Abstract
The differences in genetic susceptibility to lung adenocarcinoma and squamous cell carcinoma remain unclear. We developed a customized, targeted gene sequencing panel for efficient and sensitive identification of germline variants, including whole-gene deletion types for cancer-related drug-metabolizing enzyme genes in lung adenocarcinoma and squamous cell carcinoma. The minor allele frequencies of the variants, confirmed as clinically significant in the Japanese population, did not differ significantly from those of normal participants listed in the public database. Genotype analysis comparing lung adenocarcinoma (n = 559) and squamous cell carcinoma (n = 151) indicated that the variants of DPYD (rs190771411, Fisher’s exact test, P = 0.045; rs200562975, P = 0.045) and ALDH2 (rs568781254, P = 0.032) were associated with an increased risk of squamous cell carcinoma compared to adenocarcinoma. Conversely, whole-gene deletion of CYP2A6 was associated with adenocarcinoma but not squamous cell carcinoma. Notably, whole-gene deletion of CYP2A6 was confirmed in 22 patients with lung adenocarcinoma but not in any patients with squamous cell carcinoma. Most patients with whole-gene deletion of CYP2A6 were female non-smokers. The discovery of a whole-gene deletion of CYP2A6 in patients with lung adenocarcinoma may have an important role in clinical practice and advance our understanding of CYP2A6 germline variants and their association with carcinogenesis or their susceptibility to lung adenocarcinoma.
Similar content being viewed by others
Introduction
Individual and racial differences exist in the occurrence of adverse effects of therapeutic drugs, including anticancer drugs. Therefore, detecting variants of genes encoding drug-metabolizing enzymes is vital for understanding the variations in drug response and individual risks of adverse effects1,2,3.
Additionally, various genetic damages induced by endogenous compounds and exogenous hazards, such as environmental chemicals, may contribute to the etiology of cancer4. Approximately 30% of drug-metabolizing enzyme substrates can be metabolically enhanced5. Some genetic variants of drug-metabolizing enzymes correlate with cancer risk. However, contradictory findings have also been reported. Phase I drug-metabolizing enzymes such as cytochrome P450 (CYPs), encoded by P450 genes, metabolize pro-carcinogens into genotoxic electrophilic intermediates. Phase II drug-metabolizing enzymes bind intermediates to water-soluble derivatives to complete the detoxification cycle. Therefore, the activity and expression of genes encoding phase I and phase II drug-metabolizing enzymes are important factors in defining the toxicity or carcinogenicity of environmental chemicals, including cancer susceptibility and smoking effects4,6.
Lung cancer is one of the cancers most strongly associated with exposure to environmental factors, such as smoking and inhalation of exhaust fumes. The overall landscape of genomic abnormalities in somatic cells of lung adenocarcinoma7 and squamous cell lung carcinoma, the most common subtypes of lung cancer, has been largely revealed8,9. The mutations in lung cancer cells of smokers mainly consist of cytosine to adenine (C > A) nucleotide transversions, which arise due to the mutagenic effect of tobacco. In contrast, non-smokers usually present a predominant transition from cytosine to thymine (C > T)7. Moreover, they have fewer somatic mutations and genomic breakpoints, and a smaller fraction of the genome with chromosomal instability than smokers10. Smoking is more strongly associated with squamous cell carcinoma than adenocarcinoma. However, in terms of genetic predisposition, the difference between lung adenocarcinoma and squamous cell carcinoma in germline variants of drug-metabolizing enzymes remains unclear.
Widespread use of next-generation sequencing has enabled comprehensive investigation of genetic variants, such as drug-metabolizing enzymes, using whole-genome sequencing (WGS) and whole-exome sequencing (WES). However, genes with high homologies, such as CYP genes, still have unanalyzable genetic variants11,12. Therefore, we constructed a unique genetic variant panel that mainly covers the exon regions of 20 genes, including both lifestyle- and cancer-related genes, focusing on drug-metabolizing enzyme-coding genes that influence the therapeutic and adverse effects of anticancer drugs. Here, we compared the differences in genetic susceptibility to lung adenocarcinoma and squamous cell carcinoma in the germline of Japanese patients using a novel panel (DME panel) and next-generation sequencing.
