Abstract
Gut microbiota studies of ethnic populations reveal gut microbial biomarkers for therapeutic options and detection of the disease state. The present study aimed to analyze the gut microbiome signatures in thirty individuals from the Adi, Apatani and Nyshi tribes of Arunachal Pradesh (ten in each cohort) by sequencing the V3 and V4 regions of 16S rRNA on the Illumina MiSeq Platform. The gut microbiome was highly predominated by Firmicutes, Actinobacteria, Proteobacteria, and Bacteroidates in the three studied tribal groups. At the genus level, significant abundance of Bifidobacterium, Collinsella, Bacteroides, Prevotella, Lactobacillus, Streptococcus, Clostridium, Coprococcus, Dorea, Lachnospira, Roseburia, Ruminococcus, Faecalibacterium, Catenibacterium, Eubacterium, Citrobacter and Enterobacter were observed amongst the three tribes. The tribal communities residing in remote areas and following traditional lifestyle had higher gut microbiome diversity with a high prevalence of Prevotella and Collinsella in the Adi and Nyshi tribes, and Bifidobacterium and Catenibacterium in the Apatani tribe. Elucidating the gut microbiome of the tribal community of Arunachal Pradesh will add to the knowledge on relationships between microbial communities, dietary food factors, and the overall state of health of humans worldwide.
Similar content being viewed by others
Introduction
The human gut microbiota comprises of millions of bacterial cells belonging to several species and modulates many host processes, including metabolism, inflammation, and immune and cellular responses1. Other than this, human bone and tissue development can be influenced by human microbiota2 and a symbiotic relationship with host can also be maintained3. Diet, environment, geography, uses of antibiotics, and host genetics are some factors that could be responsible for making the gut microbiota diverse in structure4. Among these varied factors, diet and geography were found to play vital roles in determining the constitution of the human gut microbiome5,6,7,8. Recently, few studies carried out in rural preindustrialized societies including those still dependant on hunting wild animals, consumption of medicinal plants and populations residing in geographically diverse locations have revealed specific gut microbiota adaptations based on food habits and lifestyle9,10,11,12,13,14. Population based studies may help in understanding the gut microbiota as a target for medical interventions such as the role of fecal microbial transplantations in restoration of the healthy state15.
India harbors the second largest population in the world spread across multiple geographical locations with enormous diversity in ethnicity, lifestyle, and dietary habits16,17, and few studies on gut microbiome diversity in healthy individuals based on ethnicity, diet, and gender have been previously reported15,18. Firmicutes are the most common bacteria in healthy Indians' guts, followed by Bacteroidetes, Actinobacteria, Proteobacteria, Spirochetes, Verrucomicrobia, and Fusobacteria19.The geographically isolated tribal populations of India continue to follow a traditional lifestyle, diet and plant-based medicine, and are largely unexposed to modern lifestyles. They therefore provide a unique opportunity to study the human microbiome unaffected by a modern lifestyle. Previous studies have reported the gut microbiota of the ethnic tribes of India to consist mainly of the genera Prevotella, Faecalibacterium, Eubacterium, Clostridium, Blautia, Collinsella, Ruminococcus, Roseburia, Dialister, and Veillonella20,21,22,23,24. One study reported that the gut microbiota of the Ahom and Bodo ethnic groups who do not consume rice beer mainly consisted of Faecalibacterium, Roseburia, Pasteuri, Syntrophomonas, Acetivibrio, Barnesiella, Alistipes, Oxalobacter, Desulfosporosinus, Akkermensia, Anaerobranca where as Pediococcus, Leptotrichia, Fusibacter, Streptococcus, Nitrosomonas, Cystobacter, Bavaricoccus, Peptococcus, Soehngenia and Tistrella while the gut microbiota of rice beer consuming tribal people contained Fusobacterium and Succinivibrio21.
The state of Arunachal Pradesh is located in the foothills of the Himalayas in India’s North-Eastern region, and is considered to be India’s remotest state. It is home to several tribes and sub-tribes, but the gut microbiome signatures of the tribal population from Arunachal Pradesh have not yet been characterized. This study therefore seeks to explore the gut microbiome diversity of healthy individuals belonging to the Adi, Apatani, and Nyshi tribes of Arunachal Pradesh from North East India, by performing 16S metagenomics sequencing of the fecal microbiota of these tribal populations.
Methods and materials
Sample size
A total of 30 stool samples were collected through conventional sampling process based on the inclusion and exclusion criteria listed below.
