Abstract
The development of the parathyroid and the thymus from the third pharyngeal pouch depends on the activities of the Gcm2 and Foxn1 transcription factors, respectively, whose expression domains sharply demarcate two regions in the developing third pharyngeal pouch. Here, we have generated novel mouse models to examine whether ectopic co-expression of Gcm2 in the thymic epithelium and of Foxn1 in the parathyroid perturbs the establishment of organ fates in vivo. Expression of Gcm2 in the thymic rudiment does not activate a parathyroid-specific expression programme, even in the absence of Foxn1 activity. Co-expression of Foxn1 in the parathyroid fails to impose thymopoietic capacity. We conclude that the actions of Foxn1 and Gcm2 transcription factors are cell context-dependent and that they each require permissive transcription factor landscapes in order to successfully interfere with organ-specific cell fate.
Similar content being viewed by others
Introduction
In mice, several organs develop from the epithelial lining of the pharynx, such as the thyroid, the ultimobranchial bodies, the parathyroid, and the thymus. The anlagen of the parathyroid and the thymus emerge in the third pharyngeal pouch, the parathyroid from the dorso-cranial aspects of the pouch, the thymus from the caudo-ventral parts. The proper development of the future organs depends on the activities of transcription factors Gcm2 for the parathyroid1,2,3 and Foxn1 for the thymus4,5 (Fig. 1a). The expression domains of the two genes mark complementary regions in the developing third pouch even before morphological differences in the tissue become recognizable6. However, the phenotypes of mice lacking Gcm21,2,3 or Foxn14,5,7 suggest that these transcription factors are not required for the formation of organ primordia. Instead, they have been shown to play key roles at subsequent stages of development and to participate in the execution of organ-specific functional programmes3,7. In mice lacking functional Gcm2, the parathyroid rudiment forms in the third pouch, but then undergoes apoptotic regression, causing the parathyroid to disappear1,2,8. Hence, although Gcm2 is not required for the initiation of parathyroid development, it is required for the maintenance of this gland; as a result, Gcm2-null mice suffer from impaired calcium homeostasis1,8. In mammals lacking a functional Foxn1 gene, the epithelial component of the thymic rudiment forms during embryogenesis; however, in contrast to the situation of Gcm2-deficiency in the parathyroid, the Foxn1-deficient thymus-fated epithelium persists into adulthood. Although it fails to differentiate properly and hence remains in an immature non-functional state4,5,7 and is incapable of attracting haematopoietic progenitor cells, the subsequent re-introduction of Foxn1 enables the activation of the stalled differentiation programme9. Foxn1 deficiency in mammals thus effectively blocks intrathymic T cell development10,11, leading to profound immunodeficiency4,12,13. Remarkably, overexpression of Foxn1 in certain heterologous cell types, such as fibroblasts, has been reported to generate functional thymic epithelial cells14,15. This suggests that in permissive cellular environments, Foxn1 can impose a thymic epithelial cell fate.
Developmental trajectories of thymus and parathyroid. (a) In mice, the third pharyngeal pouch initially consists of undifferentiated epithelium that expresses neither Foxn1 nor Gcm2. At a later stage, two distinct domains emerge, one expressing Foxn1, the other Gcm2. Whereas the former goes on to form the epithelial component (TEC) of mediastinal and cervical thymic lobes, the latter develops into the parathyroid. Upon further (hypothetical) changes in cell states, some parathyroid-fated cells may transdifferentiate into a second kind of cervical thymus, referred to as parathyroid-fated cervical thymus (see text for details). Evidence for the existence of analogous thymus-fated parathyroid tissue has not yet been found. (b) RNA in situ hybridisation patterns identify and distinguish thymus and parathyroid tissues. In some instances, ectopic parathyroid tissue remains attached to the thymic lobe as a result of incomplete separation of parathyroid and thymus anlagen during embryogenesis, as described earlier20; in the present context, these ectopic patches of cells serve as a positive technical control for the lack of expression of Pth in the thymic epithelium, but do not represent fate-converted tissue (c.f., Fig. 2 in20). (c) RNA in situ hybridisation patterns identify and distinguish thyroid and parathyroid tissues. For (b), and (c), serial sections of E14.5 embryos were hybridized with the indicated gene-specific probes. Data representative of ten embryos.
During embryonic development, the thymic anlage (Fig. 1b) migrates to the upper anterior mediastinum, whereas the parathyroid gland becomes closely associated with the thyroid, which is marked by high levels of expression of the Ttf1 transcription factor gene (Fig. 1c)16. Interestingly, however, ectopic (cervical) thymic tissue can be found in many different locations in the neck region, sometimes even in close proximity to the parathyroid17,18 (Fig. 1c). Cervical thymi may arise either from delayed differentiation of an undifferentiated endodermal progenitor pool18,19, or from transdifferentiation of parathyroid-fated cells, after downregulation of their parathyroid differentiation programme and subsequent upregulation of Foxn1 expression19 (Fig. 1a). These transfated cervical thymi attract haematopoietic progenitors and induce the initiation of their T cell differentiation programme19. It is unclear whether transdifferentiation proceeds through intermediate Gcm2/Foxn1 double-positive and/or double-negative stages (indicated as hypothetical cell states in Fig. 1a). Considering the final outcome of these differentiation trajectories, we hypothesize that enforced Foxn1 expression in the parathyroid domain should induce transdifferentiation in post-Gcm2-expressing cells (Fig. 1a).