Results
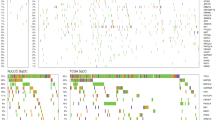
The total number of variants of the 20 target genes detected using the DME panel was 433 (Supplementary Fig. S1). The mean depth of coverage of the target regions was 455-fold that of the DME panel. All previously described to affect drug responses in Japanese populations were detectable among these genetic variants. The minor allele frequencies (MAFs) of the variants did not differ significantly from those of normal participants listed in the public database, suggesting that the DME panel is useful for comprehensively detecting germline mutations (Table 1).
The characteristics of patients with adenocarcinoma and squamous cell carcinoma of the lungs are shown in Table 2. The number of patients with squamous cell carcinoma who smoked was significantly higher (P < 0.001) than that of patients with adenocarcinoma. The proportion of patients with squamous cell carcinoma (73.5%, 111/151) who consumed alcohol was also significantly higher (P < 0.001) than that of patients with adenocarcinoma (55.4%, 309/558).
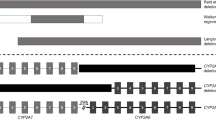
The association analysis results of individual variants of squamous cell carcinoma and adenocarcinoma of the lungs are shown in Supplementary Table S1. Two variants of DPYD (rs190771411 and rs200562975) and a variant of ALDH2 (rs568781254) were associated with an increased risk of squamous cell carcinoma compared to adenocarcinoma in the dominant model (P < 0.05) (Table 3). The characteristics of all 7 squamous cell carcinoma patients with significant variants in DPYD and ALDH2 are shown in Table 4. No distinctive items were noted. Notably, a whole-gene deletion of CYP2A6 was detected in 22 patients with adenocarcinoma but in no patient with squamous cell carcinoma (Table 5, Supplementary Fig. S2). In addition, 63.6% (14/22) of patients with a CYP2A6 whole-gene deletion were non-smokers, and 72.7% (16/22) were women. To assess its clinical effect, we analyzed the effect of the CYP2A6 whole-gene deletion in lung adenocarcinoma on overall survival (OS) using the Kaplan–Meier method. Patients with the CYP2A6 whole-gene deletion-type showed no significant (p = 0.97) difference in terms of OS compared to those with the CYP2A6 gene retain-type. Lung adenocarcinoma patients with the CYP2A6 gene retain-type had significantly (p = 0.0099) better OS compared with squamous cell carcinoma patients with the CYP2A6 gene retain-type (Fig. 1). The characteristics of all 22 adenocarcinoma patients with deletion-type of CYP2A6 gene are shown in Table 6. These patients with CYP2A6 whole-gene deletion-type on survivals showed no relationship between surgical procedure and TNM stage.
Discussion
This study presented an efficient and sensitive analysis of genetic variants, including whole-gene deletion types for drug-metabolizing enzymes and environmental- or lifestyle-related factors. Multiplex long-range PCR amplification with locus-specific primers and next-generation sequencing was also adopted for library products unique in the DME panel because of their high sequence identities to other CYPs. For example, the sequences of CYP2A6 and CYP2D6 are > 90% identical to those of CYP2A7 and CYP2D7, respectively. Although there are reports that genetic variants of CYP2A6, including whole-gene deletions, are associated with lung cancer risk13, differences in the risk for adenocarcinoma and squamous cell carcinoma of the lungs remain poorly understood. Notably, the CYP2A6 whole-gene deletion was confirmed in 22 patients with lung adenocarcinoma but in no patients having squamous cell carcinoma. In addition, patients with whole-gene deletions were primarily female non-smokers. Our results suggest that in lung adenocarcinoma, this finding may be associated with the mechanisms of carcinogens different than those activated by CYP2A6. Ariyoshi et al. demonstrated that the CYP2A6 whole-gene deletion was not found in male smokers among Japanese patients with squamous cell carcinoma (0 of 105)14, which is consistent with our results.