Exclusion criteria were as follows: pregnant women, women attempting to conceive, or nursing mothers; respondents taking any other drugs for long term (including antibiotics during the last 6 months); respondents with acute and chronic gastrointestinal tract diseases such as inflammatory bowel syndrome (IBS), food allergy and lactose intolerance; respondents with one or more co-morbidities such as type 2 diabetes, retinopathy, neuropathy, nephropathy and cardiovascular diseases; respondents with a history of organ transplantation and diseases of the oral cavity. Inclusion criteria were healthy males and females belonging to the age group 20–60 years.
Study design
Details of the dietary habits, age, and physical status of the respondents belonging to Adi, Apatani and Nyshi tribes of Arunachal Pradesh were recorded. Those individuals who were willing to participate were enrolled in the study (n = 30). Ethics approval for this study was obtained from the Human Ethical Committee, Assam University, Silchar, Assam, India (Ref. NO: IEC/AUS/2019/31/IS, Dated:13/11/2019).
Sample collection and isolation of DNA
Informed consent was obtained from the enrolled participants using a consent document prior to sample collection and each participant was provided adequate training and instructions regarding the stool collection process. Fecal samples were collected from the volunteers in RNAlater™ (Cat. no. 76104, QIAGEN, and Germany) solution in sterile stool collection tubes and stored at − 80 °C immediately after transportation to the laboratory. DNA was extracted from stool samples using the QIAamp DNA Stool Mini Kit (QIAGEN, Hilden, Germany), following manufacturer’s protocol. DNA concentration was measured using Bio Photometer Plus (Eppendorf, Hamburg, Germany) and the extracted DNA was stored at − 80 °C until further analysis25,26.
Metagenomics analysis using next-generation sequencing (NGS)
16S V3-V4 metagenome library was prepared using V3–V4 region specific primers targeting the Bacterial 16S rRNA gene from stool DNA as per the Illumina protocol27. Nextera XT Index Kit (Illumina) was used for sample indexing PCR. The amplicon size of the library was approximately 480 bp and library was purified using AMPure XP beads (Agencourt, Beckman Coulter, USA) and the library was quantified by using Quant-iT™ PicoGreen® dsDNA Reagent (Molecular Probes) at Qubit ® 2.0 Fluorimeter (Life Technologies, USA). The index PCR products were equimolar amounts of each library and were pooled and quantified using DNA High Sensitivity chip at Bioanalyzer (Agilent Technologies) after gel purification. Cluster generation and paired-end sequencing of library mixture was performed using a 500-cycle reagent kit (Illumina MiSeq) as per manufacturer’s instruction25.
Bioinformatics and statistical analysis
Bcl2Fastq tool was used for the demultiplexing of paired-end V3–V4 reads generated by the Illumina MiSeq platform. Sequence Reads were further assessed based on the quality score by using FastQC (http://www.bioinformatics.babraham.ac.uk/?/projects/fastqc/). Only high quality reads which were joined by Fastq join were considered significant and taken for subsequent analysis28. Sequences containing ambiguous bases, homopolymer run exceeding 6, mismatches in primers, or sequence length shorter than 100 bp was removed. The reads were then further analyzed using QIIME (Quantitative Insights Into Microbial Ecology) pipeline28. Operational Taxonomic Unit (OTU) were picked at 97% identity against the Green Genes database (v 13.8), and the OTUs were assigned taxonomy based on 97% similarity to the reference sequence29. UCLUST method was used to cluster query sequences against a curated chimera free 16S rRNA database (Greengenes v 13.8). All summaries of the taxonomic distributions ranging from phylum to genus were generated from the non-rarefied OTU table generated from this analysis. Generated biom file was then used for further analysis. Clustering was performed on normalized OTU abundances. OTUs representing less than 0.005% of the overall sequence were removed. To characterize diversity across individuals, Alpha-diversity was calculated using Shanon index, inverse Simpson. Beta diversity Principal coordinate analysis (PCoA) was calculated using QIIME. Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) tool was used to generate biom file for functional analysis. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database (www.kegg.jp) freely available for academic users was used for analysis of the functional compositions of bacterial communities30,31,32. Programs such as ‘adonis’ were used to perform the Permutational Multivariate Analysis of Variance, PERMANOVA (weighted and unweighted unifrac distance matrix), in the R package of ‘vegan’. A heat map was generated using relative abundance values for the top 20 taxa at all taxonomic levels (phylum to genus) across different groups using MEGAN6 (MEGAN_Community_windows-x64_6_24_1.exe). The parametric G test was used to determine a statistically significant difference between the abundance of OTUs in different sample groups by comparing the ratio of the observed OTU frequencies in the sample groups to the expected frequencies based on the extrinsic hypothesis. Kruskal–wallis H test was performed in order to compare the groups in the different cohorts based on taxa abundance.