Ectopic parathyroid tissue is also often found in scattered locations in the neck, and indeed in the mediastinum adjacent to or inside thymic lobes (Fig. 1b)20; in humans, this presents a significant challenge when extirpation of parathyroid adenomas is required21. Ectopic parathyroid tissue may not always result from aberrant migration to the final anatomical location next to the thyroid, but could, in analogy to the situation of the parathyroid-fated cervical thymi, originate from trans-fated thymic epithelium (Fig. 1a). To the best of our knowledge, direct evidence for this kind of a transfating event has not yet been reported.
To begin to experimentally address this developmental conundrum, we have generated mice in which epithelia of the two organ anlagen co-express the two transcription factors. In our scheme, the epithelium in the thymic rudiment additionally expresses Gcm2 under the control of the Foxn1 regulatory elements, whereas the parathyroid anlage additionally expresses Foxn1 under the control of the regulatory elements of the Gcm2 gene.
Results
Strategy for the ectopic expression of Gcm2 in the expression domain of Foxn1
In the first set of experiments, we focused on the ectopic expression of Gcm2 in the Foxn1 expression domain, which marks the future thymic epithelial microenvironment. Two genetic constellations were explored. In order to avoid complications arising from the known Foxn1 haploinsufficiency effect22,23, we used a transgenic strategy to additionally express Gcm2 in the Foxn1 domain. To this end, we placed the Gcm2 cDNA under the control of the complex regulatory elements of the Foxn1 gene, encompassed in a ~ 28 kb genomic fragment situated upstream of the Foxn1 gene (Fig. 2a)24. This strategy has been successfully used to express numerous cDNAs in the thymic rudiment (e.g.,10,17,18,25,26,27,28,29). In our experiments, the numbers of Gzma-expressing thymocytes within the thymus were used as a measure of thymopoietic activity. Gcm2-mediated fate changes were assessed by the expression of the parathyroid hormone (Pth) gene, one of Gcm2´s key down-stream targets and a characteristic sign of parathyroid fate30,31,32. Thus, RNA in situ hybridisation assays are a sensitive means to determine residual Foxn1-driven thymi epithelial function (Gzma) and the initiation of the parathyroid-specific expression programme (Pth).
Ectopic expression of Gcm2 in Foxn1-positive epithelial cells of Foxn1+/+ mice. (a) Schematic of the Foxn1:Gcm2 expression construct. The individual sequence elements are indicated; note that the exonic sequences following the two alternative transcriptional start sites of the Foxn1 gene (identified as exons 1a and 1b, respectively) both associate with the same splice acceptor site of exon 2. The Gcm2 cDNA is inserted in exon 2 of the Foxn1 gene. The presence of the polyadenylation sequence of the Slc13a2 gene serves as an insulator sequence suppressing insertion site-associated variegated activity of the transgene. (b), Developmental trajectories under conditions of ectopic Gcm2 expression in thymic epithelial cells; note that the development of parathyroid and parathyroid-fated cervical thymi is expected to occur normally. Data representative of two mice. (c) RNA in situ hybridisation of E14.5 tissue sections of the upper mediastinum of Foxn1:Gcm2 transgenic mice with Foxn1-, Gcm2-, Pth-, and Gzma-specific probes. Data representative of three mice. (d) RNA in situ hybridisation identifies Gzma-positive thymocytes in the wild-type thymus. (e), Number of Gzma-positive cells per thymus area (2,500 μm2) of wild-type and Foxn1:Gcm2 transgenic mice; the median value is 7 cells for the wild-type (5.7903 ± 0.4063; mean ± s.e.m.), and 1 cell for the transgenic tissue (2.1754 ± 0.2789; mean ± s.e.m.) (Welch t-test, two-sided). Two thymic lobes each were analysed. (f) RNA in situ hybridisation of E14.5 tissue sections of thymic lobes of Foxn1+/+ wildtype; Foxn1+/– heterozygous, and Foxn1+/+;Foxn1:Gcm2 and Foxn1+/–;Foxn1:Gcm2 transgenic mice with a Dll4-specific probe. (g) RNA in situ hybridisation of E14.5 tissue sections of thymic lobes of Foxn1+/+;Foxn1:Gcm2 and Foxn1+/−;Foxn1:Gcm2 transgenic mice with a Gzma-specific probe (left panel); number of Gzma-positive cells per thymus area (2,500 μm2) of Foxn1+/+;Foxn1:Gcm2 and Foxn1+/−;Foxn1:Gcm2 transgenic mice; the median value is 1 cell for Foxn1+/+;Foxn1:Gcm2 (1.6061 ± 0.1005; mean ± s.e.m.; 6 thymic lobes analysed), and 1 cell for the Foxn1+/−;Foxn1:Gcm2 tissue (0.9589 ± 0.0991; mean ± s.e.m.; 8 thymic lobes analysed) (Welch t-test, two-sided). (h), RNA in situ hybridisation of E14.5 tissue sections of thymic lobes of Foxn1+/+ wildtype and Foxn1+/+;Foxn1:Gcm2 transgenic mice with an Il7-specific probe. Note that, at this stage of development, Il7 expression in the wildtype thymus is just above the detection limit28; the results are shown here to indicate that Il7 expression is not elevated in the transgenic situation (see text). Note that results in panels (e) and (g) come from independent experiments; at these early embryonic stages, T cell development is very dynamic; therefore, slight differences in developmental time may influence the results necessitating the analysis of animals from the same litter as the best possible age-match. No positive signal could be detected with a B-cell specific probe (Cd19; not shown). The organ primordia are outlined by dashed lines. Scale bars, 0.1 mm.