CYP2A6 is an enzyme responsible for metabolizing of nicotine- and tobacco-specific carcinogens. Genetic variants of CYP2A6 are associated with changes in the activity of the CYP2A6 enzyme, which influences smoking effects and the rate at which some tobacco-specific carcinogens are metabolized, which subsequently determines the incidence of lung cancer. In smokers with lower CYP2A6 activity, tobacco-specific nitrosamines are activated at lower levels, decreasing their exposure to these activated lung carcinogens15. Considering that the whole-gene deletion of CYP2A6 is found only in lung adenocarcinoma, the potential role of CYP2A6 germline variants in lung carcinogenesis is intriguing. Its role may be explained by the following. Individuals with CYP2A6 whole-gene deletions may be less susceptible to smoking effects. Therefore, some patients may have developed lung adenocarcinomas through a pathway unrelated to the function of CYP2A6, regardless of smoking. Conversely, squamous cell carcinoma that develops in squamous epithelial cells may be directly affected by smoking in a dose-dependent manner while maintaining the function of the CYP2A6 variants.
Heterozygous or homozygous CYP2A6 deletions may be associated with a decreased occurrence of gastric cancer in females and decreased total cancer, including lung, colon, and gastric cancers in female non-smokers16. Adenocarcinoma is the most common subtype of primary lung cancer in women and is considered to be due to the later adoption of smoking by women17. Additionally, estrogen and its receptors have been identified as factors that increase the risk of lung adenocarcinoma18,19. The biological significance of CYP2A6 whole-gene deletions in lung adenocarcinoma may be the modulation of the cancer phenotype, which requires further investigation and may enhance our understanding of the oncogenic mechanism of lung adenocarcinoma. However, it remains unclear how CYP2A6 whole-gene deletions are involved in the development of lung adenocarcinoma and their interaction with xenobiotic organisms. Therefore, verifying its function using cell lines with downregulated or without CYP2A6 expression is necessary. This is currently being investigated in our laboratory. A limitation of the present study is that the absence of the CYP2A6 whole-gene deletion in patients with squamous cell carcinoma is debatable because our results were derived from a small hospital-based sample. Therefore, it will be necessary to verify the results using a larger sample.
In the present study, the ALDH2 (rs568781254) or DPYD variants (rs190771411 and rs200562975) were associated with an increased risk of squamous cell carcinoma patients compared to adenocarcinoma. However, due to the low frequency of the minor allele of the variants (MAF of 0.0029 for ALDH2 and MAF of 0.0014 for DPYD), these were not large enough to detect an association with squamous cell carcinoma. Previous Japanese studies noted that genetic variants in ALDH2 are involved in ethanol metabolism, specifically associated with the risk of esophageal cancers. The carcinogenic metabolite acetaldehyde, an ingredient in tobacco smoke and/or alcohol, is detoxified by ALDH2. Matsuo et al. reported that the ALDH2 variant interacted with cigarette smoking in the risk of lung cancer in Japanese20. Fluoropyrimidines (5-FU and its prodrug capecitabine) are widely used treat several types of cancer. Several studies have shown a link between reduced DPYD enzyme activity and increasing the risk of severe toxicity. A recent study has reported that the functional alterations of enzyme activities caused by DYPD variants were characterized21. The rs200562975of DPYD identified in the present study reportedly reduced enzymatic activity to less than 70% of wild-type in vitro21. However, none of the previous studies examined whether the DPYD variants contribute to the risk of lung cancer. Additionally, there is a lack of studies assessing the functional effect of most variants for DPYD in vivo, and inferring possible functions based on the variants is difficult. Further studies are needed to confirm our findings and expose the underlying molecular mechanism.