All statistical analyses were performed using R version 3.6.2. Microbial richness, evenness, and diversity were assessed using the R Vegan package. The co-occurrence plot was generated among the taxa for all groups using MEGAN533.
Ethics approval
The present study was approved by the Human Ethical Committee, Assam University, Silchar, Assam, India (Ref. NO: IEC/AUS/2019/31/IS, Dated:13/11/2019).
Consent to participate
The samples were collected with the participants’ consent, and the participants duly filled written informed consents.
Results
Demographic characteristics of the subjects
Age ranges of 20–60 years were chosen for participants in the current study. There were 16 female participants and 14 male participants. The mean age ± SD of the included respondents from the three tribes were: Adi tribe (39.5 ± 4.3; range 20–60), Apatani tribe (42.5 ± 4.1; range 21–60) and Nyshi tribe (41.9 ± 4.7; range 20–60). The common diet of the Adi and Nyshi tribes consisted mainly of rice with difference in consumption of cereals, millets, leaves, fish and meat. The Apatani tribe mainly consumed boiled rice, boiled vegetables, boiled fish, meat and dairy products in each serving. All three tribes also consumed fermented bamboo shoot, smoked pork and smoked fish (Supplementary Table S1).
Statistics of sequencing data
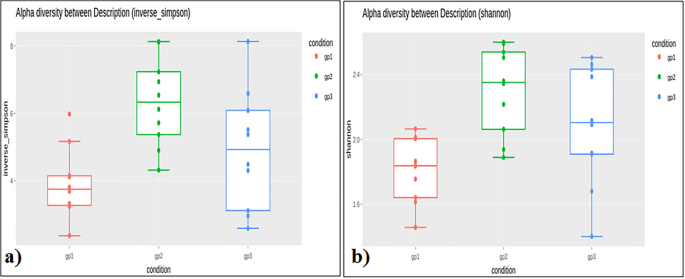
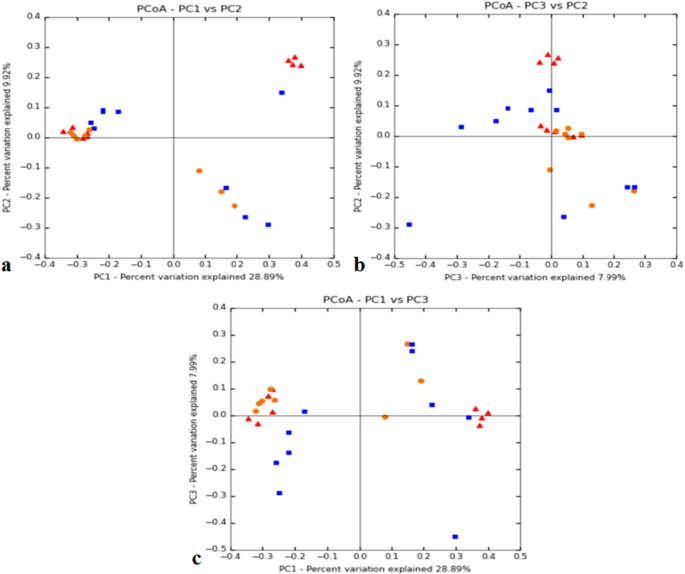
The number of reads per individual ranged from 44,090 to 70,718 for the Adi tribe; 43,377–112,936 for the Apatani tribe; and, 54,972–109,291 for the Nyshi tribe (Table 1). Alpha diversity assessment of the three studied populations significance difference amongst the tribes using inverse Simpsons diversity index (p < 0.004) and Shannon index (p < 0.003), respectively (Fig. 1a,b). Kruskal–Wallis rank sum test revealed higher diversity and abundance in Apatani tribe (group 2) than in Adi tribe (group 1) and Nyshi tribe (group 3). Beta diversity was assessed by unweighted unifrac to analyse the similarity of microbiota composition amongst tribes and significantly difference amongst the tribes was observed (R2 = 0.084, p = 0.01, Adonis analysis). Principal coordinate analysis (PCoA) indicated that the Adi and Apatani tribes have distinct gut microbiota, while the Nyshi tribe has variable microbiota with their variation in Principal Component (PC1) of 28.89% followed by 9.9% of Principal Component2(PC2) and Principal Component (PC3) of 7.9% (Fig. 2), which may be due to residing in isolated villages with different food habits and lifestyle.
