Abstract
The current study aimed to test whether the ratio of T1-weighted to T2-weighted signal intensity (T1W/T2W ratio: rT1/T2) derived from conventional MRI could act as a surrogate relaxation time predictive of IDH mutation status in histologically lower-grade gliomas. Strong exponential correlations were found between rT1/T2 and each of T1- and T2-relaxation times in eight subjects (rT1/T2 = 1.63exp−0.0005T1-relax + 0.30 and rT1/T2 = 1.27exp−0.0081T2-relax + 0.48; R2 = 0.64 and 0.59, respectively). In a test cohort of 25 patients, mean rT1/T2 (mrT1/T2) was significantly higher in IDHwt tumors than in IDHmt tumors (p < 0.05) and the optimal cut-off of mrT1/T2 for discriminating IDHmt was 0.666–0.677, (AUC = 0.75, p < 0.05), which was validated in an external domestic cohort of 29 patients (AUC = 0.75, p = 0.02). However, this result was not validated in an external international cohort derived from TCIA/TCGA (AUC = 0.63, p = 0.08). The t-Distributed Stochastic Neighbor Embedding analysis revealed a greater diversity in image characteristics within the TCIA/TCGA cohort than in the two domestic cohorts. The failure of external validation in the TCIA/TCGA cohort could be attributed to its wider variety of original imaging characteristics.
Similar content being viewed by others
Introduction
Genetic characterization is an essential diagnostic work-up in patients with glioma, as presurgical prediction of the IDH mutation status helps choose the most appropriate treatment strategy. Rapid and aggressive surgical intervention should be pursued for IDH-wildtype (IDHwt) gliomas, which have a poor prognosis1. In contrast, finding an optimal balance between maximal resection and preservation of the quality of daily life is of utmost importance for IDH-mutant (IDHmt) World Health Organization (WHO) grade 2 and 3 gliomas (lower-grade gliomas: LrGGs), as these present a more favorable prognosis1,2,3,4,5. There have been great efforts within the research community to develop noninvasive imaging techniques that can predict IDH mutation status in gliomas, including magnetic resonance spectroscopy (MRS)6,7,8, perfusion- and diffusion-weighted magnetic resonance imaging (MRI)9,10, and machine learning with conventional MRI5,11,12. Whereas these techniques rely mainly on quantitative or semi-quantitative MRI measurement, qualitative imaging features such as the “T2 fluid-attenuated inversion recovery (FLAIR) mismatch sign” have proven to be powerful imaging surrogate markers for predicting IDH mutation status in tumors that appear to be histologically LrGGs13,14,15. These qualitative imaging features are derived from radiologically quantitative measurements such as the T1- and T2-relaxation time15,16. Ideally, tissue relaxation time should be directly measured by MR relaxometry; however, this has not yet been incorporated into routine imaging protocols as it requires additional scan time.
The ratio of T1-weighted (T1W) to T2-weighted (T2W) signal intensity (T1W/T2W ratio: rT1/T2) derived from conventional MRI has been reported as quantitative imaging technique for detecting myelin integrity in the brain17,18,19 and could potentially be used as an imaging surrogate for tissue relaxation time. The aim of the present study was to test the hypothesis that rT1/T2 is predictive of IDH mutation status in histologically LrGGs.
Materials and methods
Patient cohort
The study was approved by the Institutional Review Boards of Asahikawa Medical University Hospital (approval No. 21041) and the Osaka International Cancer Institute (No. 1612065191). The requirement for written informed consent was waived for data collected retrospectively. Patients in prospectively recruited cohorts were provided with a detailed explanation and written informed consent was obtained from each patient or their family. The investigation was performed in accordance with all relevant local guidelines and regulations, and adhered to the tenets of the Declaration of Helsinki.
We first reanalyzed previously published T1- and T2-relaxometry data of histologically LrGGs16 and compared these T1- and T2-relaxometry data with rT1/T2 (the reanalyzed cohort, Fig. 1). The T1- and T2-relaxometry had been performed using MP2RAGE and multi-echo T2-weighted images of nine patients with glioma. Thus, we could access the T1- and T2-relaxometry along with the rT1/T2 data of these nine patients, and included the data of eight of these in our analyses, after excluding one patient with a K27M mutant tumor16.