Expression of Gcm2 in Foxn1-sufficient thymic epithelia
In order to assess the effects of Gcm2 expression in thymic epithelia, we first generated a condition of co-expression of Foxn1 and Gcm2 transcription factor genes (Fig. 2b), corresponding to one of the hypothetical cell states illustrated in Fig. 1a. In the Foxn1-sufficient background, Gcm2 expression can be readily detected in the transgenic thymic rudiment (Fig. 2c). The ectopic activity of the Gcm2 transcription factor gene does not perturb the expression of the Foxn1 gene (Fig. 2c). Accordingly, at a qualitative level, the Foxn1-controlled genetic network remains functional, since thymic colonisation and T cell differentiation occur in the transgenic thymi, as indicated by the presence of cells positive for the T cell-specific hybridisation probe Gzma (Fig. 2c). However, compared to wild-type thymic epithelium (Fig. 2d), the thymopoietic activity of the thymic epithelium co-expressing Gcm2 is reduced several-fold, as indicated by the presence of significantly fewer Gzma-positive cells (Fig. 2c–e). This phenotype is accompanied by significantly reduced expression levels of the Dll4 gene, the key target of Foxn1 required to initiate T cell development via Notch signalling10 (Fig. 2f). Of note, the numbers of Gzma-positive cells are further reduced when ectopic expression of Gcm2 occurs in Foxn1 heterozygous mice (Fig. 2g), indicating that the detrimental effect of Gcm2 expression on thymopoiesis is magnified under conditions of reduced Foxn1 activity. However, despite reduced Dll4 expression levels, Cd19-expressing cells indicative of B cell development are not found the transgenic thymic rudiment (not shown), presumably because no concomitant up-regulation of the Il7 gene is observed (Fig. 2h) that would have resulted in a perturbed ratio of Il7/Dll4 expression levels that we have previously found to be an important factor for thymic B cell poiesis27,28. Importantly, Gcm2 expression fails to activate its cognate differentiation programme in Foxn1-expressing thymic epithelial cells, as seen from the lack of parathormone (Pth) gene expression (Fig. 2c). Collectively, these results suggest that although Gcm2 expression impairs Foxn1-mediated thymopoietic activity, this effect is not accompanied by the concurrent activation of a parathyroid-specific differentiation programme.
Ectopic Gcm2 expression in Foxn1-deficient epithelia
Next, we considered the possibility that Gcm2 expression may not be able to overrule the function of Foxn1 in a state of co-activity of these two transcription factors. Therefore, we explored the outcome of ectopically expressing Gcm2 in Foxn1-deficient mice (Fig. 3a). Whereas the development of the parathyroid proceeds normally in this constellation (Fig. 3b), parathyroid-fated cervical thymi are not detectable. Since lack of Foxn1 activity arrests the thymic epithelium in an undifferentiated state4,5,7, Gcm2 could exert its function in the absence of an established Foxn1-mediated transcriptional programme (Fig. 3a). As expected, Gcm2 expression is readily detectable in the Foxn1-deficient epithelial rudiment without concomitant loss of Foxn1 promotor activity (Fig. 3c). Importantly, however, the Pth gene again remains transcriptionally silent in the Gcm2-expressing thymus-fated epithelia. This result suggests that Gcm2 expression does not impose the phenotype of the parathyroid onto thymic epithelia, even in the absence of the cognate transcription factor Foxn1.
Ectopic expression of Gcm2 in the thymic epithelium of Foxn1-deficient mice. (a) Developmental trajectories under conditions of ectopic Gcm2 expression in Foxn1-negative thymic epithelial cells; note that the development of parathyroid is expected to occur normally, whereas that of parathyroid-fated cervical thymi is aborted. (b) Control hybridisations of sections of the upper pharynx of the mouse shown in (c) using Gcm2- and Pth-specific probes. Data representative of five embryos. (c) RNA in situ hybridisation analysis of the thymic rudiment; note that the Foxn1-specific hybridisation probes detects sequences upstream of the insertion site of the lacZ gene in the Foxn1 knock-in allele, such that Foxn1-deficient epithelial cells can still be recognized by RNA in situ hybridisation. Tissue sections representative of three embryos. Scale bars, 0.1 mm.
