Abstract
Vitiligo is considered a disabling disease that affects physical, social, psychological, and occupational aspects of an individual's quality of life. The search for non-invasive and reliable biomarkers for vitiligo's early diagnosis, prognosis, and treatment prediction is under intensive investigation. There is currently an emerging interest in employing miRNAs as biomarkers to predict vitiligo diagnosis and prognosis, inspired by the well-preserved nature of miRNAs in serum or plasma. In the current study, we assessed a panel of 20 melanogenesis pathway-related microRNAs (miRNAs) using quantitative real-time PCR technique in 85 non-segmental vitiligo (NSV) patients compared to 85 normal controls followed by function and pathway enrichment analysis for the miRNAs with significant results. Twelve out of the 20 circulating miRNAs showed significantly higher expression levels in vitiligo patients relative to controls where miR-423 show the highest expression level followed by miR-182, miR-106a, miR-23b, miR-9, miR-124, miR-130a, miR-203a, miR-181, miR-152, and miR-320a. While six miRNAs (miR-224, miR-148a, miR-137, and miR-7, miR-148b, miR-145, miR-374b, and miR-196b) didn’t show significant expression level. The analysis of the receiver operating curve indicated that miR-423, miR-106a, and miR-182 were outstanding biomarkers with the highest areas under the curve in vitiligo. This study is the first Egyptian study to investigate a panel of miRNAs expression profile in the plasma of patients with NSV. Our results suggest that specific circulating miRNAs signature might be implicated in vitiligo pathogenesis and could potentially be used as biomarkers in vitiligo.
Similar content being viewed by others
Introduction
Vitiligo is a common, acquired discoloration of the skin affecting about 1–4% of the world population, presenting as milky-white patches over the skin and/or mucosa. Although the disease does not produce direct physical impairment, it may considerably influence the psychological well-being of the patients1,2, resulting in psychosocial distress and social stigmatization. So, vitiligo is considered a disabling disease that affects physical, social, psychological, and occupational aspects of an individual's quality of life (QoL).
Vitiligo can be classified into two major types; segmental vitiligo (SV) and non-segmental vitiligo (NSV), which accounts for nearly 90% of total vitiligo cases3. Vitiligo usually appears in childhood or early adulthood, with the highest incidence between 10 to 30 years4.
Although vitiligo has been known for thousands of years—only recently—real progress in understanding its molecular and pathological basis has emerged, which may hopefully facilitate progress in vitiligo treatment and ultimately even prevention5. The most supported etiology is the autoimmune hypothesis6. Autoantibodies against melanocytes were detected in patients' sera and have recently been used to predict disease prognosis7. Association of other autoimmune diseases, such as Hashimoto's thyroiditis, Grave's disease, and pernicious anemia with vitiligo, reinforce the autoimmune cause8. Besides immune cause, toxic damage to melanocytes due to metabolic derangements are gaining support with time9.
In all cases, vitiligo is a polygenic disease entailing complex interaction between genetic and non-genetic factors10,11. Among the genetic factors that could be implicated in the pathogenesis of vitiligo; are the microRNAs (miRNAs). MiRNAs are short (19–25 nucleotides) non-coding RNAs (ncRNAs) that are involved in the regulation of gene expression at both the post-transcriptional and translation level through imperfect binding with the 3′ untranslated region (3′UTR) of the target mRNA and subsequent degradation of mRNA or translational repression3,12,13,14. Multiple miRNAs can target a single mRNA molecule, while one miRNA can interact with multiple mRNAs15.
Various human miRNAs have been widely investigated in the past ten years. They have been found to regulate diverse physiological processes, including cell proliferation, differentiation, development, signal transduction, metabolism, apoptosis, and immune responses. Their abnormal expression is involved in developing many diseases16. It has been demonstrated that miRNAs circulate in a highly stable cell-free form in various body fluids including plasma, serum, saliva, milk, and urine, and are believed to be promising biomarkers for different diseases17.
Many studies have demonstrated the crucial role of miRNAs on the development, proliferation, and survival of cells, including the immune cells and melanocytes3,12,13,18. Deregulation of miRNAs was the underlying pathology in many inflammatory skin disorders such as atopic dermatitis, allergic contact dermatitis, and psoriasis19. Microarray analysis has elucidated the abnormal expression of multiple miRNAs in the skin and serum of patients with vitiligo12. Currently, there is an emerging interest in employing miRNAs as biomarkers to predict disease prognosis and response to treatment, inspired by the well-preserved nature of miRNAs in serum or plasma15.
In this vicinity, we have done this pilot study to investigate the differential expression of a panel of 20-miRNAs in the plasma collected from age-matched vitiligo patients and healthy controls using quantitative real-time PCR (qRT-PCR). Those 20-miRNAs are involved in the melanogenesis pathway, have putative binding sites for the most affected genes in vitiligo, and were selected using bioinformatics databases.
Subjects and methods
Study participants
The present study included one hundred and seventy participants. The study subjects were divided into two groups; (1) Study Group: 85 adult patients from both genders diagnosed with vitiligo by clinical examination and Woods's lamp recruited from the Dermatology outpatient clinic, Suez Canal University (SCU) Hospital, Ismailia, Egypt, and (2) Control Group: 85 healthy non-related participants, matched by age and gender to the study group. The clinicopathological data, including patients' age, sex, BMI, family history, past history of other autoimmune diseases (e.g., diabetes mellitus, Hashimoto's thyroiditis, Addison's disease, psoriasis), age of disease onset, disease duration, severity, and treatment were collected from the patients' history. All patients were subjected to detailed dermatological examination to determine: the site, size, pattern, and distribution of individual lesions, assessment of disease severity was performed according to the criteria of the vitiligo area severity index (VASI), vitiligo disease activity score (VADI) and the Vitiligo European Task Force (VETF) (Kawakami and Hashimoto, 2011).
Selection of the 20 circulating miRNAs understudy using bioinformatics tools
The 20 circulating miRNAs involved in the melanogenesis pathway [hsa04916] were selected using online bioinformatics tools, which are DIANA-miRPath web server (http://snf-515788.vm.okeanos.grnet.gr)20 and microrna.org21 and are listed in Table 1.
Samples collection and total RNA extraction, including small RNA
Three ml of fresh venous blood was collected in vacutainer tubes containing ethylene diamine tetra-acetic acid (EDTA) anticoagulant. Blood samples were centrifuged to separate plasma; 100 μl plasma was preserved in a 500 μl Qiazole reagent. The plasma samples were stored at − 80℃ till further analysis. The total RNA, including small RNA, was isolated from plasma using Qiagen miRNeasy mini kit and (Qiagen, Catalog no. 217004) following the protocol supplied by the manufacturer. An Eppendorf 5417C cooling microcentrifuge with adjusted temperature was used throughout the RNA extraction process. RNA purity and concentration were assessed using the NanoDrop 2000 1C at 260 and 280 nm absorbance (NanoDrop Tech., Inc. Wilmington, DE, USA). A ratio between 1.8 and 2.2 was considered acceptable for further genetic analysis. The wavelength-dependent extinction coefficient "33" was adjusted to represent the micro-component of all RNA in solution.
The circulating miRNAs relative gene expression quantification using Real-time PCR
The total RNA extracted was subjected to reverse transcription (RT) where complementary DNA (cDNA) was generated from total RNA containing miRNA with the miScript II RT Kit (Qiagen, Catalog no. 218161), in which miRNAs were polyadenylated by poly(A) polymerase and converted into cDNA by reverse transcriptase with oligo-dT priming. RT was carried out in a Veriti™ 96-Well Thermal Cycler (Applied Biosystems, USA) at 37 °C for 1 h, followed by inactivation of the reaction by briefly incubating at 95 °C.
Circulating miRNAs expression profiling was carried out using SYBR Green-based real-time PCR. The premix of cDNA was used as a template for qRT-PCR miRNA expression. Primers for the 20 miRNAs (miR-7, miR-9, miR-23b, miR-106a, miR124, miR-130a, miR-137, miR1-45, miR-148a, miR-148b, miR-152, miR-155, miR-181a, miR-182, miR-196b, miR-203a, miR-224, miR-320a, miR-374b, miR-423) are described in Table 1, and miScript SYBR Green PCR Kit (Qiagen, cat. no 218076) was used to measure the expression levels with a universal reverse primer. RNU6B and SNORD68 were used as endogenous controls to enable data analysis using the ΔΔCT method of relative quantification. The expression levels were done according to the minimum information for publication of quantitative real-time PCR experiments (MIQE) guidelines. Duplicate reactions were placed in each run, and a "no reverse transcribed" controls and a well free template were included in each run. Each plate run initially at 95 °C for 5 min, followed by 40 cycles of denaturation, annealing and extension at 95 °C (15 s), 55 °C (1 min), 72 °C (1 min) respectively.