Materials and methods
Participants
This study was conducted using blood samples from Project HOPE initiated at the Shizuoka Cancer Center (SCC; Shizuoka, Japan). The objective of this project was to improve cancer therapy22. Blood samples for germline analysis were obtained from 710 patients with lung cancer (559 adenocarcinomas and 151 squamous cell carcinomas) intraoperatively at SCC Hospital, Shizuoka, Japan, between January 2014 and January 2020. We performed deep sequencing of a custom DME panel using intraoperative blood samples.
The Institutional Review Board of SCC approved all experimental protocols (Authorization No.: 25-33). Written informed consent was obtained from all patients participating in this study. All experiments using clinical samples were performed in accordance with the approved Japanese ethical guidelines23.
Construction of an in-house custom DME panel
We analyzed the genes encoding CYP isoforms (CYP1A2, CYP2A6, CYP2B6, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, and CYP3A5), thiopurine methyltransferase (TPMT), dihydropyrimidine dehydrogenase (DPYD), N-acetyltransferase 2 (NAT2), UDP glucuronosyltransferase family member A1 (UGT1A1), catechol-O-methyltransferase (COMT), ATP binding cassette subfamily G member 2 (ABCG2), cytidine deaminase (CDA), alcohol dehydrogenase 1B (ADH1B), aldehyde dehydrogenase 2 (ALDH2), 5-methyltetrahydrofolate-homocysteine methyltransferase reductase (MTRR), and methylenetetrahydrofolate reductase (MTHFR) in this study because the variants of these genes have been reported to affect drug response in Japanese populations11,24.
The allele frequencies of each gene were compared with those obtained from the following public Japanese population databases: Human Genetic Variation Database (HGVD)25 (http://www.genome.med.kyoto-u.ac.jp) and Japanese Multi Omics Reference Panel (jMorp)12 (https://jmorp.megabank.tohoku.ac.jp/202109/).
Genomic DNA was isolated from the buffy coats of blood samples using a QIAmp DNA Blood Kit (Qiagen, Hilden, Germany). All genetic variants were analyzed using an Illumina sequencer with multiplex long-range PCR assay and Nextera DNA Flex Library Prep kit (Illumina, San Diego, CA, USA). Briefly, 50–100 ng of DNA was amplified using long-range multiplex PCR with locus-specific primers and a GXL DNA polymerase with each primer set (Supplementary Table S2). The amplicon library was prepared using the Nextera DNA Flex Library Prep kit (Illumina), and the library DNA was quantified on TapeStation using the D5000 kit (Agilent Technologies, Santa Clara, CA, USA). The libraries were subsequently used for sequencing (Supplementary Fig. S3). The sequencing data was analyzed using the pipeline described in our previous report26 and the clinical sequencing data analysis integrator (csDAI) (Mizuho-ir.co.jp/solution/research/life/infodata/csdai/index.html). The genetic variants were visualized using the Integrative Genomics Viewer27.
Statistical analyses
Fisher’s exact test, crude odds ratio (OR), and 95% confidence interval (CI) were employed to evaluate statistical differences in genotype distributions and allele frequencies of each variant between adenocarcinoma and squamous cell carcinoma in patients with lung cancer. To compare large biased populations, we performed a Fisher’s exact test A patient’s survival was analyzed using the Kaplan–Meier method and log-rank test. Statistical significance was defined at P < 0.05.
Data availability
The genotype data referenced during the current study are available in a public repository that is accessible through the NCBI (https://www.ncbi.nlm.nih.gov/), HGVD (https://www.hgvd.genome.med.kyoto-u.ac.jp/), jMorp (https://jmorp.megabank.tohoku.ac.jp/202109/), and PharmGKB (https://www.pharmgkb.org/) websites. The information on the variants between individual samples is described in Supplementary Table S1. The sequence information of the primer sets used in this study is listed in Supplementary Table S2. Although the somatic data and sample information from patients used in this study were submitted to the National Bioscience Database Center (NBDC) as ‘Controlled-Access Data’ (the accession number, hum0127 https://humandbs.biosciencedbc.jp/en/), the germline data analyzed during the current study are not available publicly, but are available from the corresponding author on reasonable request. However, all data and materials generated and/or analyzed during the current study are included in the supplementary information files of this article.