Ectopic expression of Foxn1 in the expression domain of Gcm2
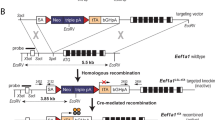
In the next set of experiments, we focused on the ectopic expression of Foxn1 in the Gcm2 expression domain, which marks the future parathyroid tissue. In order to direct the expression of the Foxn1 transcription factor to the parathyroid, we used CRISPR/Cas9-mediated homologous recombination of a Foxn1 cDNA sequence into the Gcm2 locus (Fig. 4a). We aimed at replacing a region of about 3 kb within the Gcm2 gene, spanning parts of exons 3 and almost the entire exon 6 by a ~ 4.2 kb cassette containing the Foxn1 cDNA, and a neo gene in opposite transcriptional orientation. To this end, we first identified sgRNAs that could be used to specifically and efficiently delete the relevant part of the Gcm2 gene. In this way, we successfully generated a null allele of Gcm2 (Fig. 4b). In a second step, we co-injected the selected RNPs, and a targetting construct containing the Foxn1/neo cassette into fertilised eggs. In the successfully engineered Gcm2 allele, the upstream regulatory sequences of the Gcm2 locus are preserved, as is the large intron (possibly containing regulatory elements) separating exons 2 and 3; the Foxn1 cDNA was inserted into exon 3 of Gcm2 in such a way that it preserves the cognate Foxn1 initiation codon (Fig. 4c). The 3´-untranslated region of the Foxn1 cDNA harbours a neo expression cassette flanked by loxP sites, which was removed by breeding mice heterozygous for this allele to a deleter strain expressing Cre recombinase in the germ-line33, establishing the final Gcm2:Foxn1 knock-in allele (Fig. 4d). Although the Foxn1-directed transcriptional programme is complex27,34,we have previously shown that the expression of only two of the many downstream target genes of Foxn1, those encoding the Notch ligand Dll4 and the chemokine Ccxl12 are sufficient to initiate thymopoiesis, as determined by the presence of Gzma-positive T cell lineage cells in the thymic rudiment10; hence, we considered the presence of Gzma-positive cells as a suitable marker of Foxn1 activity.
Ectopic expression of Foxn1 in the parathyroid. (a–d) Structure of Gcm2 alleles generated in this study. (a) Structure of the wild-type Gcm2 locus on mouse chromosome 13. Exons are depicted as red boxes, the 5´- and 3´-untranslated regions are shown in yellow. The initiation codon occurs in exon 2, the stop codon is situated in exon 6. The sequence contained in the fragment used for Southern blot hybridisation is indicated in black and lies outside of the homology arms that were contained in the replacement fragment. The sequences represented in the construct used for homologous recombination are indicated as blue dashed lines. (b) Structure of the Gcm2 null allele. The deleted region extends from exon 3 into exon 6 and is indicated by a dashed line. (c) Structure of Gcm2 locus after homologous recombination. The deleted region is replaced by a cassette consisting of the mouse Foxn1 cDNA (coding region depicted in dark green; the 3´-untranslated region in light green), and a Neo gene (blue), which is positioned in opposite transcriptional orientation; the latter is flanked by two loxP sites in tandem orientation (green boxes). (d) Structure of Gcm2 locus after homologous recombination and removal of the Neo cassette, which leaves one loxP site behind. (e) Developmental trajectories under conditions of ectopic Foxn1 expression in the parathyroid; note that the development of the mediastinal and cervical thymi and the hypothetical thymus-fated parathyroid are expected to occur normally. (f) RNA in situ hybridisation of adjacent E14.5 tissue sections of the pharynx of wild-type mice with probes specific for Gcm2, Foxn1, and Gzma. The organ primordia and other anatomical features are outlined by coloured dotted lines. Note that the size of the parathyroid is variable in individual sections. For orientation, the common carotid artery is indicated by red circle on one side of the section. Tissue sections representative of ten mice. (g) Hybridisation of sections equivalent to those in (f) from mice heterozygous for the Gcm2:Foxn1 knock-in allele. The Foxn1-expressing parathyroid glands are indicated by arrows. Tissue sections representative of 2 mice. Scale bars, 0.1 mm.
Expression of Foxn1 in the parathyroid
Using the Gcm2:Foxn1 knock-in allele, we first generated a condition of co-expression of Gcm2 and Foxn1 transcription factor genes in the parathyroid (Fig. 4e). Since Gcm2 haploinsufficiency has no known detrimental effects on parathyroid function, the parathyroid-fated cells will be simultaneously exposed to both Gcm2-driven and Foxn1-driven transcriptional programmes (Fig. 4e).
To determine whether Foxn1 expression could be detected in the parathyroid as a sign of successful genome engineering, mouse embryos were analysed at embryonic day 14.5 (E14.5); at this stage, the parathyroid and thymus anlagen have already separated well enough from each other in order to be able to unambiguously identify the distinct anatomical structures in the pharynx. Using an RNA in situ hybridisation assay, we compared wild-type mice (Fig. 4f) and compound heterozygotes carrying one wild-type Gcm2 allele and the Gcm2:Foxn1 knock-in allele (Fig. 4g). As expected, Foxn1 expression in the parathyroid can only be seen in the compound heterozygote—but not in wild-type mice –, whereas Gcm2 expression is seen in both genotypes (Fig. 4f,g). This result indicates that the Gcm2:Foxn1 knock-in allele is active as desired and suggests that the activity of the Foxn1 transcription factor does not repress Gcm2 gene expression. Thus, the results shown in Fig. 2c and Fig. 4g clearly indicate that Gcm2 and Foxn1 transcription factors do not cross-regulate each other, a result that is compatible with distinct signalling pathways upstream of these two genes35,36. Importantly, co-expression of cognate (Gcm2) and ectopic (Foxn1) transcription factors in the parathyroid did not induce thymopoietic activity, as shown by the lack, in the parathyroid, of immature thymocytes expressing granzyme A (Gzma) (Fig. 4f,g). In a final set of experiments, we explored the outcome of ectopically expressing Foxn1 in Gcm2-deficient mice (Fig. 5a). In this situation, a potential complication arises from the fact that the parathyroid undergoes apoptotic degeneration in Gcm2 null mice1,2,8. Hence, Foxn1 expression would have to rescue the demise of the developing parathyroid as a pre-requisite to its functional conversion to a thymopoietic tissue. However, we found that Foxn1 expression did not rescue the degeneration of the parathyroid in the absence of Gcm2, as revealed by the absence of Gcm2-expressing cells in the vicinity of the thyroid (Fig. 5b). Hence, it appears that the two transcription factors differ in their anti-apoptotic activities in cells of the parathyroid, essentially precluding the analysis of Foxn1 function in the parathyroid in the absence of Gcm2 activity.