Gene expression data analysis
Fold changes for the 20 circulating miRNAs in each patient sample relative to the corresponding control were estimated using Livak method22 based on the ΔΔCq method where ΔΔCq = (Cq Cir-miRNA − Cq SNORD68/RNU6B) vitiligo − (Cq Cir-miRNA − Cq SNORD68/RNU6B) control. Cq stands for quantification (threshold).
Function and pathway enrichment analysis of the circulating miRNAs understudy
Functional enrichment analysis of the 20 circulating miRNAs understudy was analyzed using gene analytics software (https://geneanalytics.genecards.org)23, and gProfiler software (http://biit.cs.ut.ee/gprofiler/gost)24. The gene ontology GO terms that matched our miRNAs understudy were presented in the order of the matching scores. The binomial distribution was used to test the null hypothesis that the input biomarkers were not over-represented within any SuperPath or GO term. The presented score for each biomarker is a transformation (log2) of the resulting p value, where higher scores indicated better matches. Results with p values lower than 10–50 were assigned the maximum score. The pathway enrichment analysis was conducted using the software Database for Annotation Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/)25, and DIANA Tool mirPath v.3 (http://snf-515788.vm.okeanos.grnet.gr/)20, where GO consisting of cellular components, biological processes, and molecular functions terms was searched for via pathway analysis on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, for determining the affected pathways with differential miRNA expression and their target genes.
MiRNA regulatory network construction
The targets of the statistically significant differentially expressed miRNAs (DEmiRNAs) were predicted using miRTargetLink 2.0 (Version 2.0, https://ccb-compute.cs.uni-saarland.de/)26. Statistically homogenous and significant DEGs targets were kept, and the miRNA-mRNA pairs negative correlations were also included.
Statistical analysis
Statistical Package for the Social Sciences (SPSS) for Windows software version 26.0 (Armonk, NY: IBM Corp.) was used. The study power and the sample size were calculated by G*Power version 3.1.9.2. The given power for gene expression study design option was 85% at total sample size = 170. Continuous variables were presented as means ± standard deviations, while categorical variables were presented as frequencies and percentages. Data profile was tested for outliers and normality. When appropriate, Mann–Whitney and Student t tests were used to compare between cases and controls. The Spearman correlation test was used for determining the correlation coefficient. Spearman's rank test was done for two-sided p values correlation analysis. A two-tailed p value of < 0.05 was considered statistically significant. The area under the curve (AUC) of receiver operating characteristic (ROC) was plotted to evaluate putative biomarkers' diagnostic and prognostic value.
Ethics approval and consent to participate
The study was approved by the Suez Canal University, Faculty of Medicine, Ethics Committee in Ismailia, Egypt (Approval No. 4210) and conducted according to the Declaration of Helsinki’s guidelines. Informed consent was obtained from all individual participants included in the study.
Results
Baseline characteristics and risk factors among the study population
Table 2 shows the baseline characteristics and risk factors among our study population. The age distribution among the study population ranged from 6 to 93 years. The mean age for the patients was 35.41 ± 17.7, and the mean age for the controls was 35.47 ± 18.75, with no statistically significant difference between both groups. Regarding Special habits, 56.5% of patients were passive smokers, 21.2% smokers, and 22.2% non-smokers. In comparison, 27.1% of controls were passive smokers, 20% smokers, and 37.5% non-smokers, while non-smoking showed statistically significant protective relation against vitiligo. There was no statistical difference among both groups concerning obesity grades. The family history and exposure to stress showed a highly significant correlation among our vitiligo patients.
Clinicopathological features among vitiligo patients
Table 3 shows the clinicopathological features among vitiligo patients. The patient’s skin type, premature graying of hair, trichrome, hypopigmentation, follicular re-pigmentation, Koebner’s phenomena, VIDA, and VASI score showed high statistically significant relation with vitiligo. At the same time, the duration of the disease did not appear to affect the clinicopathological features in vitiligo among the studied cohort.
The expression signature of melanogenesis pathway circulating miRNAs in vitiligo patients’
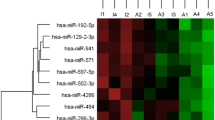
As depicted in Figs. 1 and 2, 14 circulating miRNAs showed significantly higher expression levels in vitiligo patients relative to controls where miR-423 show the highest expression level (Log2FC = 5.7) followed by miR-182 (Log2FC = 5.1), miR-106a (Log2FC = 4.5), miR-155 (Log2FC = 3.8), miR-23-b (Log2FC = 3.4), miR-9 (Log2FC = 3.1), miR-124 (Log2FC = 3.0), miR-130a (Log2FC = 2.8), miR-203a (Log2FC = 2.4), miR-181 (Log2FC = 2.4), miR-152 (Log2FC = 2.03), miR-320a (Log2FC = 1.6), miR-224 (Log2FC = 1.6), and miR-148a (Log2FC = 1.2). While miR-137 (Log2FC = 0.9), and miR-7 (Log2FC = 0.7), miR-148b (Log2FC = 0.4), miR-145 (Log2FC = -0.2), miR-374b (Log2FC = -0.5), and miR-196b (Log2FC = -0.9) didn’t show statistically significant expression levels in vitiligo patients relative to controls.
The differential expression profile of circulating miRNAs understudy in vitiligo (n = 85). Heat map illustrates the levels of all miRNAs understudy (Log2fold change) in vitiligo patients. Color grades are shown within each row, with the highest expression corresponding to deep red and the lowest to deep blue.
The relative expression level of the circulating miRNAs understudy in vitiligo. Twenty miRNAs were analyzed: miR-7, miR-9, miR-23b, miR-106a, miR124, miR-130a, miR-137, miR1-45, miR-148a, miR-148b, miR-152, miR-155, miR-181a, miR-182, miR-196b, miR-203a, miR-224, miR-320a, miR-374b, and miR-423. SNORD68 and RNU6B were used as endogenous controls. The values are represented as median (Q1 and Q3) using Whiskers and bars. Control level were set at the Log2 fold change equals 0 and all values were log-transformed. Mann–Whitney U test was used for comparison. *Significant at p value < 0.05, **Significant at p value < 0.01.
Circulating miRNAs predictive significance as biomarkers by ROC analysis
The ROC analysis including the AUC and probability levels, were presented in Table 4. The AUCs of the twenty miRNAs ranged from 0.447 to 0.976 are shown in Table 4 indicating miR-423 as the most efficient biomarker with the largest AUC (0.976 at the cutoff point of 1.57-fold; p < 0.001, sensitivity = 97.6%, and specificity = 100%), followed by miR-106a (AUC = 0.953), miR-182 (AUC = 0.941) with outstanding discrimination efficiency. MiR-23b (AUC = 0.859), miR-124 and miR-9 (AUC = 0.835) with excellent biomarker discrimination efficiency. MiR-155 (AUC = 0.771), miR-130a (AUC = 0.765), miR-181a (AUC = 0.753), miR-152 (AUC = 0.729), and miR-320a (AUC = 0.706) with acceptable biomarker discrimination power.
Correlation analysis of circulating miRNAs differential expression levels and vitiligo patients’ clinical characteristics
Concerning clinical characteristics correlations with circulating miRNAs shown in Table 5, skin type showed significant correlations with nearly all miRNAs understudy except for miR-196b, miR-224, and miR-423. While age correlated significantly with miR-203a and VASI score significantly correlated with miR-423 only.
On the other hand, the 20 selected circulating miRNAs showed various distributions among all vitiligo patients. The Spearman’s rank correlation of the 20 selected circulating miRNAs in vitiligo patients and controls was evaluated and presented in Table 6. There was a strong association between nearly all miRNAs understudy in vitiligo s with Spearman’s correlation coefficient of 0.59 and more and a two-tailed significance p < 0.001.
At the same time, many correlations were found among our studied risk factors in vitiligo as illustrated in Table 7 where age significantly increased the association with other chronic disease as CAD (r = 0.233, p = 0.032), hypertension (r = 0.348, p < 0.001), diabetes (r = 0.314, p = 0.003) and obesity (r = 0.472, p < 0.001). The duration of complaint correlated with skin type (r = 0.302, p = 0.005), obesity (r = 0.162, p = 0.138) and age (r = 0.425, p < 0.001).
Finally, regarding correlation analysis of the different clinical characteristics of vitiligo in the study shown in Table 8, VASI score significantly correlated with pre-mature graying of hair (r = 0.308, p = 0.004), hypopigmentation (r = 0.310, p = 0.004), Kopner phenomenon (r = 0.255, p = 0.018), and disease distribution (r = 0.421, p < 0.001).
Pathway and function enrichment analysis for the circulating miRNAs in vitiligo
A pathway enrichment analysis based on annotated gene targets in GO was performed to identify all the pathways targeted by DEmiRNAs in vitiligo. Different databases were used to assess the 20 miRNAs understudy regulatory functions and identify the miRNAs' molecular pathways under study. Enrichment of specific pathways revealed that the top pathways involved were proteoglycans in cancer, Hippo signaling pathway, fatty acid metabolism, protein processing in the endoplasmic reticulum, adherens junction, endocytosis, fatty acid biosynthesis, and biosynthesis of unsaturated fatty acids were found as shown in Fig. 3A,B.