Bacterial composition at different taxa of tribal groups
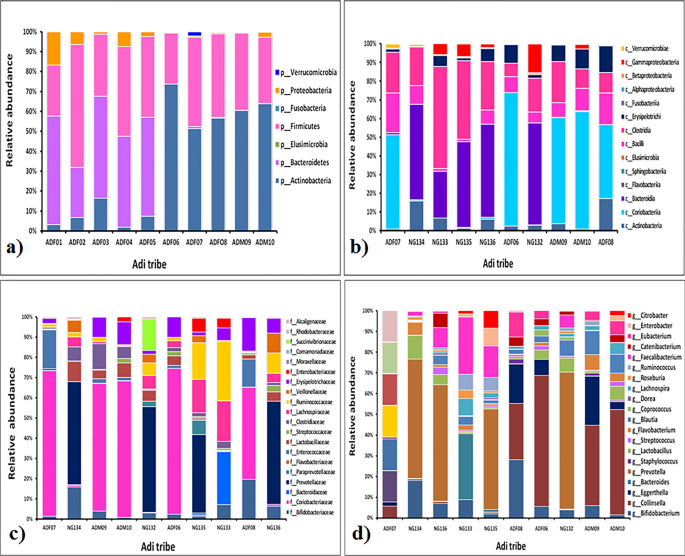
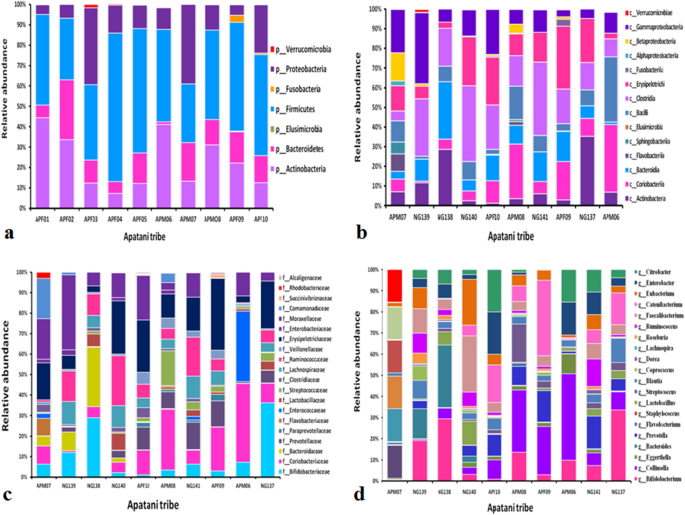
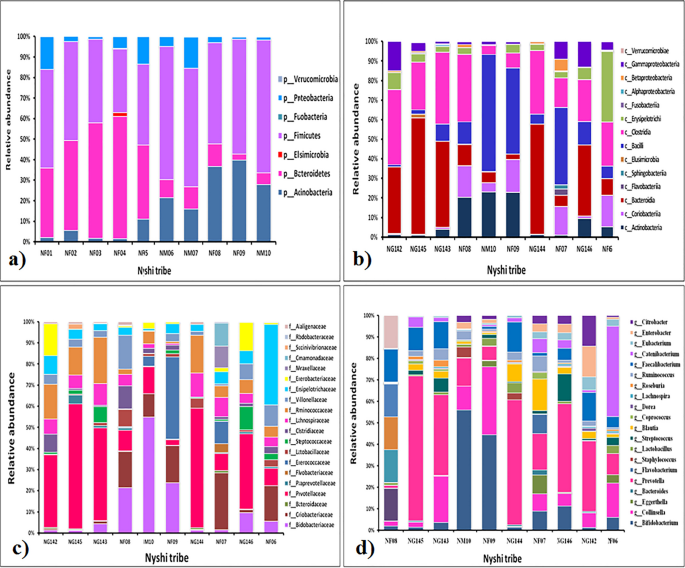
Relative abundance of fecal microbiota composition amongst the tribes were categorized at class, phylum, family and genus level. Metagenomics analysis was performed in order to determine the bacterial composition of the GI microbiota of Adi, Apatani and Nyshi tribes of Arunachal Pradesh. It was found that at the phylum level, Firmicutes, Actinobacteria, Protebacteria, Bacteroidates showed significant difference in abundance in all the tribes (p < 0.05 Kruskal–wallis H test) (Figs. 3a, 4a, 5a). At the class levels, most bacteria amongst tribes belonged to Actinobacteria, Coriobacteriia, Bacteroidia, Bacilli, Clostridia, Erysipelotrichi, Gammaproteobacteria, Betaproteobacteria, and Elusimicrobia (Figs. 3b, 4b, 5b).