Lack of parathyroid tissue in E14.5 embryos homozygous for the Gcm2:Foxn1 knock-in allele. (a) Developmental trajectories under conditions of ectopic Foxn1 expression in Gcm2-negative cells of the parathyroid; note that at this stage of development, parathyroid primordia are expected to be detectable only if Foxn1 expression rescues their apoptotic demise that occurs as a result of the lack of Gcm2 activity. (b) Failure to detect Gcm2-expressing parathyroid tissue. The sections were hybridised with a probe specific for the 5´-end of Gcm2, which is part of the chimaeric transcript emanating from the knock-in allele. The expected location of parathyroid is marked by yellow arrows; the common carotid arteries are indicated by red circles. Tissue sections representative of two mice. Scale bar, 0.1 mm.
Discussion
Our results provide important information about the roles of Foxn1 and Gcm2 transcription factors in the specification of the thymic epithelium and the parathyroid, respectively. Neither co-expression of Gcm2 and Foxn1 in the domain of the future parathyroid nor co-expression of Foxn1 and Gcm2 in the future thymic rudiment can override the endogenous differentiation programmes. Since both transcription factors are not required for the initial formation of the organ anlagen, this result may have been expected. However, we were encouraged to revisit this issue following the observation that, under special circumstances, Foxn1 can exhibit inductive activity in cell lines, leading to a stable phenotype of thymic epithelial cells even after transplantation into host mice14,15. Moreover, lineage tracing experiments have provided evidence that parathyroid-fated epithelia might change their phenotype to become thymopoietic in vivo19. Hence, we speculated that, given the functional parallels between Gcm2 and Foxn1 during normal development, overexpression of Gcm2 may have the same inductive activity in a permissive cell type.
The outcomes of our in vivo experiments, though unsuccessful when viewed from the standpoint of fate conversion, nonetheless provide important information on the developmental stability of cellular fates in the two major domains of the third pharyngeal pouch. Collectively, our data support the notions that Gcm2 and Foxn1 function as executors rather than as initiators of cell fate, when their activities are probed in their developmentally most closely related cell types.
In the case of the parathyroid, we were unable to examine a possible inductive capacity of Foxn1 in Gcm2-null mice. Hence, we cannot exclude that Gcm2 is dominant over the activity of Foxn1 and, as a consequence, blocks the conversion of the parathyroid to thymopoietic tissue. With respect to the developmental scenarios (Fig. 1a) underlying the transdifferentiation of parathyroid-fated cells to thymopietic epithelium19, we speculate that the survival of transdifferentiated Gcm2-negative, Foxn1-positive cells is possible because loss of Gcm2 occurs at a stage when its anti-apoptotic function is no longer required. If true, then our results indicate that this sensitive period ends at E15/16, when the transdifferentiated cervical thymi become readily apparent19. By contrast, the inductive capacity of Gcm2 expression could be examined in the thymic epithelium, both in the presence and in the absence of Foxn1 activity, since the thymic rudiment persists in the absence of an intact Foxn1 gene. However, in both cases, reprogramming of thymic epithelial cells to cells resembling the phenotype of parathyroid tissue failed.
Collectively, the results of the present experiments strongly suggest that both transcription factors require a permissive gene regulatory network in order to establish tissue identity. Indeed, it was shown that expression of Pth requires the cooperation of Gcm2, Mafb, and Gata3 transcription factors in cultured cells30,31,32; the latter two genes are not expressed in thymic epithelial cells37,38. We anticipate an analogous situation for Foxn1 in the parathyroid environment. Although to the best of our knowledge, no such information is yet available, we presume that Foxn1 also requires certain co-factor(s) to exert inductive capacity. The conclusion of context-dependent activity of Foxn1 is supported by the fact that although Foxn1 is expressed in thymic epithelial cells, and additionally in suprabasal keratinocytes of the skin, and in the hair follicle39,40,41, it fails to induce thymopoietic activity in the latter two cell types. Only in thymic epithelial cells does it govern the expression of the characteristic thymopoietic programme, such as the Notch ligand Dll4 and the chemokine Cxcl1210,34. By contrast, it has been reported that overexpression of Foxn1 in fibroblasts in vitro converts them to cells with thymopoietic capacity that persists even after transplantation into mice14,15. In the light of the present results, we interpret this latter finding to indicate that fibroblasts provide a permissive transcription factor landscape for Foxn1 to gain inductive activity. Of note, fibroblasts originate from a different germ layer than the derivatives of the pharyngeal endoderm, possibly facilitating the fate-changing capacity of the Foxn1 transcription factor. Future work should be directed at defining critical co-factors of Gcm2 and Foxn1, which, depending on cell type, may consist of different combinations of negative and/or positive regulators.