The GO biological processes related to vitiligo pathogenesis were found to be distinctly enriched in our analysis were gene silencing by miRNA, posttranscriptional gene silencing by RNA, posttranscriptional gene silencing, gene silencing by RNA, posttranscriptional regulation of gene expression, negative regulation of gene expression, negative regulation of macromolecule metabolic process, negative regulation of the metabolic process, regulation of gene expression, and negative regulation of biological processes as represented in Fig. 4A.
The gene ontology (GO) for the 20 circulating miRNAs under study. (A) Biological processes in descending order according to score detected using (http://biit.cs.ut.ee/gprofiler/gost) (B) Cellular component according to matching score detected using (https://geneanalytics.genecards.org) (C) Molecular function according to matching score detected using (https://geneanalytics.genecards.org).
The 20 circulating miRNAs understudy showed cellular localization in extracellular space, extracellular vesicle, RNA-induced silencing complex (RISC), extracellular exosome, apical part of the cell, and perinuclear region of cytoplasm as shown in Fig. 4B. Considering the molecular functions for the 20 miRNAs shown in Fig. 4C, the functions encompassed mRNA binding involved in posttranscriptional gene silencing, mRNA 3UTR binding, high-density lipoprotein particle binding, RNA polymerase II complex binding, and single-stranded RNA binding.
MiRNA‐mRNA regulatory network construction
Our network analysis identified the relationship between the circulating miRNAs under study and their target genes. Our miRNA-target gene network comprised the 14 significant circulating miRNAs understudy presented in Fig. 5 revealing initially 186 target genes then filtered to include strong validated targets with a minimum of 2 shared targets that revealed a final of 65 target genes using miRTargetLink 2.0 (https://ccb-web.cs.uni-saarland.de/mirtargetlink/network.php) (Fig. 5A). The circulating miRNAs understudy and their targeted genes were related to the pathways potentially involved in vitiligo, such as fatty acid metabolism, protein processing in the endoplasmic reticulum, adherens junction, endocytosis, fatty acid biosynthesis, and biosynthesis of unsaturated fatty acids.
MiRNAs-target gene network analysis. (A) The miRNAs-target gene network with significant differential expression (16 out of 20) in our study. (B) The top three miRNAs according to ROC analysis and their target gene network. MiRNAs targets used were only the validated miRNAs whether with strong or weak validity with additional filter of minimum 2 shared targets. (C) The top three miRNAs according to ROC analysis with filtering options choosing strong validated targets only with minimum shared two targets revealing CREB1 gene as a common shared target (D) The top three miRNAs according to ROC analysis with filtering options choosing strong validated targets only with minimum shared three targets revealing PTEN gene as a common shared target for miR-182 and miR-106a but not with miR-432 using miRTargetLink 2.0 (https://ccb-compute.cs.uni-saarland.de/mirtargetlink2).
According to ROC analysis, the top three miRNAs that can be used as outstanding biomarkers in vitiligo, as depicted in Table 2, were miR-423, miR-106a, and miR-182. So, we have searched for their target gene network using strong and weak validated target with a minimum of 2 shared targets filter that revealed a final of 56 target genes using miRTargetLink 2.0 (https://ccb-web.cs.uni-saarland.de/mirtargetlink/network.php) (Fig. 5B). According to our study results, Kelch Like Family Member 28 (KLHL28), RAB3A Interacting Protein (RAB3IP), Notch Receptor 2 (NOTCH2), Phosphatase and Tensin Homolog (PTEN), CAMP Responsive Element Binding Protein 1 (CREB1), Midnolin (MIDN), Nuclear Receptor Subfamily 3 Group C Member 1 (NR3C1), Proliferation and Apoptosis Adaptor Protein 15 (PEA15), Ribonucleotide Reductase Regulatory Subunit M2 (RRM2), Tyrosine 3-Monooxygenase/Tryptophan 5-Monooxygenase Activation Protein Zeta (YWHAZ), Tumor Protein P53 Inducible Nuclear Protein 1 (TP53INP1), Forkhead Box K1 (FOXK1), and Cyclin D2 (CCND2) can be considered the critical genes involved in vitiligo in relation to miRNAs understudy. Further optimization of filtering options choosing strong validated targets only with minimum shared two targets revealed that CREB1 is a common shared target between the top three miRNAs by ROC analysis shown in Fig. 5C, while increasing shared targets to three revealed the PTEN as a common shared target for miR-182 and miR-106a but not with miR-432 (Fig. 5D).
Discussion
In vitiligo, the search for non-invasive and reliable biomarkers for early diagnosis, prognosis, and treatment prediction is under intensive investigation. Although it is still a mystery how specific miRNAs regulate the melanogenesis pathway, in the current study, we identified a panel of miRNAs that could be used as potential biomarkers in vitiligo. Twelve out of the 20 circulating miRNAs showed significantly higher expression levels in vitiligo patients relative to controls where miR-423 show the highest expression level followed by miR-182, miR-106a, miR-155, miR-23b, miR-9, miR-124, miR-130a, miR-203a, miR-181, miR-152, and miR-320a. While four miRNAs (miR-224, miR-148a, miR-137, and miR-7) didn’t show significant expression level. On the other hand, four miRNAs (miR-148b, miR-145, miR-374b, and miR-196b) showed significantly lower expression levels in vitiligo patients relative to controls.
MiR-423 reported the highest over-expression level among the 20 miRNAs understudy with median log2 fold change (Q1 and Q3) of 5.7(4.06–7.3) and high discriminating power using ROC analysis (sensitivity = 97.6%, and specificity = 100%). MiR-423 has been reported to function as an oncomir in many cancers, including gastric cancer27, prostate cancer28, and glioblastoma28. In contrast, miR-423 functions as a tumor suppressor in osteosarcoma29 and ovarian cancer30. It was also initially identified as a circulating biomarker for heart disease, where Tijsen et al. reported overexpression of circulating miR-423 in patients with heart failure31. Other studies suggested that serum miR-423 plays a role in unstable angina pectoris32. Additionally, it was demonstrated that miR-423 is overexpressed in the plasma of pregnant women with preeclampsia33. Lastly, miR-423 showed to be a valid biomarker for active tuberculosis diagnosis34. Until now, miR-423 was investigated in very few studies related to non-cancerous skin diseases. Regarding vitiligo, Vaish et al. reported over-expression of miR-423 in lesional skin of vitiligo35, and Shang et al. showed an overexpression equivalent to 4.4-fold change using microarray with a significant p value in consistence with our results15. On the other hand, when Shang et al. validated their RT-PCR results, miR-423 showed non-significant results15.
The present study results showed novel findings correlating circulating miR-182, miR-106a, and miR-23b overexpression with vitiligo for the first time. MiR-182 was the second-highest over-expression level in our study with median log2 fold change (Q1 and Q3) of 5.1 (2.6–7.3) and outstanding discriminating power using ROC analysis. Although miR-182 was not previously correlated with vitiligo, it was extensively studied in melanoma. MiR-182 over-expression was reported in melanoma in many studies, and it was suggested to act via converging onto forkhead box O3 (FOXO3), melanocyte inducing transcription factor (MITF), reversion inducing cysteine rich protein with Kazal motifs (RECK), BCL2 apoptosis regulator (BCL2), and CCND2 inactivation or via epigenetic modulation of human melanoma cells36,37,38,39. These results suggest that miR-182 and its subsequent effect on vitiligo candidate genes such as MITF40 could be a useful prognostic or therapeutic biomarker in vitiligo.
Also, no previous results were reported on miR-106a differential expression role in vitiligo. In our study, miR-106a was the third-highest over-expression level with median log2 fold change (Q1 and Q3) of 4.5 (2.5–7.2) and outstanding discriminating power using ROC analysis. However, many studies reported the differential expression of miR-106a in melanoma41,42,43. Where they investigated its role as a tumor suppressor in melanoma by targeting E2F transcription factor 3 (E2F3)41. They also reported the role of its overexpression on melanoma cells via attenuating the effects caused by upregulating Connexin43 (Cx43) expression42 and its inhibition pro-oncogenic effect inhibition in melanoma cells in vitro43.
On the other hand, miR-106a was correlated with another autoimmune skin disorder, psoriasis. Miao et al. reported overexpression of serum level of miR-106a in psoriasis patients44, which was consistent with previous studies on psoriasis and autoimmune disorders45,46. Our study results provide new insight that miR-106a may have a role in vitiligo pathogenesis as it already plays a role in melanoma and psoriasis pathogenesis.
