Abstract
Leishmaniasis is one of the main infectious diseases worldwide. In the midst of all the different forms of the disease, Cutaneous Leishmania (CL) has the highest incidence in the world. Many trial vaccines have been developed with the purpose of generating long-term cell-mediated immunity to Leishmania(L) major. As there is not any multi-epitope DNA vaccine with high efficacy against L.major, the aim of this study is to design a new multi-epitope DNA vaccine in order to have effective control upon this infectious disease through the immune bioinformatics. The L.major antigens: Gp63, LACK, TSA, LmSTI1and KMP11 were selected to design a multi-epitope DNA vaccine. The initial structure of the DNA vaccine was designed, benefiting from Gen Bank's website information. Epitopes of MHC-I antigens were predicted through the Immune Epitope Database (IEDB), and the selected epitopes were used to make vaccines construct along with linkers. New multi-epitope vaccine including 459 nucleic acids designed, and inserted between BamH1 and HindIII restriction sites of pCDNA3.1 mammalian expression vector. 12 epitopes among the chosen antigens were selected by two servers (IEDB and ANTIGEN). They had high stability and high antigenic power. Physicochemical features of vaccine measured by ProtParam server, and this structure was thermostable and hydrophilic. it’s a suitable model to study on the animal and human phases. The designed vaccine is expected to be an effective candidate through development of (CL) vaccines. However, the effectiveness of this vaccine should also evaluate in vivo model.
Similar content being viewed by others
Introduction
Leishmaniasis is a disease caused by a parasite of the genus Leishmania (L), which varies from a self-healing form such as cutaneous leishmaniasis (CL) to lethal visceral leishmaniasis (VL)1. Treatment of leishmaniasis currently involves the use of a number of drugs such as pentamonial antimony, amphotericin B, miltefosine, paromomycin, which in most cases have side effects2. Vaccination is the most effective way to eradicate this infectious disease3. Structure of various types of vaccines, such as killed parasite vaccine, subunit vaccines and DNA vaccines have been studied by scientists4.There are very few Leishmania vaccines that have entered the clinical phase. Therefore, we need more studies to find a good way for the treatment5. In order to produce an effective vaccine, it is important to know the host’s immune system. To limit the CL infection, the strong produced cytokines, particularly IFN-γ, play important roles. Stimulation of T helpers, the (Th1) and (Th2) cytokines, is seen in patients with the CL infection. Among them, the Th1 has been reported to be more effective. Furthermore, the macrophages show significant capabilities in defense process. They are triggered by cytokines derived from T-cells, which are vital for controlling or aggravating the disease. Macrophages are a kind of antigen-presenting cell (APC) that distinguishes antigens of the Leishmania6.As a results, there is a critical requirement for development of a vaccine, that can bring more efficient immunity than the earlier vaccines7. Along with the various types of studied vaccines, the use of DNA vaccines has demonstrated promising outcomes. However, the use of these antigens in the form of proteins goes hand in hand by some problems, including high production price, short half-life, poor immunogenicity, low stimulation of cellular immunity, and cross-allergic reactions8. The first-generation vaccine of the CL, consists of live attenuated and killed fragmented parasites. Which is just a class of human prophylactic VL vaccine, that has entered the phase III of clinical trials until now. however, this vaccine was not successful to achieve satisfactory results9. The second-generation vaccines produced by recombinant Leishmania antigens. Among a range of approaches the LEISH-F3, a multicomponent vaccine formulated with GLA-SE adjuvant, illustrated promising results in phase I as a strong immune response inducer in healthy individuals10. More recently, a third-generation DNA vaccine that employs simian adenovirus, expressing a new synthetic gene that encode Leishmania antigens, have termed as ChAd63-KH. It has indicated potentials to be a protective and immunogenic therapeutic vaccine for the human VL, and the post kala-azar dermal leishmaniasis (PKDL) in phase I trial11. Regardless of the current progress in vaccine development, the desirable aim has not yet been attained, that is the development of a safe, effective, durable and low-cost prophylactic vaccine for human leishmaniasis12. The use of epitopes or epitope-containing peptides is advantageous, since epitopes are assessed for immune recognition and epitope specific response13. Since peptides themselves remain poorly immunogenic, the approaches that are attracting increasing interest are based on the development of peptide-based formulations in combination with potent adjuvant components (peptide, lipids, virus particles, nanoparticles etc.)14. Peptide vaccine studies, which just a few years ago were becoming more and more marginalized, are again rising as a guaranteed finding for designing more logical vaccines15. While there is an increase in the number of new peptide-base vaccines, a major challenge is how to avoid inactivation or degradation by the immune system, and enhancing the immunogenicity of those peptides. Thus, it is essential to design vaccines using diverse approaches besides the use of other adjuvants, that can help to improve the antigen immunogenicity16.