Overall study cohort. First the correlation of rT1/T2 with T1- and T2-relaxation time was investigated by reanalyzing the raw data from ref.16. Then, the study was conducted in two stages, as an exploratory cohort study followed by a validation cohort study to investigate the correlation of rT1/T2 and IDH mutation status of histologically confirmed LrGGs. IDHmt, IDH-mutant; IDHwt, IDH-wildtype; AMUH, Asahikawa Medical University Hospital; OICI, Osaka International Cancer Institute; TCIA/TCGA, Cancer Imaging Archive / Cancer Genome Atlas.
We then prepared a new set of three cohorts and conducted a two-stage study (Fig. 1). The first cohort (exploratory cohort) comprised 25 histologically and molecularly confirmed histologically LrGGs (IDHwt: 8, IDHmt: 17) treated at Asahikawa Medical University Hospital (AMUH). The second and third cohorts were used as validation cohorts. Validation cohort 1 comprised 29 patients (IDHwt: 13, IDHmt: 16) from the Osaka International Cancer Institute (OICI) and validation cohort 2 comprised 101 patients (IDHwt: 19, IDHmt: 82) from the Cancer Imaging Archive (TCIA)/Cancer Genome Atlas (TCGA) low-grade glioma collection dataset, accessed on February 1, 202020,21. Validation cohort 1 can be considered as a “domestic” and validation cohort 2 as an “international” validation cohort.
The pathological diagnosis was based on the 2016 WHO Classification of Tumors of the Central Nervous System22. IDHwt tumors in the present cohorts could not be fully characterized according to the 2021 WHO classification system, as molecular analyses for such as TERT promoter mutation, EGFR gene amplification, and + 7/− 10 chromosome copy-number alterations had not been performed. The inclusion criterion was availability of T1WI and T2WI. Excluded were patients with failed image co-registration or insufficient or atypical images (e.g., with tumoral hemorrhage). Table S1 provides detailed information regarding all three cohorts.
Genetic analysis
Two laboratories performed genetic analyses of glioma tissues: the Department of Pathology, Asahikawa Medical University, Asahikawa, Japan, for the exploratory cohort; and the Department of Biomedical Research and Innovation, Institute for Clinical Research, Osaka National Hospital, Osaka, Japan, for the validation cohort 1. Immunobiological detection of IDH1 mutation was performed for the exploratory cohort, and Sanger sequencing was performed to detect hotspot mutations of IDH1/2 (codon 132 of IDH1 and codon 172 of IDH2) for the validation cohort 13. The IDH mutation status of tumors in the TCIA/TCGA dataset were obtained from the report by Ceccarelli et al.23.
T1w/T2w (rT1/T2) image reconstruction
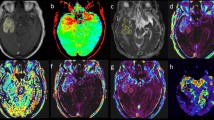
Table S1 lists the MRI acquisition parameters in detail. Most of the images in the exploratory cohort were acquired using General Electric 3 T scanners (Chicago, Illinois, USA), and those in the validation cohort 1 by Siemens 3 T scanners (Erlangen, Germany). Images in the TCIA cohort (the validation cohort 2) were acquired by 1.5 and 3 T scanners of various MRI vendors. T1WI and T2WI in Digital Imaging and COmmunication in Medicine (DICOM) format were converted into Neuroimaging Informatics Technology Initiative (NIfTI) format using Mango software (version 4.0.1; University of Texas Health Science Center, https://ric.uthscsa.edu/mango/mango.html, accessed on March 6, 2022). We used an in-house imaging software incorporating an algorithm for reconstructing rT1/T2 images from T1WI and T2WI24. The algorithm and MATLAB codes for calculating rT1/T2 can be obtained as an open-source toolbox for SPM12 developed by Ganzetti et al. (https://www.nitrc.org/projects/mrtool/, accessed on March 6, 2022)18. Details of the reconstruction analysis are previously reported by Ganzetti et al.18. Reconstruction was performed by first applying a bias field correction to the original T1WI and T2WI using SPM12 (https://www.fil.ion.ucl.ac.uk/spm/, accessed on March 6, 2022). The intensity histograms were then adjusted based on intensities extracted from non-brain tissues such as cerebrospinal fluid, bone, and soft tissues. Finally, the processed T2WI was co-registered and divided by the processed T1WI to produce an rT1/T2 image using the NIfTI “scanner-anatomical” coordinate system (Fig. 2).
Workflow across the entire study. T1WI to T2WI signal intensity ratio (rT1/T2) images were calculated from T1- and T2-weighted images, after image normalization via bias field correction and histogram matching. Voxels-of-interest (VOIs) were defined manually based on the high-intensity area of pathological lesions on T2-weighted images, followed by mean rT1/T2 measurement within the VOI.
