Abstract
Oryza australiensis is a wild rice native to monsoonal northern Australia. The International Oryza Map Alignment Project emphasises its significance as the sole representative of the EE genome clade. Assembly of the O. australiensis genome has previously been challenging due to its high Long Terminal Repeat (LTR) retrotransposon (RT) content. Oxford Nanopore long reads were combined with Illumina short reads to generate a high-quality ~ 858 Mbp genome assembly within 850 contigs with 46× long read coverage. Reference-guided scaffolding increased genome contiguity, placing 88.2% of contigs into 12 pseudomolecules. After alignment to the Oryza sativa cv. Nipponbare genome, we observed several structural variations. PacBio Iso-Seq data were generated for five distinct tissues to improve the functional annotation of 34,587 protein-coding genes and 42,329 transcripts. We also report SNV numbers for three additional O. australiensis genotypes based on Illumina re-sequencing. Although genetic similarity reflected geographical separation, the density of SNVs also correlated with our previous report on variations in salinity tolerance. This genome re-confirms the genetic remoteness of the O. australiensis lineage within the O. officinalis genome complex. Assembly of a high-quality genome for O. australiensis provides an important resource for the discovery of critical genes involved in development and stress tolerance.
Similar content being viewed by others
Introduction
The genus Oryza is made up of ~ 27 species that have been classified into 11 genome groups1,2. Two of these species (O. sativa and Oryza glaberrima), both of which belong to the AA genome clade, were independently domesticated in Asia and Africa, respectively. Together, these two domestic rice species serve as a staple food source for about 60% of the global human population. The remaining ~ 25 species occur in wild populations and have become specialised to occupy a diverse set of environments around the world3.
To date, the species and cultivars that make up the AA genome clade have provided most of the genetic variation and resources that have led to the success of domesticated rice4, while other genome clades have contributed relatively little to domestication. Encouragingly, rich diversity is apparent, even within landraces of just one O. sativa subspecies. For example, in a GWAS study for 14 agronomic traits (including tolerance to drought and degree of seed shattering) in 517 indica sub-species landraces, Huang et al.5 revealed ~ 3.6 million SNPs. Currently nine of the 11 Oryza genome assemblies available on Ensembl Plants; (accessed 26/2/2021)6 belong to the AA genome clade, one belongs to the BB genome (Oryza punctata), and one to the FF genome (Oryza brachyantha). Therefore, wild species of rice occupy a number of genomic clades that are not yet represented by an assembly and undoubtedly include traits that would be desirable in domestic cultivars. The wild species therefore represent vast reservoirs of untapped genetic variation that could be harnessed for domestic rice improvement2,3,4,7. Recently, Hiromi et al.8 provided an update to the OryzaGenome database which includes short-read sequencing data for several non-AA genome Oryza species. The availability of Oryza ‘-omics’ resources is increasing, yet the number of whole-genome assemblies for non-AA genome wild species remains low. Whole-genome assemblies of wild Oryza species will provide new sources of genes and gene variants/haplotypes, enhancing the ability of breeding programs to introduce desirable traits into domestic cultivars. However care is necessary to navigate issues such as linkage drag and suppressed recombination frequencies that may be associated with use of wild germplasm. Additionally, such projects could lead to the domestication of wild rice populations (e.g., through the targeted modification of domestication-associated traits, such as seed shattering)9. Overall, harnessing the genetics of wild Oryza species has wide application for cereal crop improvement and specifically for crop resilience. In fact, the Oryza Map Alignment Project (OMAP) and the International OMAP (IOMAP) recognised the value that the genome assemblies of wild Oryza species would provide to research in abiotic and biotic stress tolerance, and domestication, as long ago as 200310,11,12. These projects recommended sequencing the genomes of several wild rice species but progress in this respect has been slow.
Recently, several non-AA genetic resources have become available, including genomes within the O. officinalis complex13 and the genome of Oryza granulata14. O. granulata, an upland wild rice species, has a demonstrated ability to tolerate drought stress and bacterial blight, providing a valuable new source of Oryza genetic variation. However, owing to its large genome size, high repeat content, and being a short-read assembly, the O. granulata genome assembly is highly fragmented, reducing its utility. These same difficulties (i.e., genome size and repeat content) have been encountered during assembly of the EE genome, represented solely by O. australiensis, which is reported to be the largest diploid Oryza genome.