Methods
Sequence and structure retrieval
We evaluated L. major antigens and the selected antigens were Glycoprotein 63 (Gp63), Leishmania-activated C-kinas (LACK), Thiol-Specific Antioxidant Antigen (TSA), Leishmania major stress-inducible protein1 (LmSTI1), Kinetoplastid membrane protein 11 (KMP11), to design a new multi-epitope DNA vaccine. The protein sequences were obtained by using the corresponding Gen Bank server NCBI, (http://www.ncbi.nib.gov/genbank), which is the protein database of the National Center for Biotechnology Information. The sequence IDs of GP63, KMP11, LACK, LmSTI1 and TSA were ACL01096.2, XP003722616.1, AAA97577.1, AAB37318.1 and AAC31146.1 respectively. All sequences from diverse isolates downloaded from the Gene Bank in the FASTA format. The sequences of all antigenic proteins were checked manually and those unrelated sequences removed.
Antigenicity analysis
The antigenicity of the selected proteins was evaluated using two online servers, the Vaxijen and the AntigenPro at http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html and http://scratch.proteomics.ics.uci.edu/, respectively. The VaxiJen predicts the antigenicity of a protein by using a new alignment-independent method and the accuracy of this method is almost 88%. In the case of the Protein’s organism of source, a particular model selected that has a specific threshold. The VaxiJen considers a parasite protein as an antigen, when its score is more than the defined threshold which is 0.5 for the parasite data set. ANTIGENpro is a sequence-based and alignment-free predictor that has not designed for a specific pathogen group. It predicts the antigenicity of protein based on eight features sets and five machine-learning algorithms by using protein antigenicity microarray data. The accuracy of this method is near 76%.
NCBI BLASTp screening
To prevent probable cross-reactivity with human proteins, the predicted CD8 + T cell epitopes screened against human proteome by NCBI BLASTp at https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins.
T-cell epitope prediction
The specific epitopes for major histo compatibility complex (MHC-I) were predicted by using immune epitope database (IEDB) server at http://tools.iedb.org/mhci/.For this purpose, H2-Dd alleles were selected as the mouse MHC-I molecules.
Construction of leish21 vaccine
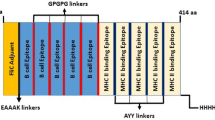
T-cell epitopes were linked together via gsgg, ggs, gsgs, ggssgg and ggsg linkers. The prediction of the secondary structure was performed by using an online server, the PSIPRED, available at (http://bioinf.cs.ucl.ac.uk/psipred). Antigenicity Prediction of the leish21, the designed vaccine, utilized by immune epitope database server (IEDB).
Physicochemical features of vaccine
Physicochemical characteristics of the Leish21 vaccine like the molecular weight (Mw), isoelectric point (pI), instability index and the half-life, were measured by the ProtParam server available at (http://web.expasy.org/protparam).