T1- and T2-relaxometry of MP2RAGE and multi-echo T2WI
Imaging was performed on a 3 T MR scanner (Prisma; Siemens Healthcare, Erlangen, Germany). T1-relaxometry was performed by converting MP2RAGE images into T1-relaxation time maps. T2-relaxometry was performed by converting multi-echo T2-weighted images into T1-relaxation time maps. In both cases, relaxometry was performed via Bayesian inference modeling (Olea Nova+; Canon Medical Systems, Tochigi, Japan). Further technical details have been reported previously16.
Voxels-of-interests (VOIs) segmentation and calculation of mean rT1/T2
Author 1, with 6 years of neurosurgical experience, performed manual segmentation of the lesions by designing voxels-of-interests (VOIs) in ITK-SNAP software (version 3.8.0, http://www.itksnap.org, accessed on March 6, 2022). VOIs were designed on T2WIs with visual identification of pathologically high-intensity areas, avoiding ambiguous and vaguely abnormal lesions as much as possible (Fig. S1). The last author, with 22 years of neurosurgical experience, then evaluated the VOIs and either confirmed their position or requested modification (which occurred for five VOIs) (Table S1). The Dice similarity coefficient ranged from 0.52 to 0.81 for these VOIs (TCIA-00067, TCIA-00071, TCIA-00110, TCIA-00111, TCIA-00113). This procedure was performed on T2WI using the NIfTI “general affine transformation” coordinate system.
Each rT1/T2 image on NIfTI “scanner-anatomical” coordinate system was then co-registered with T2WI on NIfTI “general affine transformation” coordinate system using Volume Imaging in Neurological Research, Co-Registration and ROIs included (VINCI; Max Planck Institute for Neurological Research Cologne, Germany, http://www.nf.mpg.de/vinci3/, accessed on March 6, 2022), to ensure that further analysis will be performed using the NIfTI “general affine transformation” coordinate system (Fig. S2). Three-dimensional VOIs were then applied to the rT1/T2 images for calculation of mean rT1/T2 (mrT1/T2) within the VOIs (Fig. 2). VOIs were also applied to T1- and T2-relaxation time maps when those data were available.
Image feature extraction
Image features were extracted from T1WI and T2WI according to the method described previously25. T1WI and T2WI were converted into 256-level grayscale images after cutoff of the upper 0.1% of signal. This procedure was not performed for the rT1/T2 images, as these are quantitative in nature. First-order texture features were calculated based on histograms of the 256-level grayscale within VOIs in the T1WI, T2WI, and rT1/T2 images. Second-order texture features were measured in Gray Level Co-occurrence Matrix (GLGM) and Gray Level Run Length Matrix (GLRLM) analyses. In total, 49 imaging features were extracted from each image (Table S2). Details of the extracted imaging features have been provided previously25. Texture features were used solely to compare image characteristics among the cohorts, and were not used to predict IDH mutation status. Such multiparametric variables would require a large training dataset to establish a reliable prediction model.
Statistical and t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis
Statistical analysis was performed using Prism 9 for macOS (GraphPad Software, San Diego, CA, USA). The relationship between mrT1/T2 and IDH mutation status was investigated by Mann–Whitney U test and receiver-operating characteristic (ROC) curve analysis. A p-value of less than 0.05 was considered significant. t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis was used to investigate the difference in MRI qualities and characteristics among the three cohorts. Rtsne package version 0.15 for R with default parameters was used for this analysis (Tables S3, S4, and S5)26.
Results
Correlation between relaxometry and rT1/T2
In the initial investigation of the correlation of T1- and T2-relaxation time (abbreviated as T1-relax and T2-relax, respectively) and rT1/T2 within histologically LrGG tissues, voxel-wise analysis was performed for 114,465 voxels in total. T1- and T2-relax exhibited strong exponential correlations with rT1/T2 (rT1/T2 = 1.63exp−0.0005T1-relax + 0.30 and rT1/T2 = 1.27exp−0.0081T2-relax + 0.48; R2 = 0.64 and 0.59, respectively) (Fig. 3A,B). Further comparison of the difference in rT1/T2 between IDHwt and IDHmt voxels revealed significantly higher rT1/T2 in voxels of IDHwt tumors than in those of IDHmt tumors (median value = 1.06 vs. 0.73, p < 0.0001) (Fig. 3C). These findings supported further investigation to test the hypothesis of rT1/T2 being an imaging surrogate for prediction of the IDH mutation status of histologically LrGGs.