Oryza australiensis is a perennial wild relative of rice endemic to the tropical regions of northern Australia. Due to the extremely hot and sporadically dry environment in which O. australiensis exists, this species has been described as an extremophile3. Oryza australiensis is the sole member of the EE genome clade and its genome has been estimated to be 965 Mbp in size, more than double that of O. sativa ssp. japonica15,16. The size of the O. australiensis genome is the result of the expansion of long terminal repeat (LTR) retrotransposon (RT) families (e.g., Gypsy and Copia), which comprise an estimated 65% of the genome. In comparison, the genome of O. sativa is composed of approximately 10% LTR-RT15. It is uncertain what led to the rapid genome-wide expansion of these LTR-RT families. However, it has been noted that the expansion of LTR-RTs facilitated rapid adaptive genome evolution in the Oryza genus, leading to speciation events17. These same processes may also play a role in the evolution of biotic and abiotic stress tolerances. Heat tolerance has been well-characterised in O. australiensis18, and transformation of domestic rice with a key gene (Rubisco activase) involved in heat tolerance fortifies grain yield during episodes of heat19. Further, Yichie et al.20 identified population-specific tolerance to salinity stress in O. australiensis accessions, and Hamzelou et al.21 report on drought tolerance in this species. The unique anatomy and photosynthetic efficiency of O. australiensis leaves also make it an important species for understanding how rice photosynthesis can be augmented22. In terms of food quality, the grain of O. australiensis has unique pigmentation and starch composition, distinct from Asian species23,24, that could prove to be a profitable food item in and of itself.
A highly-quality reference genome for O. australiensis could reveal novel loci for abiotic stress tolerance and help shed light on the genome evolution of the Oryza genus. However, since the conception of OMAP in 200311, and despite international sequencing efforts since12, no research group has successfully assembled a high-quality, chromosome-level genome for O. australiensis. A previous attempt to assemble the O. australiensis genome with short reads resulted in a highly fragmented assembly due to the genome size and high repeat content25. Efforts such as those by Joly-Lopez et al.25 highlight the difficulties involved in assembling highly contiguous plant genomes with significant repeat content26.
In this study, we report on the assembly of a reference-quality O. australiensis nuclear genome from Keep River (KR; Northern Territory, Australia) into 1956 contigs using Oxford Nanopore Technology (ONT) long-reads. The contigs were subsequently scaffolded into 12 pseudomolecules representing 12 chromosomes using the O. sativa Nipponbare genome assembly as a guide.
Methods
Growth conditions for plant material used for genome sequencing
Seed of four O. australiensis populations was collected from the monsoonal savannah region of northern Australia (Fig. 1), compliant with the relevant national and international guidelines and legislation for wild species collections. All seed was obtained from the Australian Tropical Crops and Forages Collection (AusTRCF) of the Australian Grains Genebank (previously the Australian Plant Genetic Resource Information System) and as part of the Australian Tropical Crops Genetic Resource Centre Collection (ATCGRC). The Keep River accession used for the reference genome assembly bears the accession number AusTRCF 318143 and was collected by Dr I. Cowie at Darwin Herbarium, Northern Territory, Australia (Voucher ID# 9583) on 2nd May 2002 from a drying creek bed in the Keep River National Park after the passing of the summer monsoon. The other three accessions used for re-sequencing (CH, D and VR) are held in the same collection with accession numbers of AusTRCF 300134, 300137 and 300131. These lines are vouchered in the Jeff Corfield Collection in Townsville (Queensland, Australia) under the accession numbers JC 2317, JC 2336 and JC 2312, respectively. CH, D and VR were collected from the sites denoted in Fig. 1 between 1995 and 1997 by Mr I. Watson. Seed was stored at 4 °C until used. Seeds were imbibed in water for one hour and then surface sterilised in a 50% sodium hypochlorite bleach solution for 30 min followed by rinsing. Sterile seeds were sown onto absorbent cotton-lined petri dishes and incubated at 34 °C for 2 days to trigger germination. Germinated seeds were sown into a fine textured krasnozem soil (sourced from Robertson, NSW, Australia) and covered in a thin layer of vermiculite. Plants were grown in glasshouses at Macquarie University (Sydney, Australia) under a 30 °C/22 °C day/night cycle in April–June 2019.
Locations of O. australiensis seed collection sites for this study. List of wild rice accessions (obtained from the Australian Grain Genebank) used in this study reported by Yichie et al.19 to demonstrate varying degrees of salt tolerance. Dots on the map show the occurrence of O. australiensis (retrieved from the Atlas of Living Australia). In-set table: accessions are ordered by salt tolerance, with Oa-KR being the most sensitive to salt, and Oa-VR being most tolerant19.