Tertiary structure prediction and evaluation
The 3D-structure of the whole protein vaccine modeled using the QUARK serve at https://zhanggroup.org. The QUARK models are built from small fragments by replica-exchange Monte Carlo simulation, through an atomic-level knowledge-based force field, and is suitable for proteins that do not have homologous templates. The multi-epitope peptide vaccine was designed and the HindIII and BamHI restriction sites were used to the N and C terminal of the DNA sequence of the peptide structure, respectively.
Results
T-cell epitope prediction
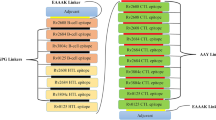
The T-cell’s epitopes prediction identified using the online server (IEDB) based on MHCI binding. Among all the predicted epitopes, only 12 epitopes with higher scores selected. We used three epitopes from each of the Gp63, KMP11 and Lack, one epitope from the LmSTI1and two epitopes from the TSA (Table1).
Construction of the Leish21 multi-epitope DNA vaccine
These epitopes with high binding affinity were fused together. The new multi-epitope DNA vaccine, including 459 nucleic acids synthesized. Kozak (GCCCC) is the essential site for initiation of replication. We used Alanine amino acid to improve stability of this structure (Fig. 1, 2). The Leish21 inserted between BamH1 and HindIII restriction sites of the pCDNA3.1 (https://www.snapgene.com/resources/plasmidfiles/?set=mammalian_expression_vectors&plasmid=pcDNA3.1_Hygro(%2B)&format=png ) of mammalian expression vector.
Antigenicity Evaluation of the leish 21 vaccine
To predict the antigenicity of this structure, VaxiJen v.2 and ANTEGENpro servers were used. The prediction by the VaxiJen was 0.58 and 0.57 by the ANTIGENpro (Table 2). According to the obtained results, the multi-epitope DNA vaccine can activate the immune response.
Secondary structure of leish21
Prediction of the secondary structure was performed by PSIPRED server with 29.93% alpha-helix, 5.44% beta strand and 64.63% random-coil which is shown in (Fig. 3).
Physicochemical parameters of evaluation
Referring to the ProtParam server, the multi-epitope DNA vaccine consists of: 147 amino acids with the molecular weight (MW) of 15.56 KD, theoretical Isoelectric point (PI) of 6.33 and 2124 number of atoms. Additionally, the predictable half-life in mammalian reticulocytes in vitro was 4.4 h. Instability index, aliphatic index and grand average of hydropathicity (GRAVY) were 44.36, 51.09 and 0.395 respectively (Fig. 4). Therefore, this structure was thermostable and hydrophilic.
Tertiary structure prediction, refinement, and validation
The 3D structures of the predicted epitopes modeled with the QUARK server. From the top 5 models predicted by this server, the one with the highest C-score (− 1.93) was selected for further works (Fig. 5). The results of the Ramachandran analysis demonstrated that 65.7% were in favored in the allowed regions. The number of residues in allowed regions were (21.3%), generously allowed regions were (6.5%) (Fig. 6a). The model quality which was performed by the ProSA-web server revealed that the z-score was −5.42. The leish21’s protein structures were also located within the normal range for the three-dimensional structures by the NMR and X-ray method (Fig. 6b). Moreover, the plot of residue scores indicated the local model quality by plotting energies for the leish21 protein structures (Fig. 6c). As most of the residues had a negative score and were below the threshold, so the leish21protein structures had the lowest problematic or erroneous parts. According to Verify3D, least 80% of the amino acids have scored ≥ 0.2 in the 3D/1D profile using, indicating to have a valid structure. (Fig. 6d)17.