References
Ma, Q. & Lu, A. Y. H. Pharmacogenetics, pharmacogenomics, and individualized medicine. Pharmacol. Rev. 63, 437–459. https://doi.org/10.1124/pr.110.003533 (2011).
Zhou, S. F., Liu, J. P. & Chowbay, B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab. Rev. 41, 89–295. https://doi.org/10.1080/03602530902843483 (2009).
Lee, W., Lockhart, A. C., Kim, R. B. & Rothenberg, M. L. Cancer pharmacogenomics: Powerful tools in cancer chemotherapy and drug development. Oncologist 10, 104–111. https://doi.org/10.1634/theoncologist.10-2-104 (2005).
Nebert, D. W. & Dalton, T. P. The role of cytochrome P450 enzymes in endogenous signaling pathways and environmental carcinogenesis. Nat. Rev. Cancer 6, 947–960. https://doi.org/10.1038/nrc2015 (2006).
Nebert, D. W., Mckinnon, R. A. & Puga, A. Human drug-metabolizing enzyme polymorphisms: Effects on risk of toxicity and cancer. DNA Cell Biol. 15, 273–280. https://doi.org/10.1089/dna.1996.15.273 (1996).
Sim, S. C., Kacevska, M. & Ingelman-Sundberg, M. Pharmacogenomics of drug-metabolizing enzymes: A recent update on clinical implications and endogenous effects. Pharmacogenomics J. 13, 1–11. https://doi.org/10.1038/tpj.2012.45 (2013).
Imielinski, M. et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120. https://doi.org/10.1016/j.cell.2012.08.029 (2012).
Herbst, R. S., Morgensztern, D. & Boshoff, C. The biology and management of non-small cell lung cancer. Nature 553, 446–454. https://doi.org/10.1038/nature25183 (2018).
Suster, D. I. & Mino-Kenudson, M. Molecular pathology of primary non-small cell cancer. Arch. Med. Res. 51, 784–798. https://doi.org/10.1016/j.arcmed.2020.08.004 (2020).
Boeckx, B. et al. The genomic landscape of nonsmall cell lung carcinoma in never smokers. Int. J. Cancer 146, 3207–3218. https://doi.org/10.1002/ijc.32797 (2020).
Ohnami, S. et al. Whole exome sequencing detects variants of genes that mediate response to anticancer drugs. J. Toxicol. Sci. 42, 137–144. https://doi.org/10.2131/jts.42.137 (2017).
Yamaguchi-Kabata, Y. et al. iJGVD: An integrative Japanese genome variation database based on whole-genome sequencing. Hum. Genome Var. 2, 1–4. https://doi.org/10.1038/hgv.2015.50.15050 (2015).
Johani, F. H., Majid, M. S. A., Azme, M. H. & Nawi, A. M. Cytochrome P450 2A6 whole-gene deletion (CYP2A64) polymorphism reduces risk of lung cancer: A meta-analysis. Tob Induc. Dis. 18, 50. https://doi.org/10.18332/tid/122465 (2020).
Ariyoshi, N. et al. Genetic polymorphism of CYP2A6 gene and tobacco-induced lung cancer risk in male smokers. Cancer Epidemiol. Biomak. Prev. 11, 890–894 (2002).
Tanner, J. A. & Tyndale, R. F. Variation in CYP2A6 activity and personalized medicine. J. Pers. Med. 7, 18. https://doi.org/10.3390/jpm7040018 (2017).
Abudushataer, M. et al. Association of CYP2A6 gene deletion with cancers in Japanese elderly: An autopsy study. BMC Cancer 20, 186. https://doi.org/10.1186/s12885-020-6663-4 (2020).