Methods
Mice
The following alleles were generated during the course of this work. The Foxn1:Gcm2 transgene was generated by cloning a 27,970-bp Foxn1 promoter fragment (Y12488, nucleotides 5680–33,650) upstream of the mouse Gcm2 cDNA (Genbank accession number NM_008104; nucleotides 35–1549; downstream of the cDNA the bovine growth hormone polyadenylation sequence (5´- GCCATCTGTTGTTTGCCCCTCCCCCGTGCCTTCCTTGACCCTGGAAGGTGCCACTCCCACTGTCCTTTCCTAATAAAATGAGGAAATTGCATCGCATTGTCTGAGTAGGTGTCATTCTATTCTGGGGGGTGGGGTGGGGCAGGACAGCAAGGGGGAGGATTGGGAAGACAATAGCAGGCATGCTGGGGATGCGGTGGGCTCTATGG) was inserted29 (Fig. 2a). The construct was linearized and injected into fertilized eggs of FVB mice to generate the FVB/N-tg(Foxn1-Gcm2)1Tbo/Mpie transgenic line. A Gcm2 null allele (FVB/N-Gcm2em1Tbo/Mpie) was generated by CRISPR/Cas9-mediated internal deletion of nucleotides 124,954 to 127,882 in Genbank accession number AC158538.2 in the Gcm2 gene using two sgRNAs (nucleotides 124,953 to 124,971; nucleotides 127,878 to 127,896 in Genbank accession number AC158538.2) as described42; note that nucleotides 125,018 to 125,035 in Genbank accession number AC158538.2 are retained in the deletion. A Gcm2:Foxn1 knock-in allele (FVB/N-Gcm2em1(Foxn1)Tbo/Mpie) was generated by homologous recombination in fertilized oocytes primed by simultaneous CRISPR/Cas9-induced internal deletion as described for the Gcm2 null allele. The left homology arm of the construct contained nucleotides 120,953 to 124,925; the right homology arm spanned nucleotides 128,029 to 134,119 in Genbank accession number AC158538.2. Sandwiched between the two homology arms are positioned mouse Foxn1 cDNA sequences (nucleotides 95 to 2,417 in Genbank accession number XM_017314286.2), followed by a loxP site (nucleotides 7,256–7,320 in Genbank accession number EF591490.1), a poly(A) signal sequence (nucleotides 6,608 to 6,661 in Genbank accession number AJ627603.1), a Neo expression cassette (nucleotides 2,400 to 3,915 in Genbank accession number MK802894.1), a loxP site (nucleotides 52,030 to 52,063 in CP049847.1.). The structures of these two new alleles are described in Fig. 4. The germline Cre-deleter line (B6.C-Tg(CMV-cre)1Cgn/J33 and the Foxn1-deficient line (Foxn1tm1Tbo)7 were described earlier. Timed matings were used to retrieve embryos at embryonic day (E)14.5; the day of the plug was counted as day E0.5 of gestation. Mice were kept in the animal facility of the Max Planck Institute of Immunobiology and Epigenetics under specific pathogen-free conditions.
Ethics declaration. All experimental protocols were approved by the ethics committee of the Max Planck Institute of Immunbiology and Epigenetics, under license permit 35–9185.81/G-14/57 issued by the Regierungspräsidium Freiburg, Germany. All methods and procedures were carried out in accordance with the relevant guidelines and regulations. All methods are reported in accordance with ARRIVE guidelines. The following information is given in the manuscript: genotypes of animals, groups of animals being compared, sample size, outcome measures, statistical methods, and relevant experimental procedures. No animal was excluded from the study, randomization was not performed, and experimenters were not blinded.
Genotyping
The following primer combinations were used to genotype the mice used in this study. Gcm2 wild-type allele: 5´-ccatcttttcccaggaaccaa; 5´-agttggtgacagggtatccgc (amplicon size 465 bp). Gcm2 null allele: 5´- ccatcttttcccaggaaccaa; 5´-ttgtcaaagctaaagggctgc (amplicon size 200 bp, spanning the deletion). Validation of Gcm2:Foxn1 knock-in allele by long range PCR: first round, 5´-aaggtgcttctgttcagagggatgc; 5´-atggaaggcaggctgagaagaacag (amplicon size 5,346 bp); second round, 5´-acccacagatgcctcaggtaag; 5´-tgcaccaagcctctgctggga (amplicon size 4,587 bp) under conditions optimal for long-range PCR (Roche). Presence of the knock-in cassette and its correct insertion into the Gcm2 locus can also be ascertained by Southern blot hybridization using a probe spanning nucleotides 119,822 to 120,317 in Genbank accession number AC158538.2. In subsequent matings, the Gcm2:Foxn1 knock-in allele was traced using: 5´-cccagactcccctgccccac; 5´-ggtgtcctctctggagggccg (amplicon size 84 bp for the inserted cDNA, 481 bp for the endogenous Foxn1 gene which serves as internal control for the reaction); in addition, the absence of an internal Gcm2 sequence (replaced by the knock-in cassette) was examined by 5´-ctttgccatcttttcccaggaac; 5´-caccatttctgccctcactttg (amplicon size 291 bp). Cre: 5´-tgcatgatctccggtattga; 5´-cgtactgacggtgggagaat (amplicon size 374 bp); Foxn1:lacZ knock-in allele: multiplex assay using 5´-tgcaccaagcctctgctggga; 5´-ctgtgaactcagccatactc; 5´-tcgccttcttgacgagttct (wild-type amplicon size 500 bp; knock-in amplicon size 230 bp). The Foxn1:Gcm2 transgene can be detected using primers 5´-cattgtagctggctttcttcgag and 5´-cttgtcacagatggctggcctcagc (amplicon size 330 bp).