MiR-23b showed an overexpression level with median log2 fold change (Q1 and Q3) of 3.4 (1.3–5.8) and an excellent discriminating power using ROC analysis. No results were previously reported about its role in vitiligo. MiR-23b regulates normal physiological function, cellular immunity, and cell differentiation47 and has been shown to have a potent anti-inflammatory role in tissue-resident cells through its inhibitory actions on nuclear factor kappa B subunit 1 (NF-κB)48,49. Vellaichamy et al. reported underexpression of miR-23b in their in vivo model of post-inflammatory hyperpigmentation50, which is consistent with the downregulation of miR-23b reported in immune-mediated conditions such as lupus and rheumatoid arthritis49. Regarding melanoma, miR-23b was consistently downregulated in previous publications51,52,53 and showed that miR-23b targeted the Nicotinamide Phosphoribosyltransferase (NAMPT) gene53. MiR-23b was reported to have a significant role in autoimmune disorders via its downregulation by the effect of Interleukin 17A (IL-17A)54,55. Among autoimmune diseases, miR-23b proved to have a role in rheumatoid arthritis (RA), where Liu et al. and Abdeen et al. reported higher expression levels of miR-23b in RA patients56,57. Thus, elucidating the specific mechanism for miR-23b expression regulation can provide insights into its precise role in the pathogenesis of vitiligo.
MiR-155 was extensively studied in all autoimmune disorders, including vitiligo. In the present study, miR-155 showed an overexpression level with median log2 fold change (Q1 and Q3) of 3.8 (1.0–6.1) and high discriminating power using ROC analysis. Consistent with our results, Šahmatova et al. and Issa et al. reported overexpression of miR-155 in the epidermis and the peripheral blood of vitiligo patients, respectively12,58. Previous studies described the functions of miR-155 in vitiligo pathogenesis by targeting melanogenesis-associated genes such as tyrosinase related protein 1 (TYRP1), tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon (YWHAE), Syndecan Binding Protein (SDCBP), and SRY-Box transcription factor 10 (SOX10), thereby causing their inactivation35. In addition, the overexpression of miR-155 alters the levels of interferon-regulated genes such as interferon regulatory factor 1 (IRF1), suppressor of cytokine signaling 1 (SOCS1), and Interferon induced transmembrane protein 1 (IFITM1) in melanocytes and encourages interferon gamma (IFN-γ) and tumor necrosis factor alpha (TNF-α) expression confirming its role as proinflammatory miRNA3,14,59. Vaish et al. reported that miR-155 over-expression might contribute to vitiligo pathogenesis and recommended using an antagomiR for miR-155 and thus suppressing vitiligo progression35.
MiR-9 showed an overexpression level with median log2 fold change (Q1 and Q3) of 3.1 (0.9–5.1) and good discriminating power using ROC analysis. MiR-9 is considered one of the miRNAs associated with oxidative stress in vitiligo (Li et al.). MiR-9 expression is affected by oxidative stress and is responsible for mediating ROS pathogenic effect in vitiligo60. Sirtuin 1 (SIRT1) gene is known to be a target for miR-961. Previous studies have indicated that SIRT1 protects against stress-related diseases by interacting with forkhead box protein (FOXO), NFκB, protein P53 (p53), and peroxisome proliferator-activated receptor-gamma coactivator 1-alpha (PGC-1α), which regulate various cellular processes, including inflammation and stress responses62,63. A recent Egyptian study by Raia et al. reported overexpression of miR-9 in both serum and tissues of vitiligo patients64. They also reported a statistical correlation between miR-9 and patients’ VASI scores64. However, Su et al., Shi et al. and Kadir et al. reported downregulation of miR-9 resulting from inducing Interleukin 10 (IL-10) involved in UVB mediated vitiligo repigmentation where IL-10 is known to be an essential immunoregulatory element that is decreased in vitiligo3,65,66.
MiR-124 was identified as a brain-enriched miRNA, but it is expressed in a wide range of human/animal tissues participating in the pathogenesis of several disorders67. MiR-124 plays different roles in various pathologic conditions and suppresses acute stress and inflammatory responses68. In the present study, miR-124 showed an overexpression level with median log2 fold change (Q1 and Q3) of 3.0 (0.3–5.4) and an excellent discriminating power using ROC analysis. It was not previously reported to have a role in vitiligo, but it was reported to be implicated in other autoimmune disorders. Regarding RA, the miR-124 level was underexpressed in RA tissues as reported by Nakamachi et al. and directly downregulating the production of CDK-2 and MCP-168. In addition, Nakamachi et al. concluded that miR-124 might be a promising therapeutic agent for RA and other autoimmune diseases68. MiR-124 was also downregulated in different cancers69, and it functions as a tumor suppressor in melanoma70.
MiR-130a can function as either an oncogene or tumor suppressor in many human diseases76. In the present study, miR-130a showed an overexpression level with median log2 fold change (Q1 and Q3) of 2.76 (0.2–4.9) and a good discriminating power using ROC analysis. It has not been previously investigated in vitiligo, but few studies elucidated its role in cancer and autoimmune disease. A previous study on miR-130a revealed that it could stop cancer metastasis by enhancing antitumor host immunity77. Another study reported the oncogenic role of miR-130a in triggering tumor growth and malignant cell survival by targeting phosphatase and tensin homolog78. At the same time, Wu et al. indicated that miR-130a acts as a tumor suppressor to reduce uveal melanoma metastasis by activating the Wnt/β-catenin signaling pathway by targeting the oncogene Ubiquitin Specific Peptidase 6 (USP6)79. From the perspective of autoimmune diseases, Wade et al. reported an overexpression level of miR-130a in psoriatic patients with higher disease activity80. Wade et al. also stated that miR-130a was among the highest accuracy miRNAs in the stratification of patient response that distinguished good/moderate and non-responders in early psoriatic arthritis80.
In the present study, miR-203a showed an overexpression level with median log2 fold change (Q1 and Q3) of 2.4 (0.24–5.0) and a fair discriminating power using ROC analysis. MiR-203a is a skin-specific miRNA, significantly expressed only in differentiated suprabasal keratinocytes by asymmetric cell division via a driven transcriptional activation mechanism81. MiR-203a has been reported to target significant pigmentation genes like Tyrosinase (TYR)82,83, MITF84, SRY-Box Transcription Factor 9 (SOX9), TYRP1, RAB27a, Myosin VA (MYO5a), and Fascin Actin-Bundling Protein 1 (FSCN1)59. MiR-203 role in vitiligo has been investigated in very few studies, where it did not reveal conclusive results12. Šahmatova et al. results were in contrast to ours as they did not detect any differences in miR-203a expression levels in the skin of control subjects and patients with vitiligo12. In response to narrowband ultraviolet B (NB-UVB), the classical pathway of the p53/αMSH/MC1R/MITF cascade is activated in the keratinocytes while Transforming Growth Factor Beta 1 (TGF-β1) ligand secretion via the miR-203/c-Jun pathway is suppressed, which stimulates the differentiation and proliferation of melanocytes85. NB-UVB also stimulated heparan sulfate-binding growth factors via activating heparanase release, which facilitate the melanogenesis process86,87. MiR-203a overexpression has a critical role in psoriasis pathogenesis13,85,88,89 as it has a role in regulating the keratinocyte differentiation and proliferation as it directly targets and negatively regulates several epidermal genes90. Further, miR-203 with its known function as a key regulator of epidermal differentiation, has been shown to hinder stemness by decreasing the expression of the p63 and thus, affecting melanosome transport mechanisms and subsequently melanogenesis91.
MiR-181a showed overexpression in the present study with median log2 fold change (Q1 and Q3) of 2.4 (− 0.04 to 4.3) and adequate discriminating power using ROC analysis. MiR-181a has not been previously overexpressed in vitiligo, but it has been reported as a central metabolic regulator during development and homeostasis92. It has been implicated in various diseases93,94,95. Among its essential targets is the PTEN gene, where the overexpression of miR-181a acts as the PTEN gene suppressor and is associated with a variety of solid tumors, leukemias, and obesity93,94,95. So, it is currently considered one of the principal guardians of cellular and metabolic regulation96.
MiR-148a, miR-148b, and miR-152 represent members of the miR-148/-152 family97. In the present study, miR-148a and miR-152 were significantly differentially expressed with median Log2 fold change (Q1 and Q3) of 1.2 (− 1.2 to 3.5) and 2.0 (−0.08 to 3.8), respectively. At the same time, miR-148b did not show a significant expression level in vitiligo patients compared to controls. These three miRNAs were not previously investigated in vitiligo. The miR-148/-152 family plays a crucial role in different physiologic and pathological processes, including tumorigenesis and autoimmune diseases. The expression of these miRNA family members is regulated by hypermethylation, transcription factors, cytokines, long non-coding RNAs (lncRNAs), and signal transduction pathways. Their differential expression pattern in autoimmune diseases and tumors suggests that they might be used as prognostic biomarkers and/or as therapeutic targets for these diseases97.