Hutchinson, B. D., Shroff, G. S., Truong, M. T. & Ko, J. P. Spectrum of lung adenocarcinoma. Semin. Ultrasound CT MR 40, 255–264. https://doi.org/10.1053/j.sult.2018.11.009 (2018).
Hsu, L. H., Chu, N. M. & Kao, S. H. Estrogen, estrogen receptor and lung cancer. Int. J. Mol. Sci. 18, 1713. https://doi.org/10.3390/ijms18081713 (2017).
Rodriguez-Lara, V., Hernandez-Martinez, J. M. & Arrieta, O. Influence of estrogen in non-small cell lung cancer and its clinical implications. J. Thorac. Dis. 10, 482–497. https://doi.org/10.21037/jtd.2017.12.61 (2018).
Park, J. Y. et al. Impact of smoking on lung cancer risk is stronger in those with the homozygous aldehyde dehydrogenase 2 null allele in a Japanese population. Carcinogenesis 31, 660–665. https://doi.org/10.1093/carcin/bgq021 (2010).
Hishinuma, E. et al. Importance of rare DPYD genetic polymorphisms for 5-fluorouracil therapy in the Japanese population. Front. Pharmacol. 13, 930470. https://doi.org/10.3389/fphar.2022.930470 (2022).
Yamaguchi, K. et al. Implementation of individualized medicine for cancer patients by multiomics-based analyses—the Project HOPE—. Biomed. Res. 35, 407–412. https://doi.org/10.2220/biomedres.35.407 (2014).
Ministry of Health, Labour and Welfare, Japanese ethical guidelines for human genome/gene analysis research 2017. http://www.mhlw.go.jp/stf/seisakunitsuite/bunya/hokabunya/kenkyujigyou/i-kenkyu/index.html (2019).
Kurose, K., Sugiyama, E. & Saito, Y. Population differences in major functional polymorphisms of pharmacokinetics/pharmacodynamics-related genes in Eastern Asians and Europeans: Implications in the clinical trials for novel drug development. Drug Metab. Pharmacokinet. 27, 9–54. https://doi.org/10.2133/dmpk.DMPK-11-RV-11 (2012).
Higasa, K. et al. Human genetic variation database, a reference database of genetic variations in the Japanese population. J. Hum. Genet. 61, 547–553. https://doi.org/10.1038/jhg.2016.12 (2016).
Nagashina, T. et al. Japanese version of The Cancer Genome Atlas, JCGA, established using fresh frozen tumors obtained from 5143 cancer patients. Cancer Sci. 111, 687–699. https://doi.org/10.1111/cas.14290 (2020).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. https://doi.org/10.1038/nbt.1754 (2011).
Acknowledgements
We thank the members of Shizuoka Cancer Center Hospital and Research Institute for their support and suggestions. This study was supported by the Shizuoka Prefectural Government, Japan. We would like to thank Editage (www.editage.jp) for English language editing.
Author information
Authors and Affiliations
Contributions
Principal investigator: Su.O. Planning and design: Su.O., A.N., S.O. M.I. and K.Y. Ascertainment and diagnostic evaluations of subjects for genetic analysis: M.I., M.M., Su.O., Y.O., H.K. and T.T. Genetic analyses and technical assistance: Su.O., A.N., S.N., F.K., Y.S., A.S., K.M., K.O., K.H., Y.A., T.N., K.U., S.O. and K.Y. Statistical analyses and database search: Su.O. and T.N. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ohnami, S., Naruoka, A., Isaka, M. et al. Comparison of genetic susceptibility to lung adenocarcinoma and squamous cell carcinoma in Japanese patients using a novel panel for cancer-related drug-metabolizing enzyme genes. Sci Rep 12, 17928 (2022). https://doi.org/10.1038/s41598-022-22914-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-22914-6
This article is cited by
-
Associating CYP2A6 structural variants with ovarian and lung cancer risk in the UK Biobank: replication and extension
European Journal of Human Genetics (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.