RNA in situ hybridisation
Gene-specific probes for Gcm2, Pth, Gzma, and Foxn1 and conditions for RNA in situ hybridisation have been described25,26; details of Dll4 and Il7 probes can be found in (ref. 10); B cells were detected using a Cd19 probe corresponding to nucleotides 1155–1886 of Genbank accession number NM_001357091.1. The Gcm2 probe recognizing the 5´-end of the mRNA (that is, upstream of the insertion site in the knock-in allele) consists of nucleotides 1229 in Genbank accession number XM_017315354.
Image analysis
Images were acquired on Zeiss microscopes (Axioplan 2 or Imager Z1 with ApoTome attachment) equipped with AxioCam MRc 5 cameras.
Statistical analysis and reproducibility
Two tailed t-tests were used to determine the significance levels of the differences between the means of two independent samples, considering equal or unequal variances as determined by the F-test.
Data availability
All data supporting the conclusions of this paper are included.
References
Günther, T. et al. Genetic ablation of parathyroid glands reveals another source of parathyroid hormone. Nature 406, 199–203 (2000).
Liu, Z., Yu, S. & Manley, N. R. Gcm2 is required for the differentiation and survival of parathyroid precursor cells in the parathyroid/thymus primordia. Dev. Biol. 305, 333–346 (2007).
Yamada, T. et al. Gcm2 regulates the maintenance of parathyroid cells in adult mice. PLoS ONE 14, e0210662 (2019).
Nehls, M., Pfeifer, D., Schorpp, M., Hedrich, H. & Boehm, T. New member of the winged-helix protein family disrupted in mouse and rat nude mutations. Nature 372, 103–107 (1994).
Blackburn, C. C. et al. The nu gene acts cell-autonomously and is required for differentiation of thymic epithelial progenitors. Proc. Natl. Acad. Sci. USA 93, 5742–5746 (1996).
Gordon, J., Bennett, A. R., Blackburn, C. C. & Manley, N. R. Gcm2 and Foxn1 mark early parathyroid- and thymus-specific domains in the developing third pharyngeal pouch. Mech. Dev. 103, 141–143 (2001).
Nehls, M. et al. Two genetically separable steps in the differentiation of thymic epithelium. Science 272, 886–889 (1996).
Yuan, Z., Opas, E. E., Vrikshajanani, C., Libutti, S. K. & Levine, M. A. Generation of mice encoding a conditional null allele of Gcm2. Transgenic Res 23, 631–641 (2014).
Bleul, C. C. et al. Formation of a functional thymus initiated by a postnatal epithelial progenitor cell. Nature 441, 992–996 (2006).
Calderón, L. & Boehm, T. Synergistic, context-dependent, and hierarchical functions of epithelial components in thymic microenvironments. Cell 149, 159–172 (2012).
Itoi, M., Kawamoto, H., Katsura, Y. & Amagai, T. Two distinct steps of immigration of hematopoietic progenitors into the early thymus anlage. Int. Immunol. 13, 1203–1211 (2001).
Romano, R. et al. FOXN1: a master regulator gene of thymic epithelial development program. Front. Immunol. 4, 187 (2013).
Du, Q. et al. FOXN1 compound heterozygous mutations cause selective thymic hypoplasia in humans. J. Clin. Invest. 129, 4724–4738 (2019).
Bredenkamp, N. et al. An organized and functional thymus generated from FOXN1-reprogrammed fibroblasts. Nat. Cell Biol. 16, 902–908 (2014).
Oh, J., Wang, W., Thomas, R. & Su, D. M. Thymic rejuvenation via FOXN1-reprogrammed embryonic fibroblasts (FREFs) to counteract age-related inflammation. JCI Insight 5, e140313 (2020).
Civitareale, D., Lonigro, R., Sinclair, A. J. & Di Lauro, R. A thyroid-specific nuclear protein essential for tissue-specific expression of the thyroglobulin promoter. EMBO J. 8, 2537–2542 (1989).
Terszowski, G. et al. Evidence for a functional second thymus in mice. Science 312, 284–287 (2006).
Corbeaux, T. et al. Thymopoiesis in mice depends on a Foxn1-positive thymic epithelial cell lineage. Proc. Natl. Acad. Sci. USA 107, 16613–16618 (2010).