Voxel-wise comparisons between rT1/T2 and each of T1- and T2-relaxation times, according to reanalysis of data published previously (the reanalyzed cohort)16. The data consisted of 114,465 voxels from eight patients. T1- and T2-relaxation times both exhibited strong exponential correlations with rT1/T2 (A and B). Voxels in IDHwt tumors (26,969 voxels) showed significantly higher rT1/T2 than those in IDHmt tumors (87,496 voxels) (median value = 1.06 vs. 0.73, p < 0.0001) (C)16.
Determination of the cut-off value of rT1/T2 to differentiate the IDH mutation status of LrGG
The exploratory cohort from AMUH was used to explore and determine the cut-off value of mean rT1/T2 (mrT1/T2) for differentiating IDHwt and IDHmt tumors. As shown in Fig. 4A, mrT1/T2 was significantly higher in IDHwt tumors than in IDHmt tumors (median value = 0.74 vs. 0.66, p < 0.05). ROC curve analysis revealed that the optimal cut-off value of mrT1/T2 for discriminating IDHmt was 0.666–0.677, with sensitivity of 58.8% and specificity of 87.5% (AUC = 0.75, p < 0.05) (Fig. 4B).
Exploration of rT1/T2 cut-off for IDH mutation in the exploratory cohort from AMUH (study 1). (A) There was statistically significant difference in mean rT1/T2 between the IDH-wildtype and IDH-mutant groups (p = 0.0495 < 0.05) (A). ROC curve for detecting IDH-mutant LrGGs shows AUC of 0.75, p = 0.0486. The red star indicates an ideal cut-off for mrT1/T2 of < 0.677, with sensitivity of 58.8% and specificity of 87.5% (B).
Validation of the cut-off value of rT1/T2 in the domestic and TCIA/TCGA datasets
The determined mrT1/T2 cut-off values were then validated using a domestic cohort (the validation cohort 1 from OICI) and an international public dataset (TCIA/TCGA cohort). The OICI cohort validated a statistically significant difference in mrT1/T2 between IDHwt and IDHmt tumors (median value = 0.83 vs. 0.63, p = 0.02) (Fig. 5A). ROC analysis of the OICI cohort showed AUC of 0.75 (p = 0.02) with sensitivity of 56.3% and specificity of 69.2%, using the previously obtained cut-off value for detecting IDHmt tumors (Fig. 5B).
Validation of rT1/T2 cut-off for IDH mutation in the validation cohort 1 (study 2 using the domestic OICI cohort). There was statistically significant difference in mean rT1/T2 between the IDH-wildtype and IDH-mutant groups (p = 0.022, p < 0.05) (A). ROC curve for detecting IDH-mutant LrGGs shows AUC of 0.75, p = 0.023 (p < 0.05), with sensitivity of 56.3% and specificity of 69.2% (B).
In the validation cohort 2 (the TCIA/TCGA cohort), there was no significant difference in mrT1/T2 between IDHwt and IDHmt tumors (p = 0.08) (Fig. 6A). ROC analysis of the validation cohort 2 (the TCIA/TCGA cohort) showed sensitivity of 42.7% and specificity of 73.7% using the previously obtained cut-off value for detecting IDHmt LrGGs (AUC = 0.63, p = 0.08) (Fig. 6B).
Revalidation of rT1/T2 cut-off for IDH mutation in the validation cohort 2 (study 3 using the international cohort from TICA/TCGA). There was no significant difference in mean rT1/T2 between the IDH-wildtype and IDH-mutant groups (p = 0.083) (A). ROC curve for detecting IDH-mutant LrGGs shows AUC of 0.63, (p = 0.082), with sensitivity of 42.9% and specificity of 73.7% (B).
