 48 h post transfection. In brief, 30 pmol siRNA was complexed with 12 μl Lipofectamine RNAiMAX (13,778,030, Invitrogen) according to the manufacturer’s protocol (Thermo Fisher Scientific).
Functional assays in tissue culture
100 µl cell suspension of SKNAS, SKNBE2 and NB69 (1 × 104 cells/well) was seeded in 96-well culture plates (Corning Incorporated). After culturing to 80% confluence the supernatant was removed and transfection media was added to the cells. 48 h post transfection, cell proliferation was measured using the MTS/MPS Cell Titer 96® AQueous One solution Cell Proliferation Assay (MTS) (G3582, Promega) and detecting the color variation (FLUOstar Omega, BMG Labtech) as per the manufacturer’s recommendations. The absorbance values were normalized to the mock transfection and expressed as a percentage. All experiments were repeated three times.
For the cell viability imaging assays, cells were stained with ready probes cell viability imaging kit (blue/green) (R37111, Invitrogen) or Cell event Caspase 3/7 staining kit (R37609; Invitrogen), according to the manufacturer’s instructions and imaged on EVOS M7000 to determine whether they were live (blue) or dead (green) or had active caspase 3/7 (green). The result was subsequently analyzed using EVOS analysis software (Version 1.4.998.659) to determine cell number. The relative number of cells was subsequently determined by normalizing against the control transfections.
Cell cycle analysis was performed using the Cell-clock cell cycle assay (C1000, Biocolor). Images were subsequently analyzed using ImageJ image analysis as per the manufacturer’s instructions. The data presented is the average of three biological replicates. Each experiment series was repeated in triplicate.
Quantitative PCR (qPCR) analysis
The total RNA was extracted using the mirVana™ miRNA Isolation Kit (AM1560, Thermo Fisher Scientific) and reverse transcribed in a 2-step process with TaqMan MicroRNA Assays Reverse Transcription Kit (4,366,597, Applied Biosystems). miRNA expression was quantified using hsa-miR-548l (002,904, Thermo Fischer Scientific), or hsa-miR-34a-5p (478,048, Thermo Fischer Scientific) and normalized to RNU6B (001,093, Thermo Fischer Scientific) using the Livak method54. RNA from transfected NB cell lines was extracted using the RNeasy plus mini kit® (Qiagen) according to the manufacturer’s protocol. The RNA concentration was quantified by NanoDrop (NanoDrop Technologies) and 10 ng of RNA was reverse transcribed into double stranded cDNA on a T-professional Basic Gradient thermal cycler (Biometra) using the TaqMan® MicroRNA Reverse Transcription Kit (4,366,596, Applied Biosystems). cDNA corresponding to 20 ng of RNA for each qPCR reaction was used. qPCR was performed on a Pikoreal qPCR System (Thermo Fisher Scientific) in triplicate.
Statistical analysis
Data presented are plotted as either mean ± SD, Tukeys box and whisker plots showing IQR, line at the median, + at the mean with whiskers ± 1.5-fold of interquartile range or scatter plot from at least 3 independent experiments. For all multi-group analyses, differences were determined by one-way ANOVA test followed by Holm-Šídák's multiple comparison test. For comparisons between two unpaired groups a Mann–Whitney U test was used. For comparisons between paired samples a paired t-test was used: with a p value < 0.05 considered significant. *p < 0.05, **p < 0.01, ***p < 0.001. All analyses were conducted using GraphPad Prism version 9.2.0 for Windows, (GraphPad Software, www.graphpad.com).
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
References
Pugh, T. J. The genetic landscape of high-risk neuroblastoma. Nat. Genet., 279 (2013).
Caren, H. et al. High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc. Natl. Acad. Sci. US https://doi.org/10.1073/pnas.0910684107 (2010).
Cohn, S. L. et al. The international neuroblastoma risk group (INRG) classification system: An INRG task force report. J. Clin. Oncol. 27, 289–297. https://doi.org/10.1200/jco.2008.16.6785 (2009).
Hill-Kayser, C. E. et al. Outcomes after proton therapy for treatment of pediatric high-risk neuroblastoma. Int. J. Radiat. Oncol. Biol. Phys. 104, 401–408. https://doi.org/10.1016/j.ijrobp.2019.01.095 (2019).
Salloum, R. et al. Relapsed perinatal neuroblastoma after expectant observation. Pediatr. Blood Cancer 62, 160–162. https://doi.org/10.1002/pbc.25218 (2015).
Mlakar, V. et al. 11q deletion in neuroblastoma: a review of biological and clinical implications. Mol. Cancer 16, 114. https://doi.org/10.1186/s12943-017-0686-8 (2017).
Keane, S., Améen, S., Lindlöf, A. & Ejeskär, K. Low DLG2 gene expression, a link between 11q-deleted and MYCN-amplified neuroblastoma, causes forced cell cycle progression, and predicts poor patient survival. Cell Commun. Signal 18, 65. https://doi.org/10.1186/s12964-020-00553-6 (2020).