Li, J., Liu, Z., Xiao, S. & Manley, N. R. Transdifferentiation of parathyroid cells into cervical thymi promotes atypical T-cell development. Nat. Commun. 4, 2959 (2013).
Liu, Z. et al. Thymus-associated parathyroid hormone has two cellular origins with distinct endocrine and immunological functions. PLoS Genet. 6, e1001251 (2010).
Planas, G. et al. What to do when the parathyroid gland is not found during a transcervical exploration?. Interact Cardiovasc. Thorac. Surg. 33, 82–84 (2021).
Kojima, A., Saito, M., Hioki, K., Shimanura, K. & Habu, S. NFS/N-nu/+ mice can macroscopically be distinguished from NFS/N-+/+ littermates by their thymic size and shape. Exp. Cell Biol. 52, 107–110 (1984).
Bosticardo, M. et al. Heterozygous FOXN1 Variants Cause Low TRECs and Severe T Cell Lymphopenia, Revealing a Crucial Role of FOXN1 in Supporting Early Thymopoiesis. Am. J. Hum. Genet. 105, 549–561 (2019).
Schorpp, M., Hofmann, M., Dear, T. N. & Boehm, T. Characterization of mouse and human nude genes. Immunogenetics 46, 509–515 (1997).
Swann, J. B., Krauth, B., Happe, C. & Boehm, T. Cooperative interaction of BMP signalling and Foxn1 gene dosage determines the size of the functionally active thymic epithelial compartment. Sci Rep. 7, 8492 (2017).
Swann, J. B., Happe, C. & Boehm, T. Elevated levels of Wnt signaling disrupt thymus morphogenesis and function. Sci. Rep. 7, 785 (2017).
Swann, J. B., Nusser, A., Morimoto, R., Nagakubo, D. & Boehm, T. Retracing the evolutionary emergence of thymopoiesis. Sci. Adv. 6, eabd9585 (2020).
Swann, J. B. et al. Conversion of the thymus into a bipotent lymphoid organ by replacement of Foxn1 with its paralog, Foxn4. Cell Rep. 8, 1184–1197 (2014).
Bleul, C. C. & Boehm, T. BMP signaling is required for normal thymus development. J. Immunol. 175, 5213–5221 (2005).
Han, S. I., Tsunekage, Y. & Kataoka, K. Gata3 cooperates with Gcm2 and MafB to activate parathyroid hormone gene expression by interacting with SP1. Mol. Cell Endocrinol. 411, 113–120 (2015).
Kamitani-Kawamoto, A. et al. MafB interacts with Gcm2 and regulates parathyroid hormone expression and parathyroid development. J. Bone Miner. Res. 26, 2463–2472 (2011).
Grigorieva, I. V. et al. Gata3-deficient mice develop parathyroid abnormalities due to dysregulation of the parathyroid-specific transcription factor Gcm2. J. Clin. Invest. 120, 2144–2155 (2010).
Schwenk, F., Baron, U. & Rajewsky, K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23, 5080–5081 (1995).
Zuklys, S. et al. Foxn1 regulates key target genes essential for T cell development in postnatal thymic epithelial cells. Nat. Immunol. 17, 1206–1215 (2016).
Bain, V. E. et al. Tissue-specific roles for sonic hedgehog signaling in establishing thymus and parathyroid organ fate. Development 143, 4027–4037 (2016).
Gordon, J., Patel, S. R., Mishina, Y. & Manley, N. R. Evidence for an early role for BMP4 signaling in thymus and parathyroid morphogenesis. Dev. Biol. 339, 141–154 (2010).
Sultana, D. A. et al. Gene expression profile of the third pharyngeal pouch reveals role of mesenchymal MafB in embryonic thymus development. Blood 113, 2976–2987 (2009).
Wei, Q. & Condie, B. G. A focused in situ hybridization screen identifies candidate transcriptional regulators of thymic epithelial cell development and function. PLoS ONE 6, e26795 (2011).
Mecklenburg, L., Tychsen, B. & Paus, R. Learning from nudity: lessons from the nude phenotype. Exp. Dermatol. 14, 797–810 (2005).
Schlake, T. The nude gene and the skin. Exp. Dermatol. 10, 293–304 (2001).
Meier, N., Dear, T. N. & Boehm, T. Whn and mHa3 are components of the genetic hierarchy controlling hair follicle differentiation. Mech. Dev. 89, 215–221 (1999).
Iwanami, N. et al. Epigenetic protection of vertebrate lymphoid progenitor cells by Dnmt1. iScience 23, 101260 (2020).
Acknowledgements
This work was supported by the Max-Planck Gesellschaft. We thank Benoît Kanzler for his help with the preparation of transgenic mouse lines, Brigitte Krauth, Dagmar Diekhoff, Christiane Happe, and Stephanie Birmelin for their technical help, Jeremy Swann and Anja Nusser for helpful discussions and advice.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
All authors designed experiments and analysed data. T.B. conceived the study, and wrote the paper with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nagakubo, D., Hirakawa, M., Iwanami, N. et al. Limits to in vivo fate changes of epithelia in thymus and parathyroid by ectopic expression of transcription factors Gcm2 and Foxn1. Sci Rep 12, 13554 (2022). https://doi.org/10.1038/s41598-022-17844-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-17844-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.