MiR-320a was significantly overexpressed in vitiligo patients than normal controls with median log2 fold change (Q1 and Q3) of 1.6 (− 0.3 to 3.3) in the present study. No previous studies reported a differential expression level of miR-320a in vitiligo. MiR-320a is a member of the miRNA320 family explicitly expressed in epithelial tissues and is involved in skin development, functional maintenance, and homeostasis; thus, it is an essential regulatory factor in the skin98. A study by Wei et al. found that miR-320 expression is significantly underexpressed in psoriatic lesions than in healthy control skin tissues; miR-320 is postulated to participate in psoriasis development by regulating surviving gene expression98. MiR-320 also serves as a potential biomarker for glucose and lipid metabolism-associated diseases like diabetes, atherosclerosis, adiposity, and nonalcoholic fatty liver disease99.
MiR-224 showed significant expression level in vitiligo patients compared to normal controls with median log2 fold change (Q1 and Q3) of 1.59 (− 0.5 to 3.8) in the present study. MiR-224 is differentially expressed in biological processes, including cell proliferation, migration, and invasion, in various malignancies100,101,102,103,104. In consistence with our results, Wang et al. reported significant overexpression of miR-224 in the peripheral blood mononuclear cells (PBMC) patients with non-segmental vitiligo. Thus, they suggested that specific miRNAs signatures in PBMC are a part of the vitiligo-associated immune response, and miRNA may serve as novel drug targets for vitiligo therapy14. Also, Rashed et al. reported overexpression of miR-224 in lesional skin of vitiligo compared to normal controls105.
MiR-137 regulates melanocyte differentiation by repressing MITF expression71 and exerts antitumor effects in melanoma cells by regulating MITF, c-MET, Y-box-binding protein 1, and enhancer of zeste homolog 2 (EZH2)72. In the present study, miR-137 showed non-significant underexpression with median log2 fold change (Q1 and Q3) of 0.9 (− 1.6 to 2.7). In contrast to our results, Dong et al. found overexpression of miR-137 in their study that targets c-KIT in melanocytes73. Overexpression of miR-137 may reduce the expression of Tyrosinase Related Protein 2 (TYRP2) and c-KIT and reduce the increase in melanin production caused by ultraviolet (UV) treatment74. It is suggested that miR-137 can inhibit melanogenesis in mouse skin melanocytes by inhibiting the expression of c-KIT and TYRP2 in the SCF/c-KIT signaling pathway75. Interestingly, miR-137 showed an effect on coat color, demonstrating that changes in the expression of a specific miRNA may significantly affect melanogenesis; however, till now, this is evident only in animal models73.
Different studies revealed that miR-145 plays a role in melanogenesis via targeting genes that are essential for melanogenesis, including Rho Associated Coiled-Coil Containing Protein Kinase 1 (ROCK1), Eukaryotic Translation Initiation Factor 2 Alpha Kinase 1 (EIF2AK1), and Calcium/Calmodulin Dependent Protein Kinase ID (CAMK1D) and the pigmentation process as MITF, TYR, and TYRP117,61,106, our results reported non-significant differential expression of miR-145 which was underexpressed in our study. In contrast to our results, Issa et al. reported overexpression of miR-145 in the peripheral blood of vitligo patients compared to controls58. Another study also described a decrease in cell proliferation and initiation of apoptosis via caspase-3 and caspase-7 induction due to miR-145 overexpression in non-lesional skin of patients with vitiligo via targeting ROCK1, CAMK1D, and EIF2AK161,106. Furthermore, Dyndoot et al. reported underexpression of miR-145 in cultured pigment cells after induction of pigmentation59. In line with our results, Vaish et al.35 reported that miR-145 was downexpressed in vitiligo lesional skin.
Although previous studies reported overexpression of miR-196b in patients with vitiligo, miR-196b showed non-significant underexpression levels in our study15. Moreover, polymorphisms involving miR-196 were suggested to play a significant role in the pathogenesis and the prognosis of vitiligo15.
In conclusion, to better understand the molecular mechanism of vitiligo pathogenesis, it is fundamental to have more information regarding miRNAs and their functions. However, little is known about miRNA-based regulations in vitiligo pathogenesis. This study is the first Egyptian study to analyze the melanogenesis pathway miRNAs-related expression profile in the plasma of patients with NSV. The data raise the possibility that miRNAs may be involved in the pathogenesis of NSV. Based on these results, we suggest that specific circulating miRNAs signature might be a part of the immune response implicated in vitiligo pathogenesis, and miRNAs could potentially be used as biomarkers for skin pigmentation disorders, including vitiligo.
Further multicentric studies with a larger number of cases are needed to emphasize whether the dysregulated miRNAs described in the present study can be utilized as diagnostic, prognostic, and/or therapeutic markers and/or targets for vitiligo. Finally, as there was an overlap in the miRNAs signature pinpointed from different studies, we recommend more observational and experimental research on the studied miRNAs. Observational studies should try to standardize the laboratory methods and address larger cohorts to elucidate miRNAs' role in vitiligo pathogenesis. Experimental studies, whether in vitro or in vivo animal models, should investigate the effect on the melanogenesis process followed by functional and clinical validation studies using miRNA inhibitors or mimetics.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- 3′UTR:
-
3′ Untranslated region
- AUC:
-
Area under the curve
- BCL2:
-
BCL2 apoptosis regulator
- CAMK1D:
-
Calcium/calmodulin dependent protein kinase ID
- CCND2:
-
Cyclin D2
- CREB1:
-
CAMP responsive element binding protein 1
- Cx43:
-
Connexin43
- DAVID:
-
Database for Annotation Visualization and Integrated Discovery
- DEmiRNAs:
-
Differentially expressed miRNAs
- Distri.:
-
Distribution
- DM:
-
Diabetes mellitus
- E2F3:
-
E2F transcription factor 3
- EDTA:
-
Ethylene diamine tetra-acetic acid
- EIF2AK1:
-
Eukaryotic translation initiation factor 2 alpha kinase 1
- EZH2:
-
Enhancer of zeste homolog 2
- FC:
-
Fold change
- FH:
-
Family history
- FOXK1:
-
Forkhead box K1
- FOXO:
-
Forkhead box protein
- FOXO3:
-
Forkhead box O3
- FSCN1:
-
Fascin actin-bundling protein 1
- GO:
-
Gene ontology
- HTN:
-
Hypertension
- IFITM1:
-
Interferon induced transmembrane protein 1
- IFN-γ:
-
Interferon gamma
- IL-10:
-
Interleukin 10
- IL-17A:
-
Interleukin 17A
- IRF1:
-
Interferon regulatory factor 1
- KEGG:
-
Encyclopedia of genes and genomes
- KLHL28:
-
Kelch like family member 28
- LncRNAs:
-
Long non-coding RNAs
- MIDN:
-
Midnolin
- MIQE:
-
Minimum information for publication of quantitative real-time PCR experiments
- miRNAs:
-
MicroRNAs
- MITF:
-
Melanocyte inducing transcription factor
- MYO5a:
-
Myosin VA
- NAMPT:
-
Nicotinamide phosphoribosyltransferase
- NB-UVB:
-
Narrowband ultraviolet B
- ncRNAs:
-
Non-coding RNAs
- NF-κB:
-
Nuclear factor kappa B subunit 1
- NOTCH2:
-
Notch receptor 2
- NR3C1:
-
Nuclear receptor subfamily 3 group C member 1
- NSV:
-
Non-segmental vitiligo
- P53:
-
Protein P53
- PBMC:
-
Peripheral blood mononuclear cells
- PEA15:
-
Proliferation and apoptosis adaptor protein 15
- PGC-1α:
-
Peroxisome Proliferator-activated receptor-gamma coactivator 1-alpha
- Pre_Gray:
-
Pre greying
- PTEN:
-
Phosphatase and tensin homolog
- QoL:
-
Quality of life
- qRT-PCR:
-
Quantitative real-time PCR
- RA:
-
Rheumatoid arthritis
- RAB3IP:
-
RAB3A interacting protein
- RECK:
-
Reversion inducing cysteine rich protein with Kazal motifs
- RISC:
-
RNA-induced silencing complex
- ROC:
-
Receiver operating curve
- ROCK1:
-
Rho associated coiled-coil containing protein kinase 1
- RRM2:
-
Ribonucleotide reductase regulatory subunit M2
- RT:
-
Reverse transcription
- SCU:
-
Suez Canal University
- SDCBP:
-
Syndecan binding protein
- SIRT1:
-
Sirtuin 1
- SOCS1:
-
Suppressor of cytokine signaling 1
- SOX9:
-
SRY-box transcription factor 9
- SOX10:
-
SRY-box transcription factor 10
- SPSS:
-
Statistical package for the social sciences
- SV:
-
Segmental vitiligo
- TGF-β1:
-
Transforming growth factor beta 1
- TNF-α:
-
Tumor necrosis factor alpha
- TP53INP1:
-
Tumor protein P53 inducible nuclear protein 1
- TYR:
-
Tyrosinase
- TYRP1:
-
Tyrosinase related protein 1
- TYRP2:
-
Tyrosinase related protein 2
- USP6:
-
Ubiquitin specific peptidase 6
- UV:
-
Ultraviolet
- VADI:
-
Vitiligo disease activity score
- VASI:
-
Vitiligo area severity index
- VETF:
-
Vitiligo European Task Force
- YWHAE:
-
Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon
- YWHAZ:
-
Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta
References
Sehgal, V. & Srivastava, G. Vitiligo: Compendium of clinico-epidemiological features. Indian J. Dermatol. Venereol. Leprol. 73(3), 149. https://doi.org/10.4103/0378-6323.32708 (2007).