Mandriota, S. J. et al. Ataxia-telangiectasia mutated (ATM) silencing promotes neuroblastoma progression through a MYCN independent mechanism. Oncotarget 6, 18558–18576. https://doi.org/10.18632/oncotarget.4061 (2015).
Ando, K. et al. Expression of TSLC1, a candidate tumor suppressor gene mapped to chromosome 11q23, is downregulated in unfavorable neuroblastoma without promoter hypermethylation. Int. J. Cancer 123, 2087–2094. https://doi.org/10.1002/ijc.23776 (2008).
Lau, N. C., Lim, L. P., Weinstein, E. G. & Bartel, D. P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858–862. https://doi.org/10.1126/science.1065062 (2001).
Lee, R. C. & Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 294, 862–864. https://doi.org/10.1126/science.1065329 (2001).
Friedman, R. C., Farh, K. K., Burge, C. B. & Bartel, D. P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. https://doi.org/10.1101/gr.082701.108 (2009).
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. https://doi.org/10.1016/s0092-8674(04)00045-5 (2004).
Behm-Ansmant, I. et al. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 20, 1885–1898. https://doi.org/10.1101/gad.1424106 (2006).
Krutzfeldt, J., Poy, M. N. & Stoffel, M. Strategies to determine the biological function of microRNAs. Nat. Genet. 38(Suppl), S14-19. https://doi.org/10.1038/ng1799 (2006).
Peng, Y. & Croce, C. M. The role of MicroRNAs in human cancer. Signal Transduct. Target Ther. 1, 15004. https://doi.org/10.1038/sigtrans.2015.4 (2016).
Lu, J. et al. MicroRNA expression profiles classify human cancers. Nature 435, 834–838. https://doi.org/10.1038/nature03702 (2005).
Hammond, S. M. MicroRNAs as oncogenes. Curr. Opin. Genet. Dev. 16, 4–9. https://doi.org/10.1016/j.gde.2005.12.005 (2006).
Kent, O. A. & Mendell, J. T. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene 25, 6188–6196. https://doi.org/10.1038/sj.onc.1209913 (2006).
Wang, Z. et al. Reduction of miR-21 induces SK-N-SH cell apoptosis and inhibits proliferation via PTEN/PDCD4. Oncol. Lett. 13, 4727–4733. https://doi.org/10.3892/ol.2017.6052 (2017).
Roth, S. A. et al. MicroRNA-193b-3p represses neuroblastoma cell growth via downregulation of Cyclin D1, MCL-1 and MYCN. Oncotarget 9, 18160–18179. https://doi.org/10.18632/oncotarget.24793 (2018).
Siaw, J. T. et al. 11q Deletion or ALK activity curbs DLG2 expression to maintain an undifferentiated state in neuroblastoma. Cell Rep. 32, 108171. https://doi.org/10.1016/j.celrep.2020.108171 (2020).
Chin, C. H. et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8 Suppl 4, S11, doi:https://doi.org/10.1186/1752-0509-8-S4-S11 (2014).
Bindea, G., Galon, J. & Mlecnik, B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics 29, 661–663. https://doi.org/10.1093/bioinformatics/btt019 (2013).
Upton, K. et al. Epigenomic profiling of neuroblastoma cell lines. Sci. Data 7, 116. https://doi.org/10.1038/s41597-020-0458-y (2020).
Sussman, R. T. et al. CAMKV is a candidate immunotherapeutic target in MYCN amplified neuroblastoma. Front. Oncol. 10, 302. https://doi.org/10.3389/fonc.2020.00302 (2020).
Zeid, R. et al. Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nat. Genet. 50, 515–523. https://doi.org/10.1038/s41588-018-0044-9 (2018).
Shen, X. et al. YAP promotes the proliferation of neuroblastoma cells through decreasing the nuclear location of p27(Kip1) mediated by Akt. Cell Prolif. 53, e12734. https://doi.org/10.1111/cpr.12734 (2020).
Misiak, D. et al. The MicroRNA landscape of MYCN-amplified neuroblastoma. Front. Oncol. 11, 647737. https://doi.org/10.3389/fonc.2021.647737 (2021).
Caren, H. et al. High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc. Natl. Acad. Sci. USA 107, 4323–4328. https://doi.org/10.1073/pnas.0910684107 (2010).
Tonini, G. P., Nakagawara, A. & Berthold, F. Towards a turning point of neuroblastoma therapy. Cancer Lett 326, 128–134. https://doi.org/10.1016/j.canlet.2012.08.017 (2012).
Poon, C. L., Mitchell, K. A., Kondo, S., Cheng, L. Y. & Harvey, K. F. The hippo pathway regulates neuroblasts and brain size in drosophila melanogaster. Curr. Biol. 26, 1034–1042. https://doi.org/10.1016/j.cub.2016.02.009 (2016).
Johnsen, J. I., Dyberg, C. & Wickstrom, M. Neuroblastoma-A neural crest derived embryonal malignancy. Front. Mol. Neurosci. 12, 9. https://doi.org/10.3389/fnmol.2019.00009 (2019).