Discussion
While leishmaniasis is one of the main health issues in many countries, development of protective vaccines against it, is becoming a big concern. Peptide vaccines are potentially a new technology to struggle with the Leishmania infection. Bioinformatics procedures for the analysis of proteins have become an essential instrument for vaccine researches13,17,18. The prediction of epitopes is a significant facility for designing new vaccines4.Nowadays use of immune-informatics software, have led to save a great deal of time to design vaccine constructs. Numerous studies have carried out for epitope predictions on microorganisms such as viruses, parasites and bacteria17,19. Selecting several epitopes from protein antigens has several advantages over designing vaccines using full–length proteins. One of the advantages is that expressing full length of some proteins to obtain soluble protein form is difficult due to their large molecular weight, so it is better to use epitopes rather than whole protein. Moreover, some regions in the outer part of antigenic proteins have similar sequences with the host proteins and cause autoimmunity and cross- reaction with auto-antigens. Certain regions of the full-length protein also may have immunosuppressive properties. For instance, by inducing regulatory T cells, the parasite’s immune escape happens, and even leads to delay in the elimination and persistence of the parasite. By size controlling of the protein that can induce a stronger immune response, epitopes from more antigens can be used. This makes it possible to consider a large number of combinations of B cell and T cell epitopes, which can trigger a powerful humoral immune response with the help of CD4 Th cells20. So far, there is not a definite treatment report for the disease caused by L. major. The greatest strategy is vaccine development that can trigger the immune system with respect to intracellular parasite fragments and activation of TC and TH21. In addition to diminishing the intensity of reinfection, immunity against Leishmania through activating CD4+ T cells and CD8+ T cells play an important role in limiting the infection22. Multiple epitope-based vaccines have potential advantages when compared with vaccines using single or combinations of recombinant proteins. Vaccine design can be performed by selecting the most antigenic CD4+ and CD8+ T-cell epitopes from immunogenic Leishmania proteins, and therefore the avoided epitopes were those which presented low immunogenicity or detrimental immune dominance in the whole proteins23. Additionally, positives of multiple epitope-based vaccines consist of safety, low manufacturing price, easy quality control process, (and the chance of rationally engineering of the epitopes, to increase their immunogenicity24. In the present study, a new multiple epitope DNA vaccine has designed for the L. major to be considered as an important choice in reducing this infectious disease. Previous studies pointed out the efficacy of vaccines based on LACK combined with other components, including IL-12 and IL-18 in a murine model. The Leish21 should be tested for the immunogenicity and the protective effects in rodent models and maybe for protection of human in the future. One of the selected genes is LACK, which has various functions, such as signal transduction, DNA replication, RNA synthesis and cell cycle control in eukaryotes, and it is expressed in promastigotes and amastigotes. Vaccination by LACK facilitates the initiation of the protective response against CL, due to shifting from Th2 cytokine responses to Th1. The reason for this effective response is that the IFN-γ production is inducted by interleukin 12. Hence, it has been as an appropriate method for improvement of vaccines for leishmaniosis25. The ‘GGGS’ constructed linkers causes increasing humoral and cellular immune responses in BALB/c mice. Former studies indicated that in addition to the epitopes, the linker between epitopes is crucial in the efficiency of vaccines, with G-rich linkers being particularly competent for multi-epitope vaccine constructure. G-rich linkers have the capacity to improve sequence flexibility without affecting the function of proteins they attach to. In addition, glycine and serine are high flexibility, and have less side branches compared to other amino-acids. The presence of alcoholic factor, (OH) in serin, can be dissolved in the hypotonic solutions. Immunogenicity of the DNA vaccines on Leishmaniasis evaluated in BALB/c mice, and the immune response provoked in animals26. Several physiological characteristics have compared based on previous studies and this study. Aliphatic index of a protein is defined as the relative volume occupied by aliphatic side chains (alanine, valine, isoleucine and leucine). It may be considered as a positive factor for increasing the thermal stability of globular proteins. In a study conducted by Khatoon and Daneshi, it showed that the stability of protein against heat was much higher than other studies. In our study it has shown that the molecular weight of the design construct is lower than the protein size of other studies because in this study, fewer and more specific epitopes were used, which caused the molecular weight to be low27(Table 3)
It should be noted that comparison between former studies and ours about the assessment of the designed vaccine shows that the molecular weight of the Leish21 is lower than others, and it makes an important advantage for us. Because the possibility of loop production is much lower than other vaccines. In addition, selected epitopes in this study have the highest score in the IEDB. Also, different antigens were used to design this vaccine, which this method is better than the way single-antigen vaccines are designed because it increases the efficacy of a vaccine. Aliphatic index of our vaccine has a lower quantity in comparison with the Khatoon et. al.’s, and this is because of the different molecular weight of their structures. About the vaccine-efficacy comparison with other designed vaccines, it should say that as different vaccines have positives and negatives, while they are not tested in clinical phase a proper comparison is not possible. An ideal vaccine against Leishmaniasis should have a number of potentialities, like: being immune, immunization against different types with limited number of injections for a longer duration, lower production cost, and possibility to be used both for prevent and treatment28. To design an effective vaccine for cutaneous leishmaniasis, bioinformatics methods, online servers, and numerous software were utilized for prediction of T cell epitops. Thus, more investigations by using in silico and in vivo patterns must be done in the future studies for assessing potency of the polytopes as a final vaccine choice.