Ongenae, K., Van Geel, N., De Schepper, S. & Naeyaert, J. M. Effect of vitiligo on self-reported health-related quality of life. Br. J. Dermatol. 152(6), 1165–1172. https://doi.org/10.1111/j.1365-2133.2005.06456.x (2005).
Shi, Y.-L. et al. MicroRNA expression profiling identifies potential serum biomarkers for non-segmental vitiligo. Pigment Cell Melanoma Res. 26(3), 418–421. https://doi.org/10.1111/pcmr.12086 (2013).
Aboul-Fettouh, N. & Pandya, A. G. Epidemiology of Vitiligo (Medical and Surgical Management, Vitiligo, 2018). https://doi.org/10.1002/9781118937303.ch3.
Spritz, R. A. The genetics of vitiligo. J. Investig. Dermatol. 131, E18–E20. https://doi.org/10.1038/skinbio.2011.7 (2011).
Seneschal, J., Boniface, K., D’Arino, A. & Picardo, M. An update on Vitiligo pathogenesis. Pigment Cell Melanoma Res. 34(2), 236–243. https://doi.org/10.1111/pcmr.12949 (2021).
Dey-Rao, R. & Sinha, A. A. Vitiligo blood transcriptomics provides new insights into disease mechanisms and identifies potential novel therapeutic targets. BMC Genom. 18(1), 109. https://doi.org/10.1186/s12864-017-3510-3 (2017).
Hadi, A., Wang, J. F., Uppal, P., Penn, L. A. & Elbuluk, N. Comorbid diseases of vitiligo: A 10-year cross-sectional retrospective study of an urban US population. J. Am. Acad. Dermatol. 82(3), 628–633. https://doi.org/10.1016/j.jaad.2019.07.036 (2020).
Dell’anna, M. L., & Picardo, M.,. A review and a new hypothesis for non-immunological pathogenetic mechanisms in vitiligo. Pigment Cell Res. 19(5), 406–411. https://doi.org/10.1111/j.1600-0749.2006.00333.x (2006).
Czajkowski, R. & Męcińska-Jundziłł, K. Current aspects of vitiligo genetics. Postepy Dermatologii i Alergologii 31(4), 247–255. https://doi.org/10.5114/pdia.2014.43497 (2014).
Spritz, R. A. & Andersen, G. H. L. Genetics of vitiligo. Dermatol. Clin. 35(2), 245–255. https://doi.org/10.1016/j.det.2016.11.013 (2017).
Šahmatova, L. et al. MicroRNA-155 is dysregulated in the skin of patients with vitiligo and inhibits melanogenesis-associated genes in melanocytes and keratinocytes. Acta Derm. Venereol. 96(6), 742–747. https://doi.org/10.2340/00015555-2394 (2016).
Sonkoly, E. et al. MicroRNA-203 functions as a tumor suppressor in basal cell carcinoma. Oncogenesis 1(3), e3–e3. https://doi.org/10.1038/oncsis.2012.3 (2012).
Wang, Y. et al. Differential expression analysis of mi RNA in peripheral blood mononuclear cells of patients with non-segmental vitiligo. J. Dermatol. 42(2), 193–197. https://doi.org/10.1111/1346-8138.12725 (2015).
Shang, Z. & Li, H. Altered expression of four mi RNA (miR-1238-3p, miR-202-3p, miR-630 and miR-766-3p) and their potential targets in peripheral blood from vitiligo patients. J. Dermatol. 44(10), 1138–1144. https://doi.org/10.1111/1346-8138.13886 (2017).
Cui, T.-T. et al. miR-196a-2 rs11614913 polymorphism is associated with vitiligo by affecting heterodimeric molecular complexes of Tyr and Tyrp1. Arch. Dermatol. Res. 307(8), 683–692. https://doi.org/10.1007/s00403-015-1563-1 (2015).
Mansuri, M. S., Singh, M. & Begum, R. miRNA signatures and transcriptional regulation of their target genes in vitiligo. J. Dermatol. Sci. 84(1), 50–58. https://doi.org/10.1016/j.jdermsci.2016.07.003 (2016).
Rahman, R. & Hasija, Y. An integrated genomic analysis on miRNAs and SNPs associated with vitiligo to reveal potential drug candidates. Can. J. Biotechnol. 1(Special Issue), 45. https://doi.org/10.24870/cjb.2017-a32 (2017).
Mannucci, C. et al. Involvement of microRNAs in skin disorders: A literature review. Allergy Asthma Proc. 38(1), 9–15. https://doi.org/10.2500/aap.2017.38.4013 (2017).
Vlachos, I. S. et al. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 43(W1), W460–W466. https://doi.org/10.1093/nar/gkv403 (2015).
Betel, D., Wilson, M., Gabow, A., Marks, D. S. & Sander, C. The microRNA org resource: Targets and expression. Nucleic Acids Res. 36(suppl_1), D149–D153. https://doi.org/10.1093/nar/gkm995 (2008).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25(4), 402–408. https://doi.org/10.1006/meth.2001.1262 (2001).
Fuchs, B.-A. et al. GeneAnalytics: An integrative gene set analysis tool for next generation sequencing, RNAseq and microarray data. Omics J. Integr. Biol. 20(3), 139–151. https://doi.org/10.1089/omi.2015.0168 (2016).
Reimand, J., Kull, M., Peterson, H., Hansen, J. & Vilo, J. Profiler-a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35(suppl_2), W193–W200. https://doi.org/10.1093/nar/gkm226 (2007).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37(1), 1–13. https://doi.org/10.1093/nar/gkn923 (2009).
Kern, F. et al. miRTargetLink 2.0-interactive miRNA target gene and target pathway networks. Nucleic Acids Res. 49(W1), W409–W416. https://doi.org/10.1093/nar/gkab297 (2021).
Liu, J. et al. miRNA423-5p regulates cell proliferation and invasion by targeting trefoil factor 1 in gastric cancer cells. Cancer Lett. 347(1), 98–104. https://doi.org/10.1016/j.canlet.2014.01.024 (2014).
Lin, H. et al. Inhibition of miR-423-5p suppressed prostate cancer through targeting GRIM-19. Gene 688, 93–97. https://doi.org/10.1016/j.gene.2018.11.021 (2019).
Wang, X. et al. MiR-423-5p inhibits osteosarcoma proliferation and invasion through directly targeting STMN1. Cell. Physiol. Biochem. 50(6), 2249–2259. https://doi.org/10.1159/000495085 (2018).
Tang, X. et al. miR-423-5p serves as a diagnostic indicator and inhibits the proliferation and invasion of ovarian cancer. Exp. Ther. Med. 15, 4723–4730. https://doi.org/10.3892/etm.2018.6015 (2018).
Tijsen, A. J. et al. MiR423-5p as a circulating biomarker for heart failure. Circ. Res. 106(6), 1035–1039. https://doi.org/10.1161/CIRCRESAHA.110.218297 (2010).
Rizzacasa, B. et al. MiR-423 is differentially expressed in patients with stable and unstable coronary artery disease: A pilot study. PLoS ONE 14(5), e0216363. https://doi.org/10.1371/journal.pone.0216363 (2019).
Guo, L., Liu, Y., Guo, Y., Yang, Y. & Chen, B. MicroRNA-423-5p inhibits the progression of trophoblast cells via targeting IGF2BP1. Placenta 74, 1–8. https://doi.org/10.1016/j.placenta.2018.12.003 (2018).
Tu, H. et al. Elevated pulmonary tuberculosis biomarker miR-423-5p plays critical role in the occurrence of active TB by inhibiting autophagosome-lysosome fusion. Emerg. Microb. Infect. 8(1), 448–460. https://doi.org/10.1080/22221751.2019.1590129 (2019).
Vaish, U. et al. Author Correction: Micro RNAs upregulated in Vitiligo skin play an important role in its aetiopathogenesis by altering TRP1 expression and keratinocyte-melanocytes cross-talk. Sci. Rep. 10(1), 2166. https://doi.org/10.1038/s41598-020-58949-w (2020).
Segura, M. F. et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc. Natl. Acad. Sci. U.S.A. 106(6), 1814–1819. https://doi.org/10.1073/pnas.0808263106 (2009).
Yan, D. et al. Role of microRNA-182 in posterior uveal melanoma: Regulation of tumor development through MITF, BCL2 and cyclin D2. PLoS ONE 7(7), e40967. https://doi.org/10.1371/journal.pone.0040967 (2012).