Impact of original image characteristics on rT1/T2 reconstruction
We further investigated the impact of the image features in the original T1WI and T2WI for reconstructing rT1/T2 images. t-SNE analysis of the original T1WI and T2WI revealed clusters of image features in the exploratory cohort (the AMUH cohort) and the validation cohort (the OICI cohort), whereas those in the validation cohort 2 (the TCIA/TCGA cohort) were scattered across both the t-SNE1 and t-SNE axes (Fig. 7A). Reconstruction of rT1/T2 from T1WI and T2WI resulted in more dense clustering of all data points (Fig. 7B), bringing data points of the AMUH and OICI cohorts closer. However, data points obtained from the TCIA/TCGA cohort remained scattered compared to the two domestic cohorts. Further analysis revealed that the scattered data points of the TCIA/TCGA cohort were partially attributed to the variety of institutions consisting of the entire cohort (Fig. S3A). Reconstructing rT1/T2 from T1WI and T2WI was insufficient to cluster all data points, which was particularly apparent in data from Henry Ford Hospital (Fig. S3B).
t-SNE analyses of both the original T1WI and T2WI features and the rT1/T2 features of the entire cohort were unable to discriminate the IDH mutation status (Fig. S4).
Discussion
Along with others, we have reported that the T1- and T2-relaxation properties of tumor tissue tightly correlate with the IDH mutation status of LrGGs16,27. These observations were obtained either by employing conventional MR relaxometry techniques16,28 or by newly developed magnetic resonance fingerprinting technology29. However, neither technology has been incorporated into routine LrGG imaging studies as yet due to additional requirements in terms of scan time16 or sequence and postprocessing optimization30,31. In contrast, conventional structural MRI (i.e., T1WI and T2WI) can partially represent the relaxation properties of a tissue following radiofrequency-induced nuclear magnetic resonance spin polarization. For example, the T2-FLAIR mismatch sign utilizes the qualitative imaging features represented on conventional structural MRI derived from quantitative tissue relaxation properties, which enable detection of IDH-mutant 1p/19q intact tumors13,14,15,16.
The present study aimed to explore the possibility of retrieving quantitative data from qualitative conventional structural MRI by utilizing rT1/T2, a method initially proposed as an imaging surrogate of myelin content within the brain17. Although the exact pathological mechanism is unknown19,32, the relatively high sensitivity of rT1/T2 for detecting multiple sclerosis suggests that it might be suitable for characterizing tissue microstructure within the brain33. As shown in Fig. 3, a good correlation was demonstrated between rT1/T2 and the T1- and T2-relaxation times of tissues. Furthermore, in the two domestic cohorts of patients with histologically LrGGs, we explored and validated that the mean value of rT1/T2 within the lesion could differentiate IDHwt from IDHmt tumors with sensitivity of 56%–59% and specificity of 69–88% (Figs. 4 and 5). These encouraging findings supported the hypothesis that rT1/T2 could be usable for the presurgical determination of IDH mutation status in histologically LrGGs, without performing direct measurement of the tissue relaxation properties.
However, the results obtained in the domestic cohorts (the validation cohort 1) were not reproduced in the TCIA/TCGA cohort (the validation cohort 2) (Fig. 6). Although the trend of lower rT1/T2 in IDHmt tumors than in IDHwt tumors was reproduced, the difference was much smaller compared with that found in the domestic cohorts. We consider that this discrepancy is due to differences in image quality and image characteristics between the domestic cohorts (the exploratory and validation 1 cohort) and the TCIA/TCGA cohort (the validation cohort 2). As expected, there was a greater diversity of imaging features in the TCIA/TCGA cohort than in the domestic cohorts, presumably because the TCIA/TCGA cohort contained images from four different medical institutions within the United States (Fig. 7 and Fig. S3)34. Table S1 shows that each institution had distinct preferences in MRI acquisition parameters, which could have impacted the final analysis. This argument is confirmed by the results of a recent multicenter clinical investigation of rT1/T2 in multiple sclerosis, which employed various MR scanners of different field strengths. A wide variation in rT1/T2 was observed throughout the entire cohort, presumably due to the heterogenic nature of the input data35. Thus, to use rT1/T2 for predicting IDH mutation status, it might be necessary to use a preset MRI acquisition protocol or an institution-dependent cut-off value.