Conclusion
In this study, immune bioinformatics tools were employed to develop a new multi-epitopes vaccine which obtained from five immunogenic proteins: KMP11, GP63, LACK, LmSTI1 and TSA of the L. major. Leish21 need to further be tested on murine models and requiring more studies on this vaccine.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary material.
References
Ashwin, H. et al. Characterization of a new Leishmania major strain for use in a controlled human infection model. Nat. Commun. 12(1), 1–12 (2020).
Yadav, S. et al. Design of a multi-epitope subunit vaccine for immune-protection against Leishmania parasite. Pathog. Glob. Health 114(8), 471–481 (2020).
Greenwood, B. The contribution of vaccination to global health: Past, present and future. Philos. Trans. R. Soc. B: Biol. Sci. 369(1645), 20130433 (2014).
Karimi, M. et al. Construction of a synthetic gene encoding the multi-epitope of toxoplasma gondii and demonstration of the relevant recombinant protein production: A vaccine candidate. Galen Med. J. 9, 1708 (2020).
Rabienia, M. et al. Exploring membrane proteins of Leishmania major to design a new multi-epitope vaccine using immunoinformatics approach. Eur. J. Pharm. Sci. 152, 105423 (2020).
Do Nascimento, N. R. F. et al. In vitro and in vivo leishmanicidal activity of a ruthenium nitrosyl complex against Leishmania (Viannia) braziliensis. Acta trop. 192, 61–65 (2019).
Lakshmi, B. S., Wang, R. & Madhubala, R. Leishmania genome analysis and high-throughput immunological screening identifies tuzin as a novel vaccine candidate against visceral leishmaniasis. Vaccine 32(30), 3816–3822 (2014).
Solana, J. C. et al. Subcutaneous immunization of leishmania HSP70-II null mutant line reduces the severity of the experimental Visceral leishmaniasis in BALB/c mice. Vaccines 8(1), 141 (2020).
Teixeira, M. C. A. et al. An experimental protocol for the establishment of dogs with long-term cellular immune reactions to Leishmania antigens. Mem. Inst. Oswaldo Cruz 106(2), 182–189 (2011).
Coler, R. N. et al. From mouse to man: Safety, immunogenicity and efficacy of a candidate leishmaniasis vaccine LEISH-F3+ GLA-SE. Clin. Transl. Immunol. 4(4), e35 (2015).
Osman, M. et al. A third generation vaccine for human visceral leishmaniasis and post kala azar dermal leishmaniasis: First-in-human trial of ChAd63-KH. PLoS Negl. Trop. Dis. 11(5), e0005527 (2017).
Iborra, S. et al. Vaccine candidates against leishmania under current research. Expert Rev. Vaccines 17(4), 323–334 (2018).
Ebrahimi, M. et al. Designing and modeling of multi-epitope proteins for diagnosis of Toxocara canis infection. Int. J. Pept. Res. Ther. 26(3), 1–10 (2020).