Liu, S., Howell, P. M. & Riker, A. I. Up-regulation of miR-182 expression after epigenetic modulation of human melanoma cells. Ann. Surg. Oncol. 20(5), 1745–1752. https://doi.org/10.1245/s10434-012-2467-3 (2013).
Ding, J., Zhu, X., Chen, X., Guan, J. & Li, H. MicroRNA-182 suppresses malignant melanoma proliferation by targeting RECK. Clin. Lab. https://doi.org/10.7754/Clin.Lab.2019.190646 (2020).
Passeron, T. & Ortonne, J.-P. Physiopathology and genetics of vitiligo. J. Autoimmun. 25, 63–68. https://doi.org/10.1016/j.jaut.2005.10.001 (2005).
Luan, W. et al. Long non-coding RNA H19 promotes glucose metabolism and cell growth in malignant melanoma via miR-106a-5p/E2F3 axis. J. Cancer Res. Clin. Oncol. 144(3), 531–542. https://doi.org/10.1007/s00432-018-2582-z (2018).
Wang, J.-L., Li, H., Zhang, J.-B., Zhang, C.-H. & Hou, X.-Q. Suppression of connexin 43 expression by miR-106a promotes melanoma cell proliferation. Eur. Rev. Med. Pharmacol. Sci. 23(3), 965–971. https://doi.org/10.26355/eurrev_201902_16983 (2019).
Palkina, N. V., Komina, A. V., Aksenenko, M. B. & Ruksha, T. G. The pro-oncogenic effect of miR-106a microRNA inhibition in melanoma cells in vitro. Cell Tissue Biol. 11(1), 1–8. https://doi.org/10.1134/s1990519x17010072 (2017).
Miao, X., Tong, X., Hu, J. & Wang, J. Diagnostic value of miR-106a-5p in patients with psoriasis and its regulatory role in inflammatory responses. In Research Square. https://doi.org/10.21203/rs.3.rs-60976/v1 (2020).
Delić, D. et al. Integrated microRNA/mRNA expression profiling of the skin of psoriasis patients. J. Dermatol. Sci. 97(1), 9–20. https://doi.org/10.1016/j.jdermsci.2019.11.003 (2020).
Torri, A. et al. Extracellular MicroRNA signature of human helper T cell subsets in health and autoimmunity. J. Biol. Chem. 292(7), 2903–2915. https://doi.org/10.1074/jbc.m116.769893 (2017).
Viswanathan, V., Fields, J. & Boman, B. M. The miRNA23b-regulated signaling network as a key to cancer development—implications for translational research and therapeutics. J. Mol. Med. 92(11), 1129–1138. https://doi.org/10.1007/s00109-014-1208-4 (2014).
Yu-Xin, G. et al. The role of miR-23b in cancer and autoimmune disease. J. Oncol. https://doi.org/10.1155/2021/6473038 (2021).
Hu, R. & O’Connell, R. M. MiR-23b is a safeguard against autoimmunity. Nat. Med. 18(7), 1009–1010. https://doi.org/10.1038/nm.2849 (2012).
Vellaichamy, G. et al. An in vivo model of postinflammatory hyperpigmentation and erythema: Clinical, colorimetric and molecular characteristics. Br. J. Dermatol. 186(3), 508–519. https://doi.org/10.1111/bjd.20804 (2022).
Kozubek, J. et al. In-depth characterization of microRNA transcriptome in melanoma. PLoS ONE 8(9), e72699. https://doi.org/10.1371/journal.pone.0072699 (2013).
Gencia, I. et al. A preliminary study of microRNA expression in different types of primary melanoma. Bosn. J. Basic Med. Sci. 20(2), 197–208. https://doi.org/10.17305/bjbms.2019.4271 (2020).
Lv, R., Yu, J. & Sun, Q. Anti-angiogenic role of microRNA-23b in melanoma by disturbing NF-κB signaling pathway via targeted inhibition of NAMPT. Future Oncol. (Lond. Engl.) 16(10), 541–458. https://doi.org/10.2217/fon-2019-0699 (2020).
Zhu, S. et al. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-α. Nat. Med. 18(7), 1077–1086. https://doi.org/10.1038/nm.2815 (2012).
Song, X. & Qian, Y. The activation and regulation of IL-17 receptor mediated signaling. Cytokine 62(2), 175–182. https://doi.org/10.1016/j.cyto.2013.03.014 (2013).
Liu, X. et al. Circulating microRNA-23b as a new biomarker for rheumatoid arthritis. Gene 712(143911), 143911. https://doi.org/10.1016/j.gene.2019.06.001 (2019).
Abdeen, H. M. et al. Micro RNA-23b as a potential biomarker in rheumatoid arthritis disease activity and severity: Clinical, laboratory, and radiological cross-sectional study. Egypt. Rheumatol. Rehabil. https://doi.org/10.1186/s43166-021-00090-1 (2021).
Issa, Y. W. & Salih, S. M. Impact of miR-155, miR-145 and miR-328 on pigmentary Process in Iraqi Patients with vitiligo. Gene Rep. 21(100955), 100955. https://doi.org/10.1016/j.genrep.2020.100955 (2020).
Dynoodt, P. et al. Identification of miR-145 as a Key regulator of the pigmentary process. J. Investig. Dermatol. 133(1), 201–209. https://doi.org/10.1038/jid.2012.266 (2013).
Li, L. The role of MicroRNAs in vitiligo: Regulators and therapeutic targets. Ann. Dermatol. 32(6), 441–451. https://doi.org/10.5021/ad.2020.32.6.441 (2020).
Mansuri, M. S. et al. Micro RNA profiling reveals differentially expressed microRNA signatures from the skin of patients with nonsegmental vitiligo. Br. J. Dermatol. 171(5), 1263–1267. https://doi.org/10.1111/bjd.13109 (2014).
Wu, Q. et al. Long noncoding RNA FOXD3-AS1 promotes colon adenocarcinoma progression and functions as a competing endogenous RNA to regulate SIRT1 by sponging miR-135a-5p. J. Cell. Physiol. 234(12), 21889–21902. https://doi.org/10.1002/jcp.28752 (2019).
Saunders, L. R. et al. miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging 2(7), 415–431. https://doi.org/10.18632/aging.100176 (2010).
Raia, N. M. A., Shaker, O. G., Hassan, Z. M. & Abd Elrahim, T. A. Is there a relation between long non-coding RNA MALAT-1 and miRNA-9 in Egyptian patients with Vitiligo?. Exp. Dermatol. 31(3), 381–383. https://doi.org/10.1111/exd.14487 (2022).
Su, M. et al. miR-9 regulates melanocytes adhesion and migration during vitiligo repigmentation induced by UVB treatment. Exp. Cell Res. 384(1), 111615. https://doi.org/10.1016/j.yexcr.2019.111615 (2019).
Kidir, M., Karabulut, A. A., Ercin, M. E. & Atasoy, P. Regulatory T-cell cytokines in patients with nonsegmental vitiligo. Int. J. Dermatol. 56(5), 581–588. https://doi.org/10.1111/ijd.13564 (2017).
Ghafouri-Fard, S. et al. An update on the role of miR-124 in the pathogenesis of human disorders. Biomed. Pharmacother. 135(111198), 111198. https://doi.org/10.1016/j.biopha.2020.111198 (2021).
Nakamachi, Y., Saegusa, J. & Kawano, S. MicroRNA-124: A promising therapeutic agent for various human diseases, including rheumatoid arthritis. RNA & DISEASE, 4. http://www.smartscitech.com/index.php/rd (2017).
Shen, C. et al. MiR-124 functions as a melanoma tumor suppressor by targeting RACK1. OncoTargets Ther. 12, 9975–9986. https://doi.org/10.2147/OTT.S225120 (2019).
Gao, M., Chang, Y., Wang, X., Ban, C. & Zhang, F. Reduction of COX-2 through modulating miR-124/SPHK1 axis contributes to the antimetastatic effect of alpinumisoflavone in melanoma. Am. J. Transl. Res. 9(3), 986–998 (2017).
Segura, M. F. et al. Melanoma MicroRNA signature predicts post-recurrence survival. Clin. Cancer Res. 16(5), 1577–1586. https://doi.org/10.1158/1078-0432.CCR-09-2721 (2010).
Luo, C. et al. MiR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. J. Invest. Dermatol. 133(3), 768–775. https://doi.org/10.1038/jid.2012.357 (2013).
Dong, C. et al. Coat color determination by miR-137 mediated down-regulation of microphthalmia-associated transcription factor in a mouse model. RNA 18(9), 1679–1686. https://doi.org/10.1261/rna.033977.112 (2012).
Zhang, Z., Shen, W., Liu, W. & Lyu, L. Role of miRNAs in melanin metabolism: Implications in melanin-related diseases. J. Cosmet. Dermatol. https://doi.org/10.1111/jocd.14762 (2022).