The most abundant families found in the Adi, Apatani and Nyshi tribes of Arunachal Pradesh were Prevotellaceae, Bifidobacteriaceae, Coriobacteriaceae, Bacteroidaceae, Lactobacillaceae, Streptococcaceae, Lachnospiraceae, Ruminococcaceae, Veillonellaceae, Erysipelotrichaceae, Succinivibrionaceae, Enterobacteriaceae, and Rhodobacteraceae (p < 0.05 Kruskal–wallis H test)(Figs. 3c, 4c, 5c). At the genus level, significant abundance of Bifidobacterium, Collinsella, Bacteroides, Prevotella, Lactobacillus, Streptococcus, Clostridium, Coprococcus, Dorea, Lachnospira, Roseburia, Ruminococcus, Faecalibacterium, Catenibacterium, Eubacterium, Citrobacter and Enterobacter were observed amongst the three tribes (p < 0.034 Kruskal–wallis H test) (Figs. 3d, 4d, 5d). PERMANOVA analysis also revealed that the different tribal populations have significantly different microbial composition variance at phylum level (F = 3.113, p = 0.03), class level (F = 2.801, p = 0.024), family level (F = 2.790,p = 0.001) and genus levels (F = 2.523, p = 0.001).
On comparing amongst tribes, it was observed that Prevotella and Collinsella were highly prevalent in the Adi (p < 0.032) (Group 1) and Nyshi (p < 0.04) (Group 3) tribes than in the Apatani tribe. On the other hand, Bifidobacterium and Catenibacterium (p < 0.034, p < 0.021) were significantly more abundant in the Apatani than the Adi and Nyshi tribes.
For understanding the interactions of different gut bacteria in the different groups, we performed a co-occurrence network analysis by using MEGAN tool. We observed that the genera Lactococcus, Actinomyces, Enterococcus, Enterococcus and Coprococcus (p < 0.01) in the Adi tribe, Ruminococcus, Enterococcus, Streptococcus, Coprococcus and Staphylococcus (p < 0.004) in Apatani tribe, and Citrobacter, Enterobacter, Enterococcus, Coprococcus and Actinomyces (p < 0.01) in the Nyshi tribe were positively associated with each other (Supplementary Fig. S1). Hierarchical clustering analysis revealed that the Adi, Apatani and Nyshi tribes exhibited distinctly separate clusters at the genus level (Supplementary Fig. S2).
Differentially altered bacterial abundance in three tribes
While comparing the gut microbiota of the Adi, Apatani and Nyshi tribes using G-test analysis at the species level, it was observed that Coprococcus eutactus (p = 0.01), Eubacterium biforme (p = 0.001), Bacteroides uniformis (p = 0.01), Ruminococcus flavefaciens (p = 0.023), Bifidobacterium adolescentis (p = 0.001), Bacteroides ovatus (p = 0.023), and Streptococcus anginosus (p = 0.02) were less abundant in the Adi Tribe. The prevalence of Prevotella copri (p = 0.001), Prevotella stercorea (p = 0.001), Bacteroides fragilis (p = 0.004), Veillonella dispar (p = 0.01), Lactobacillus ruminis (p = 0.013), and Prevotella nanceiensis (p = 0.02) were significantly reduced in the Apatani tribe, while Bifidobacterium longum, (p = 0.001) and Dorea formicigenerans (p = 0.017) were significantly elevated in abundance in the Apatani tribe. Similarly, Prevotella copri (p = 0.001), Collinsella aerofaciens (p = 0.05) andAcinetobacter johnsonii (p = 0.05) was found to be significantly reduced in the Nyshi tribe (Table 2).
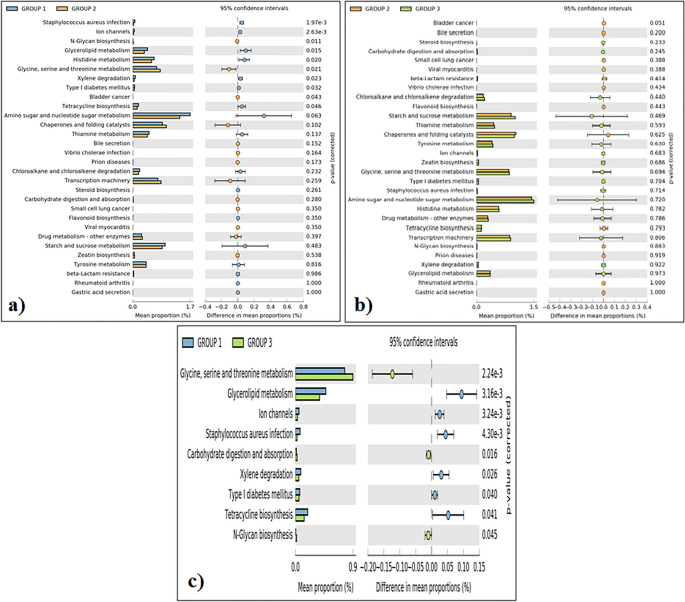
Functional potential of differentially enriched pathways by PICRUSt analysis
Functional prediction analysis revealed that the gut microbiota of the Adi tribe (group 1) has an abundance of genes relevant to Staphylococcus aureus infection, ion channels, glycerolipid metabolism, histidine metabolism, xylene degradation, type I diabetes mellitus and tetracycline biosynthesis (Fig. 6a). The gut microbiota of the Apatani tribe (group 2) has an abundances of genes related to N-glycan biosysthesis, glycine, serine and theronine metabolism, and bladder cancer (Fig. 6b). Similarly, the gut microbiota of the Nyshi tribe (group 3) has high abundances of genes realted to glycine, serine and theronine metabolism, carbohydrate digestion and absorption followed by to N-glycan biosysthesis (Fig. 6c).