Several limitations of the present study must be addressed. First, the sample sizes of the two domestic cohorts were relatively small, and the distributions of IDHwt and IDHmt differed among the three cohorts. This distribution difference might be responsible for the failure to validate the cut-off value in the TCIA/TCGA cohort. Ideally, a cohort with a larger sample size and an even distribution of tumors with IDH mutation status is necessary to validate our findings. Second, the overall sensitivity and specificity were not high enough to be acceptable in routine clinical use. The present retrospective study was unable to integrate other imaging modalities for analysis due to insufficient data; however, combining rT1/T2 with diffusion and perfusion MRI might increase diagnostic accuracy, and such research should be pursued in the future. For example, a recent study proposed the concept of combining diffusion and perfusion MRI for IDH mutation detection, but requires external validation36. Third, some of the procedures performed during rT1/T2 reconstruction are highly complicated, as mentioned by Ganzetti et al.18, especially image intensity normalization. The normalization method used in the present study calibrated image intensity using non-brain tissues, including cerebrospinal fluid, bone, and soft tissues; however, other methods such as deep learning could offer potential solutions to improve rT1/T2 analysis37. Finally, recent rapid developments in deep learning and artificial intelligence have augmented the diagnostic precision of IDH mutation status in glioma, with reported pooled sensitivity and specificity of 0.88 and 0.8612, which are much higher than those in the present study12. The most appropriate machine learning algorithm is yet to be determined. Therefore, the simplicity of rT1/T2 reconstructed by T1WI and T2WI alone remains appealing, withstanding the need for further improvements and validations.
Conclusions
The findings of the present study showed that rT1/T2 strongly correlates with T1- and T2-relaxation times in histologically LrGGs. The mean value of rT1/T2 was able to discriminate IDHwt and IDHmt tumors in two domestic cohorts, with statistical significance. However, this result was not validated in the original TCIA/TCGA cohort due to the wide variety of imaging characteristics in the cohort.
Data availability
The datasets used and analyzed in the current study are available from the corresponding author upon reasonable request.
References
Eckel-Passow, J. E. et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N. Engl. J. Med. 372, 2499–2508 (2015).
Suzuki, H. et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 47, 458–468 (2015).
Arita, H. et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol. Commun. 4, 79 (2016).
Arita, H. et al. IDH1/2 mutation detection in gliomas. Brain Tumor Pathol. 32, 79–89 (2015).
Arita, H. et al. Lesion location implemented magnetic resonance imaging radiomics for predicting IDH and TERT promoter mutations in grade II/III gliomas. Sci. Rep.-UK 8, 11773 (2018).
Choi, C. et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat. Med. 18, 624–629 (2012).
Andronesi, O. C. et al. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci. Transl. Med. 4, 116ra4 (2012).
Andronesi, O. C. et al. Detection of oncogenic IDH1 mutations using magnetic resonance spectroscopy of 2-hydroxyglutarate. J. Clin. Invest. 123, 3659–3663 (2013).
Leu, K. et al. Perfusion and diffusion MRI signatures in histologic and genetic subtypes of WHO grade II–III diffuse gliomas. J. Neuro-oncol. 134, 177–188 (2017).
Xing, Z. et al. Noninvasive assessment of IDH mutational status in world health organization grade II and III astrocytomas using DWI and DSC-PWI combined with conventional MR imaging. Am. J. Neuroradiol. 38, 1138–1144 (2017).
Zhao, J. et al. Diagnostic accuracy and potential covariates for machine learning to identify IDH mutations in glioma patients: Evidence from a meta-analysis. Eur. Radiol. 30, 4664–4674 (2020).
Jian, A. et al. Machine learning for the prediction of molecular markers in glioma on magnetic resonance imaging: A systematic review and meta-analysis. Neurosurgery 89, 31–44 (2021).
Patel, S. H. et al. T2–FLAIR mismatch, an imaging biomarker for IDH and 1p/19q status in lower-grade gliomas: A TCGA/TCIA project. Clin. Cancer Res. 23, 6078–6085 (2017).
Broen, M. P. G. et al. The T2-FLAIR mismatch sign as an imaging marker for non-enhancing IDH-mutant, 1p/19q-intact lower-grade glioma: A validation study. Neuro Oncol. 20, 1393–1399 (2018).
Kinoshita, M. et al. Impact of inversion time for FLAIR acquisition on the T2-FLAIR mismatch detectability for IDH-mutant, non-CODEL astrocytomas. Front. Oncol. 10, 596448 (2021).
Kinoshita, M. et al. T2-FLAIR mismatch sign is caused by long T1 and T2 of IDH-mutant, 1p19q non-codeleted astrocytoma. Magn. Reson. Med. Sci. 20, 119–123 (2021).
Glasser, M. F. & Essen, D. C. V. Mapping human cortical areas in vivo based on myelin content as revealed by T1- and T2-weighted MRI. J. Neurosci. 31, 11597–11616 (2011).