Jiang, S., Yu, X. & Dong, C. MiR-137 affects melanin synthesis in mouse melanocyte by repressing the expression of c-Kit and Tyrp2 in SCF/c-Kit signaling pathway. Biosci. Biotechnol. Biochem. 80(11), 2115–2121. https://doi.org/10.1080/09168451.2016.1200455 (2016).
Zhang, H., Jiang, L.-H., Sun, D.-W., Li, J. & Ji, Z.-L. The role of miR-130a in cancer. Breast Cancer (Tokyo, Japan) 24(4), 521–527. https://doi.org/10.1007/s12282-017-0776-x (2017).
Ishii, H. et al. miR-130a and miR-145 reprogram Gr-1+ CD11b+ myeloid cells and inhibit tumor metastasis through improved host immunity. Nat. Commun. 9(1), 1–5. https://doi.org/10.1038/s41467-018-05023-9 (2018).
Wei, H. et al. miR-130a Deregulates PTEN and Stimulates Tumor Growth. Can. Res. 77(22), 6168–6178. https://doi.org/10.1158/0008-5472.can-17-0530 (2017).
Wu, S., Han, M. & Zhang, C. Overexpression of microRNA-130a represses uveal melanoma cell migration and invasion through inactivation of the Wnt/β-catenin signaling pathway by downregulating USP6. Cancer Gene Ther. https://doi.org/10.1038/s41417-021-00377-7 (2021).
Wade, S. M., McGarry, T., Wade, S. C., Fearon, U. & Veale, D. J. Serum MicroRNA signature as a diagnostic and therapeutic marker in patients with psoriatic arthritis. J. Rheumatol. 47(12), 1760–1767. https://doi.org/10.3899/jrheum.190602 (2020).
Jackson, S. J. et al. Rapid and widespread suppression of self-renewal by microRNA-203 during epidermal differentiation. Development (Cambridge, England) 140(9), 1882–1891. https://doi.org/10.1242/dev.089649 (2013).
Wu, D. T., Chen, J. S., Chang, D. C. & Lin, S.-L. Mir-434-5p mediates skin whitening and lightening. Clin. Cosmet. Investig. Dermatol. 1, 19–35. https://doi.org/10.2147/ccid.s4181 (2008).
Rambow, F. et al. miR-330-5p targets tyrosinase and induces depigmentation. J. Invest. Dermatol. 134(11), 2846–2849. https://doi.org/10.1038/jid.2014.231 (2014).
Kim, K.-H. et al. Novel inhibitory function of miR-125b in melanogenesis. Pigment Cell Melanoma Res. 27(1), 140–144. https://doi.org/10.1111/pcmr.12179 (2014).
Sonkoly, E. et al. Protein kinase C-dependent upregulation of miR-203 induces the differentiation of human keratinocytes. J. Invest. Dermatol. 130(1), 124–134. https://doi.org/10.1038/jid.2009.294 (2010).
Fritz, G. & Kaina, B. Activation of c-jun N-terminal kinase 1 by UV irradiation is inhibited by wortmannin without affecting c- jun expression. Mol. Cell. Biol. 19(3), 1768–1774. https://doi.org/10.1128/mcb.19.3.1768 (1999).
Garmyn, M. & Degreef, H. Suppression of UVB-induced c-fos and c-jun expression in human keratinocytes by N-acetylcysteine. J. Photochem. Photobiol. B Biol. 37(1–2), 125–130. https://doi.org/10.1016/s1011-1344(96)07340-x (1997).
Sonkoly, E. et al. MicroRNAs: Novel regulators involved in the pathogenesis of psoriasis?. PLoS ONE 2(7), e610. https://doi.org/10.1371/journal.pone.0000610 (2007).
Dopytalska, K., Ciechanowicz, P., Wiszniewski, K., Szymańska, E. & Walecka, I. The role of epigenetic factors in psoriasis. Int. J. Mol. Sci. 22(17), 9294. https://doi.org/10.3390/ijms22179294 (2021).
Xiuli, Y. & Honglin, W. MiRNAs flowing up and down: The concerto of psoriasis. Front. Med. 8, 646796. https://doi.org/10.3389/fmed.2021.646796 (2021).
Noguchi, S. et al. MicroRNA-203 regulates melanosome transport and tyrosinase expression in melanoma cells by targeting kinesin superfamily protein 5b. J. Invest. Dermatol. 134(2), 461–469. https://doi.org/10.1038/jid.2013.310 (2014).
Kuchen, S. et al. Regulation of MicroRNA expression and abundance during lymphopoiesis. Immunity 32(6), 828–839. https://doi.org/10.1016/j.immuni.2010.05.009 (2010).
Calin, G. A. et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 353(17), 1793–1801. https://doi.org/10.1056/NEJMoa050995 (2005).
Carracedo, A., Alimonti, A. & Pandolfi, P. P. PTEN level in tumor suppression: How much is too little?. Can. Res. 71(3), 629–633. https://doi.org/10.1158/0008-5472.can-10-2488 (2011).
Jones, K. B. et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Can. Res. 72(7), 1865–1877. https://doi.org/10.1158/0008-5472.CAN-11-2663 (2012).
Williams, A., Henao-Mejia, J., Harman, C. C. D. & Flavell, R. A. miR-181 and metabolic regulation in the immune system. Cold Spring Harb. Symp. Quant. Biol. 78, 223–230. https://doi.org/10.1101/sqb.2013.78.020024 (2013).
Friedrich, M. et al. The role of the miR-148/-152 family in physiology and disease. Eur. J. Immunol. 47, 2026–2038. https://doi.org/10.1002/eji.201747132 (2017).
Wang, Y., Yu, X., Wang, L., Ma, W. & Sun, Q. MiR-320b is down-regulated in psoriasis and modulates keratinocyte proliferation by targeting AKT3. Inflammation 41(6), 2160–2170. https://doi.org/10.1007/s10753-018-0859-7 (2018).
Du, H., Zhao, Y., Yin, Z., Wang, D. W. & Chen, C. The role of miR-320 in glucose and lipid metabolism disorder-associated diseases. Int. J. Biol. Sci. 17(2), 402–416. https://doi.org/10.7150/ijbs.53419 (2021).
Hu, K. & Liang, M. Upregulated microRNA-224 promotes ovarian cancer cell proliferation by targeting KLLN. In Vitro Cell. Dev. Biol. Anim. 53(2), 149–156. https://doi.org/10.1007/s11626-016-0093-2 (2017).
Huang, Y. et al. Over-expressed miR-224 promotes the progression of cervical cancer via targeting RASSF8. PLoS ONE 11(9), e0162378. https://doi.org/10.1371/journal.pone.0162378 (2016).
Liu, F., Liu, Y., Shen, J., Zhang, G. & Han, J. MicroRNA-224 inhibits proliferation and migration of breast cancer cells by down-regulating Fizzled 5 expression. Oncotarget 7(31), 49130–49142. https://doi.org/10.18632/oncotarget.9734 (2016).
Cui, R. et al. MicroRNA-224 is implicated in lung cancer pathogenesis through targeting caspase-3 and caspase-7. Oncotarget 6(26), 21802–21815. https://doi.org/10.18632/oncotarget.5224 (2015).
Wan, Y. et al. Dysregulated microRNA-224/apelin axis associated with aggressive progression and poor prognosis in patients with prostate cancer. Hum. Pathol. 46(2), 295–303. https://doi.org/10.1016/j.humpath.2014.10.027 (2015).
Rashed, L. A. & Ibrahim, H. A. Expression of MicroRNA-224-3p in vitiligo. J. Pak. Assoc. Dermatol. 31(4), 680–684 (2021).
Mansuri, M. S. et al. The catalase gene promoter and 5ʹ-untranslated region variants lead to altered gene expression and enzyme activity in vitiligo. Br. J. Dermatol. 177(6), 1590–1600. https://doi.org/10.1111/bjd.15681 (2017).
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). This research was funded by the Egyptian Science, Technology and Innovation Funding Authority (STIFA)- Grant number 34908 for conducting this study.
Author information
Authors and Affiliations
Contributions
Conceptualization: H.Y.A., A.E., E.A.M., and E.A.T.; Methodology: H.Y.A., N.Y.A., N.Z.T., and A.H.A.G.; Formal analysis and investigation: H.Y.A., N.Z.T., N.Y.A., and N.R.A.. Writing—original draft preparation: H.Y.A., N.Z.T., N.Y.A., and N.R.A.; Writing—review and editing: H.Y.A., N.R.A., E.A.M., N.Y.A., N.Z.T., A.H.A.G., E.A.T., and A.E.; Funding acquisition: H.Y.A., E.A.T., and A.E.; Supervision: A.E., A.H.G., and E.A.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Abdallah, H.Y., Abdelhamid, N.R., Mohammed, E.A. et al. Investigating melanogenesis-related microRNAs as disease biomarkers in vitiligo. Sci Rep 12, 13526 (2022). https://doi.org/10.1038/s41598-022-17770-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-17770-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.