Relative abundances of genes related to different selected function amongst the tribal people (a), Adi tribe and Apatani tribe; (b) Apatani tribe and Nyshi tribe; (c) Adi tribe and Nyshi tribe (Group 1: Adi tribe, Group 2: Apatani tribe, Group3: Nyshi tribe). The significant differences observed between the three groups at 95% confidence level and p < 0.05 are reported.
Discussion
Population specific microbiota studies have revealed the distribution of specific microbial communities in their gut. It is very important to understand the role of microbiome in human health34. The human gut microbiome composition is unique and stable in each individual living in different geographical locations. Tribal ethnic groups consume different diets and live in isolated villages and environmental conditions from the rest of the world. However, our knowledge on the gut microbiome and genetic diversity of healthy tribals living in such distinct and isolated parts of the state of Arunachal Pradesh in North East India is limited. Therefore, the present study analyzed the gut microbiome composition of healthy respondents belonging to the Adi, Apatani and Nyshi tribes of Arunachal Pradesh. The ethnic tribes of India mostly consume rich food along with dietary fibres including fruits, vegetables and grains. However, the Apatani tribe consumes boiled rice, boiled vegetables, boiled fish, meat and also dairy products in each serving. The common food of the Adi and Nyshi tribes was rice with difference in consumption of cereals, millets, leaves, fish and meat. All three tribes also consume fermented bamboo shoot, smoked pork and smoked fish. Gut microbiota analysis in our study revealed that the gut microbiome of the Adi, Apatani and Nyshi tribes are dominated by the genera Bifidobacterium, Collinsella, Bacteroides, Prevotella, Lactobacillus, Streptococcus, Clostridium, Coprococcus, Dorea, Lachnospira, Roseburia, Ruminococcus, Faecalibacterium, Oscillospira, Dialister, Catenibacterium, Eubacterium, Citribacter and Enterobacter. In the present study it was observed that Prevotella, Collinsella were highly prevalent in Adi tribe and Nyshi tribe while lower number was observed in the Apatani tribe. Furthermore, Bifidobacterium was observed to be much higher in abundance in the Apatani tribe than the Adi and Nyshi tribe. Presence of Bifidobacterium and Lactobacillus may be due to consumption of fermented foods, and the high altitude may also influence the composition of human gut microbiota35. A recent study22 stated that Bifidobacterium and Lactobacillus exhibited higher abundance in the Lepcha, Bhutia and Nepali populations of Sikkim due to consumption of milk and fermented food products. Interestingly, a study conducted from the urban and rural areas in Delhi found a high abundance of Prevotella, Lactobacillus, Lachnospira, and Roseburia which are similar to the present study36.