Fernandez, L. A. et al. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 23, 2729–2741. https://doi.org/10.1101/gad.1824509 (2009).
Zhang, L. et al. The hippo pathway effector YAP regulates motility, invasion, and castration-resistant growth of prostate cancer cells. Mol. Cell Biol. 35, 1350–1362. https://doi.org/10.1128/mcb.00102-15 (2015).
Steinhardt, A. A. et al. Expression of yes-associated protein in common solid tumors. Hum. Pathol. 39, 1582–1589. https://doi.org/10.1016/j.humpath.2008.04.012 (2008).
Yang, C., Tan, J., Zhu, J., Wang, S. & Wei, G. YAP promotes tumorigenesis and cisplatin resistance in neuroblastoma. Oncotarget 8, 37154–37163. https://doi.org/10.18632/oncotarget.16209 (2017).
Huang, T. T., Lampert, E. J., Coots, C. & Lee, J. M. Targeting the PI3K pathway and DNA damage response as a therapeutic strategy in ovarian cancer. Cancer Treat. Rev. 86, 102021. https://doi.org/10.1016/j.ctrv.2020.102021 (2020).
Tran, H. et al. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science 296, 530–534. https://doi.org/10.1126/science.1068712 (2002).
Hagenbuchner, J. et al. Nuclear FOXO3 predicts adverse clinical outcome and promotes tumor angiogenesis in neuroblastoma. Oncotarget 7, 77591–77606. https://doi.org/10.18632/oncotarget.12728 (2016).
Liu, X. et al. Deregulated Wnt/beta-catenin program in high-risk neuroblastomas without MYCN amplification. Oncogene 27, 1478–1488. https://doi.org/10.1038/sj.onc.1210769 (2008).
Alshareef, A. et al. High expression of β-catenin contributes to the crizotinib resistant phenotype in the stem-like cell population in neuroblastoma. Sci. Rep. 7, 16863. https://doi.org/10.1038/s41598-017-17319-9 (2017).
Regan, P. L. et al. Hsp90 inhibition increases p53 expression and destabilizes MYCN and MYC in neuroblastoma. Int. J. Oncol. 38, 105–112 (2011).
Rodrigo, M. A. M. et al. Transcriptomic Landscape of Cisplatin-Resistant Neuroblastoma Cells. Cells 8, doi:https://doi.org/10.3390/cells8030235 (2019).
Dharap, A., Pokrzywa, C., Murali, S., Pandi, G. & Vemuganti, R. MicroRNA miR-324-3p induces promoter-mediated expression of RelA gene. PLoS ONE 8, e79467. https://doi.org/10.1371/journal.pone.0079467 (2013).
Keane, S., Martinsson, T., Kogner, P. & Ejeskär, K. The loss of DLG2 isoform 7/8, but not isoform 2, is critical in advanced staged neuroblastoma. Cancer Cell Int. 21, 170. https://doi.org/10.1186/s12935-021-01851-w (2021).
Wang, Y. et al. Functionalized graphene oxide triggers cell cycle checkpoint control through both the ATM and the ATR signaling pathways. Carbon 129, 495–503 (2018).
Sanmartin, E. et al. Deletion of 11q in Neuroblastomas Drives Sensitivity to PARP Inhibition. Clin. Cancer Res. 23, 6875–6887. https://doi.org/10.1158/1078-0432.CCR-17-0593 (2017).
Sethupathy, P., Corda, B. & Hatzigeorgiou, A. G. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA 12, 192–197. https://doi.org/10.1261/rna.2239606 (2006).
Lewis, B. P., Burge, C. B. & Bartel, D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20. https://doi.org/10.1016/j.cell.2004.12.035 (2005).
service integration into miRNA functional analysis workflows. Paraskevopoulou, M. D. et al. DIANA-microT web server v5.0. Nucleic Acids Res. 41, W169-173. https://doi.org/10.1093/nar/gkt393 (2013).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198. https://doi.org/10.1093/nar/gkz369 (2019).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. https://doi.org/10.1006/meth.2001.1262 (2001).
Acknowledgements
We thank the Swedish Childhood Cancer Fund and Assar Gabrielsson Found for financial support.
Funding
Open access funding provided by University of Skövde.
Author information
Authors and Affiliations
Contributions
S.J. have collected data, performed the analyses, prepared Figs. 1, 2 and 3 and wrote the manuscript. S.K. developed the experimental methodology, performed the assays, analysed, organised and interpreted the data, prepared Figs. 4, 7 and 8 and wrote the manuscript. E.B. and Z.L.P. participated in the bioinformatics analysis of data and design. Z.L.P. has also prepared Figs. 5 and 6. K.E. designed the study, provided technical and material support, and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jurcevic, S., Keane, S., Borgmästars, E. et al. Bioinformatics analysis of miRNAs in the neuroblastoma 11q-deleted region reveals a role of miR-548l in both 11q-deleted and MYCN amplified tumour cells. Sci Rep 12, 19729 (2022). https://doi.org/10.1038/s41598-022-24140-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-24140-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.