Barati, M. et al. Regulatory T Cells in bioactive peptides-induced oral tolerance; a two-edged sword related to the risk of chronic diseases: A systematic review. Nutr. Cancer 73(6), 956 (2021).
Skwarczynski, M. & Toth, I. Peptide-based synthetic vaccines. Chem. Sci. 7(2), 842–854 (2016).
Reed, S. G., Orr, M. T. & Fox, C. B. Key roles of adjuvants in modern vaccines. Nat. Med. 19(12), 1597–1608 (2013).
Mamaghani, A. J. et al. Designing diagnostic kit for toxoplasma gondii based on GRA7, SAG1, and ROP1 antigens: An in silico strategy. Int. J. Pept. Res. Ther. 26(4), 2269 (2020).
Aghamolaei, S., et al., (2020) Design and expression of polytopic construct of cathepsin-L1, SAP-2 and FhTP16. 5 proteins of fasciola hepatica. J. helminthol. 94.
Mamaghani, A. J. et al. Candidate antigenic epitopes for vaccination and diagnosis strategies of toxoplasma gondii infection: A review. Microb. Pathog. 137, 103788 (2019).
Bemani, P., Amirghofran, Z. & Mohammadi, M. Designing a multi-epitope vaccine against blood-stage of Plasmodium falciparum by in silico approaches. J. Mol. Graph. Model. 99, 107645 (2020).
Zahedifard, F. et al. Anti-leishmanial activity of brevinin 2R and its lauric acid conjugate type against L. major: In vitro mechanism of actions and in vivo treatment potentials. PLoS Negl. Trop. Dis. 13(2), e0007217 (2019).
Basu, R., Roy, S. & Walden, P. Hla class I—restricted t cell epitopes of the kinetoplastid membrane protein—11 presented by leishmania donovani—infected human macrophages. J. Infect. Dis. 195(9), 1373–1380 (2007).
Joshi, S. et al. Immunogenicity and protective efficacy of T-cell epitopes derived from potential Th1 stimulatory proteins of leishmania (Leishmania) donovani. Front. Immunol. 10, 288 (2019).
Martins, V. T. et al. A recombinant fusion protein displaying murine and human MHC class I-and II-specific epitopes protects against Leishmania amazonensis infection. Cell. Immunol. 313, 32–42 (2017).
Ghaffarifar, F. et al. Enhancement of immune response induced by DNA vaccine cocktail expressing complete LACK and TSA genes against Leishmania major. APMIS 121(4), 290–298 (2013).
Yang, Y. et al. In silico design of a DNA-based HIV-1 multi-epitope vaccine for Chinese populations. Hum. Vaccin. Immunother. 11(3), 795–805 (2015).
Salehi-Sangani, G. et al. Immunization against leishmania major infection in BALB/c mice using a subunit-based DNA vaccine derived from TSA, LmSTI1, KMP11, and LACK predominant antigens. Iran. J. Basic Med. Sci. 22(12), 1493 (2019).
Khatoon, N., Pandey, R. K. & Prajapati, V. K. Exploring leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Sci. Rep. 7(1), 1–12 (2017).
Acknowledgements
The authors would like to appreciate the vice chancellor of research, Shiraz University of Med-ical Sciences, and Cellular and Molecular Biology Research Center, Shahid Beheshti University of Medical Sciences for all supports.
Funding
Shiraz University of Medical Sciences and Cellular and Molecular Biology Research Center, Shahid Beheshti University of Medical Sciences were supported of the study. This article is a part of PhD thesis of Sama Rashidi grant number 23364.
Author information
Authors and Affiliations
Contributions
All authors discussed the results and contributed to this study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rashidi, S., Faraji, S.N., Mamaghani, A.J. et al. Bioinformatics analysis for the purpose of designing a novel multi-epitope DNA vaccine against Leishmania major. Sci Rep 12, 18119 (2022). https://doi.org/10.1038/s41598-022-22646-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-22646-7
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.