Prevalence of bacteria like Faecalibacterium, Lachnospira, and Bifidobacterium contribute anti-inflammatory and antimicrobial activities that act as barrier effect37,38. The present study observed a higher number of Bacteroides as well as lower number of proteobacteria in the Adi and Nyshi tribes than in the Apatani tribe where higher number of proteobacteria was observed. Increase of Bacteroidetes and proportionally reduced proteobacteria may be linked with energy consumption and high animal fat diet consumption17. Prevotellaceae members belonging to genus such as Prevotella which are associated with the food highly rich in carbohydrates derived from plant and fibre in the Indian diet5,39. Prevotella occurred in high abundance in the gut of Mongolian, Amerindian and Malawian populations due to their carbohydrate rich food habits. In the present study it was observed that Prevotella copri, Eubacterium biforme were found in higher relative abundance in the Adi, Apatani and Nyshi tribes from Arunachal pradesh which is similar with the previous study32. Few studies on Prevotella copri in central Indian individuals showed that it was involved in branch-chain amino acids (BCAA) and threonine-independent isoleucine biosynthetic pathway3. The present study revealed that at phylum level, Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria are commonly present in all the tribal groups. Similarly, previous study reported that the gut microbiome of Indian communities is dominated by Firmicutes followed by Bacteroidetes, Actinobateria and Proteobacteria, along with Prevotella, Bifidobacterium, Bacteroides, Eubacterium and Faecalibacterium were most prevalent in Indian subjects which is consistent with the present findings19,40. Few studies of the Tibetan Malawian, Amerindian and West African populations have also revealed high abundance of Prevotella14,35,39. Presence of Bacteroides, Ruminococcus, and Faecalibacterium were also observed as the most dominant bacteria in Kerala (South India) populations23,41. Similarly, another report from Delhi and Pune Indian populations reported Actinobacteria, Firmicutes, Bacteroidetes and Proteobacteria as major phyla and Prevotella and Megasphaera as the dominant genera36. Furthermore, studies from Manipur, Telangana, Assam, and Sikkim have also shown high abundance of Firmicutes, Bacteroidetes and Actinobacteria22. While the diet of Indian tribes is mainly dependent on carbohydrate and plant rich food, few reports also describe the abundance of Prevotella associated with the animal-based diet6,19 and the Bacteroides prevalence may be due to consumption of protein rich animal diet39,42.The present study observed that healthy tribal people of Adi, Apatani and Nyshi tribes living in Arunachal Pradesh highly prevalent with Firmicutes members which is consistent with a previous study19.Further, functional genomics analysis revealed that the highly abundant genera encoded several genes with diverse functions. On comparing, it was observed that the microbiota of the Apatani and Nyshi tribes mostly encode genes linked with N-glycan biosysthesis, glycine, serine and theronine metabolism. However, most interestingly, the microbiota of the Adi tribe is predicted to have genes highly linked with Staphylococcus aureus infection, ion channels, glycerolipid metabolism, histidine metabolism, xylene degradation, type I diabetes mellitus and tetracycline biosynthesis. Such occurrences in these tribes may be due to far away settlement from industrialization and agricultural pollution19.
Conclusion
In conclusion, the present study elucidated the gut microbiome of the tribal community of Arunachal Pradesh particularly the Adi, Apatani and Nyshi tribes, revealing the presence of beneficial and some harmful bacteria with key genera belonging to Bifidobacterium, Bacteroides, Prevotella, Lactobacillus, Ruminococcus, Faecalibacterium and the phylum Firmicutes, Actinobacteria, Protebacteria, Bacteroidates. The dominance of Firmicutes and Bacteroides in the gut flora of tribal communities of Arunachal Pradesh living in distinct and remote geographical areas was the most significant observation as their presence may be due to rice-based food habits and consumption of traditionally prepared beverages.
Data availability
The datasets generated during/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Sekirov, I., Russell, S. L., Antunes, L. C. M. & Finlay, B. B. Gut microbiota in health and disease. Physiol. Rev. 90, 859–904 (2010).
Sjogren, K. et al. The gut microbiota regulates bone mass in mice. J. Bone Miner. Res. 27, 1357–1367 (2012).
Pulipati, P. et al. The Indian gut microbiota-Is it unique? Indian J. Gastroenterol. 39, 133–140 (2020).
Wu, G. D. et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108 (2011).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Gupta, V. K., Paul, S. & Dutta, C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front. Microbiol. 8, 1162 (2017).
Yadav, D., Ghosh, T. S. & Mande, S. S. Global investigation of composition and interaction networks in gut microbiomes of individuals belonging to diverse geographies and age-groups. Gut Pathog. 8, 1–21 (2016).
Clemente, J. C. et al. The microbiome of uncontacted Amerindians. Sci. Adv. 2015, e1500183 (2015).
Martinez, L. et al. The gut microbiota of rural papua new guineans: Composition, diversity patterns, and ecological processes. Cell Rep. 11, 527–538 (2015).
Obregon-Tito, A. J. et al. Subsistence strategies in traditional societies distinguish gut microbiomes. Nat. Commun. 6, 6505 (2015).
Rampelli, S. et al. Metagenome sequencing of the hazda hunter-gatherer gut microbiota. Curr. Biol. 25, 1682–1693 (2015).
Schnorr, S. L. et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 5, 3654 (2014).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Borody, T. J., Brandt, L. J. & Paramsothy, S. Therapeutic faecal microbiota transplantation: Current status and future developments. Curr. Opin. Gastroenterol. 30, 97–105 (2014).
Hullar, M. A. & Fu, B. C. Diet, the gut microbiome, and epigenetics. Cancer J. 20, 170 (2014).