Ganzetti, M., Wenderoth, N. & Mantini, D. Whole brain myelin mapping using T1- and T2-weighted MR imaging data. Front. Hum. Neurosci. 8, 671 (2014).
Pareto, D. et al. Ratio of T1-weighted to T2-weighted signal intensity as a measure of tissue integrity: Comparison with magnetization transfer ratio in patients with multiple sclerosis. Am. J. Neuroradiol. 41, 461–463 (2020).
The_Cancer_Genome_Atlas. https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga (2020). Accessed 1 February 2020.
The_Cancer_Imaging_Archive. https://public.cancerimagingarchive.net/ncia/login.jsf. (2020). Accessed 1 February 2020.
International_Agency_for_Research_on_Cancer. WHO Classification of Tumours of the Central Nervous System (International Agency for Research on Cancer, 2016).
Ceccarelli, M. et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164, 550–563 (2016).
Yamamoto, S. et al. Prediction and visualization of non-enhancing tumor in glioblastoma via T1w/T2w-ratio map. Brain Sci. 12, 99 (2022).
Sasaki, T. et al. Radiomics and MGMT promoter methylation for prognostication of newly diagnosed glioblastoma. Sci. Rep.-UK 9, 14435 (2019).
van der Maaten, L. J. P. & Hinton, G. E. Visualizing data using t-SNE. J. Mach. Learn Res. 9, 2579–2605 (2008).
Springer, E. et al. MR fingerprinting—A radiogenomic marker for diffuse gliomas. Cancers 14, 723 (2022).
Kinoshita, M. et al. Magnetic resonance relaxometry for tumor cell density imaging for glioma: An exploratory study via 11C-methionine PET and its validation via stereotactic tissue sampling. Cancers 13, 4067 (2021).
Ma, D. et al. Magnetic resonance fingerprinting. Nature 495, 187–192 (2013).
Poorman, M. E. et al. Magnetic resonance fingerprinting Part 1: Potential uses, current challenges, and recommendations. J. Magn. Reson. Imaging 51, 675–692 (2019).
McGivney, D. F. et al. Magnetic resonance fingerprinting review part 2: Technique and directions. J. Magn. Reson. Imaging 51, 993–1007 (2019).
Preziosa, P. et al. Neurite density explains cortical T1-weighted/T2-weighted ratio in multiple sclerosis. J. Neurol. Neurosurg. Psychiatry. JNNP. https://doi.org/10.1136/jnnp-2020-324391 (2021).
Boaventura, M. et al. T1/T2-weighted ratio in multiple sclerosis: A longitudinal study with clinical associations. Neuroimage Clin. 34, 102967 (2022).
Choi, Y. S. et al. Fully automated hybrid approach to predict the IDH mutation status of gliomas via deep learning and radiomics. Neuro Oncol. 23, 304–313 (2020).
Margoni, M. et al. Exploring in vivo multiple sclerosis brain microstructural damage through T1w/T2w ratio: A multicentre study. J. Neurol. Neurosurg. Psychiatry 93, 741–752 (2022).
Pruis, I. J. et al. Noninvasive differentiation of molecular subtypes of adult nonenhancing glioma using MRI perfusion and diffusion parameters. Neuro-oncol. Adv. 4, vdac023 (2022).
Hu, Z. et al. MIL normalization—Prerequisites for accurate MRI radiomics analysis. Comput. Biol. Med. 133, 104403 (2021).
Acknowledgements
This research was funded by the Japan Society for the Promotion of Science (Nos. 19K09526, 22K09200), the Japan Agency for Medical Research and Development (Japan Cancer Research Project, Nos. 19188187 and 21459042), the Takeda Science Foundation, the MSD Life Science Foundation, and the Okawa Foundation for Information and Telecommunications. All scientific grants were for MK.
Author information
Authors and Affiliations
Contributions
M.K. conceived the experiment; T.S., M.S., H.S., M.S., N.M., S.H., R.A., Y.K., M.T., and K.N. collected data; T.S., S.Y., M.S., T.U., Y.K., M.T., K.N. and M.K. conducted the experiment; T.S. and M.K. wrote the manuscript; and H.K. and M.K. supervised the research project. All authors have reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sanada, T., Yamamoto, S., Sakai, M. et al. Correlation of T1- to T2-weighted signal intensity ratio with T1- and T2-relaxation time and IDH mutation status in glioma. Sci Rep 12, 18801 (2022). https://doi.org/10.1038/s41598-022-23527-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-23527-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.