Li, L. & Zhao, X. Comparative analyses of fecal microbiota in Tibetan and Chinese Han living at low or high altitude by barcoded 454 pyrosequencing. Sci. Rep. 5, 14682 (2015).
Chong, C. W. et al. Effect of ethnicity and socioeconomic variation to the gut microbiota composition among pre-adolescent in Malaysia. Sci. Rep. 5, 13338 (2015).
Das, B. et al. Analysis of the gut microbiome of rural and urban healthy Indians living in sea level and high altitude areas. Sci. Rep. 8, 1–5 (2018).
Bhute, S. S. et al. Gut Microbial diversity assessment of indian type-2-diabetics reveals alterations in Eubacteria, Archaea, and Eukaryotes. Front. Microbiol. 8, 214 (2017).
Deb, D., Das, S., Adak, A. & Khan, M. R. Traditional rice beer depletes butyric acid-producing gut bacteria Faecalibacterium and Roseburia along with fecal butyrate levels in the ethnic groups of Northeast India. 3 Biotech 10, 1–1 (2020).
Dehingia, M. et al. Gut bacterial diversity of the tribes of India and comparison with the worldwide data. Sci. Rep. 5, 1–2 (2015).
Dhakan, D. B. et al. The unique composition of Indian gut microbiome, gene catalog, and associated fecal metabolome deciphered using multiomics approaches. Gigascience 8, 1–20 (2019).
Kulkarni, A. S., Kumbhare, S. V., Dhotre, D. P. & Shouche, Y. S. Mining the core gut microbiome from a sample Indian population. Indian J. Microbiol. 59, 90–95 (2019).
Dubey, A. K. et al. LogMPIE, pan-India profiling of the human gut microbiome using 16S rRNA sequencing. Sci. Data 5, 180232 (2018).
Ghosh, T. S. et al. Gut microbiomes of Indian children of varying nutritional status. PLoS ONE 9, e95547 (2014).
Yang, C. Y. et al. Oral microbiota community dynamics associated with oral squamous cell carcinoma staging. Front. Microbiol. 9, 862 (2018).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
DeSantis, T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Keohane, D. M. et al. Microbiome and health implications for ethnic minorities after enforced lifestyle changes. Nat. Med. 26, 1089–1095 (2020).
Huson, D. H., Auch, A. F., Qi, J. & Schuster, S. C. MEGAN analysis of metagenomic data. Genome. Res. 17, 377–386 (2007).
Cho, I. & Blaser, M. J. The human microbiome: At the interface of health and disease. Nat. Rev Genet. 13, 260–270 (2012).
Li, K. et al. Comparative analysis of gut microbiota of native Tibetan and Han populations living at different altitudes. PLoS ONE 11, e0155863 (2016).
Bhute, S. et al. Molecular characterization and meta-analysis of gut microbial communities illustrate enrichment of prevotella and megasphaera in Indian subjects. Front. Microbiol. 7, 660 (2016).
Lievin, V. et al. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut 47, 646–652 (2000).
Serafini, F. et al. Evaluation of adhesion properties and antibacterial activities of the infant gut commensal Bifidobacterium bifidum PRL2010. Anaerobe 21, 9–17 (2013).
De Filippo, C. et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. 107, 14691–14696 (2010).
Kaur, K., Khatri, I., Akhtar, A., Subramanian, S. & Ramya, T. N. Metagenomics analysis reveals features unique to Indian distal gut microbiota. PLoS ONE 15, e0231197 (2020).
Kaoutari, A. E., Armougom, F., Gordon, J. I., Raoult, D. & Henrissat, B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 11, 497–504 (2013).
De Filippis, F. et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut Microbiota 65, 1812–1821 (2016).
Acknowledgements
We gratefully acknowledge the Indian Council of Medical Research (ICMR) Vide RFC No. RBMCH/NER/10/2019-2021 for financial support.
Funding
This study was funded by the Indian Council of Medical Research (ICMR) Vide RFC No. RBMCH/NER/10/2019-2021.
Author information
Authors and Affiliations
Contributions
P.H. performed the experiments and bioinformatics analysis. I.S., I.C., Y.C. prepared the experimental design and analysis tools. M.U. helped in the sample collection process. I.S., P.H. and I.C. wrote the manuscript, and all the authors read the manuscript thoroughly and approved it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hazarika, P., Chattopadhyay, I., Umpo, M. et al. Elucidating the gut microbiome alterations of tribal community of Arunachal Pradesh: perspectives on their lifestyle or food habits. Sci Rep 12, 18296 (2022). https://doi.org/10.1038/s41598-022-23124-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-23124-w
This article is cited by
-
Exploring the influences of geographical variation on sequence signatures in the human gut microbiome
Journal of Genetics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.