DNA extraction
Roughly 1 g of healthy, fully expanded leaf material was collected from ~ 2-month-old O. australiensis plants (one plant per genotype) and ground in a mortar and pestle under liquid nitrogen until a fine powder was achieved. High molecular-weight DNA was extracted at the Australian National University (ANU; Canberra, Australia) following the method of Jones et al.27.
Nanopore library preparation, sequencing, and base calling
The sequencing library was prepared as per the manufacturer’s (Oxford Nanopore Technologies, Oxford, United Kingdom) instructions for genomic DNA by ligation method (SQK-LSK109) with small modifications. The manufacturer recommends using 0.2 pmol of DNA, however this is optimised for DNA fragments 8 kbp in length. As we were working with substantially longer (~ 20 kbp) DNA fragments, we loaded 3 μg (0.23 pmol) of DNA.
The library was run on FLO-MIN106 (R9.4.1 revD) flowcells as per the manufacturer’s instructions. To obtain a good level of coverage of the O. australiensis KR genome, the same library preparation for this sample was loaded onto three separate flow cells (i.e., the genomic DNA from one plant was sequenced on three flowcells). The raw signal intensities obtained from the flow cells were used by guppy (version 3.0.3) from ONT to perform base calling.
Illumina sequencing
Aliquots (containing 2.24 ng DNA) of the same DNA samples from the four O. australiensis genotypes described above were prepared for Illumina short read sequencing. Short-read libraries were created using the Illumina Nextera tagment DNA enzyme (TDE1) according to Jones et al.28. Libraries were size selected for 350–600 bp fragments using a PippinHT (Sage Science). Sequencing was performed on an Illumina NovaSeq 6000 S4 flow cell 300 cycles (150 bp paired end), being multiplexed with other projects. Sequencing was performed at the Biomolecular Resource Facility, ANU.
PacBio Iso-Seq library preparation and sequencing
Young grains were harvested during the milky period of development from mature O. australiensis KR plants from the same collection as the individual used for ONT and Illumina sequencing (i.e., different plants from the same population; germinated as above and grown in a glasshouse at 30 °C with ambient light at The Plant Accelerator, University of Adelaide, Adelaide, South Australia from February to June 2020). Some developing grain still possessed their pedicel/rachilla and pollen-containing anthers and these were included in the samples to increase the diversity of transcripts in the RNA library.
The growth zones of ten 115-day-old O. australiensis KR plants were excised and used for RNA extraction. The growth zone is defined here as ~ 10 mm from the base of the youngest leaves emanating from the central cylinder of the plants. It was necessary to remove surrounding leaf sheathes to access the central cylinder.
A separate cohort of O. australiensis KR plants was germinated as above, except germination was achieved with an incubation temperature of 30 °C. Germinated seeds were transferred to a growth rack partially submerged in a ½ Hoaglands solution (pH 5.01) spiked with 25 mg/L ferrous sulphate. After seven days of growth, root tips (5 mm from the end of the root), older seminal roots with young lateral roots, and healthy fully expanded leaves were harvested, snap frozen in liquid nitrogen and stored at − 80 °C. The growth solution was changed once during this time.
An additional cohort of O. australiensis KR seed was germinated at 30 °C and submerged in water in the dark to promote coleoptile growth. After six days under these conditions, etiolated coleoptiles were harvested as above.
Total RNA was extracted from the six tissues (7-day-old leaves, 7-day-old root tips, 7-day-old mature root sections, 6-day-old hypoxic coleoptiles, reproductive tissue, and 118-day-old growth zones) with the Sigma Spectrum Plant Total RNA Kit according to the manufacturer’s instructions, with minor modifications for the developing grain sample as per Betts et al.29. RNA samples were dried in RNA Stable tubes as per the manufacturer’s instructions and shipped to DNA Link (Seoul, South Korea). RNA quality and concentration of the samples were checked with a Nanodrop and with a Bioanalyzer 2100 instrument (Agilent, Santa Clara, USA). Only low-quality RNA (RIN < 7) could be extracted from the older seminal roots so this sample was omitted from further analyses.
RNA samples were barcoded upon conversion to cDNA libraries (to facilitate multiplexing) following the manufacturer’s instructions of the SMARTer PCR cDNA Synthesis kit. cDNA libraries were amplified by PCR and then an equal concentration of cDNA from each was pooled into a single library. The library was prepared for SMRTbell sequencing with the Express Template Prep 2.0 kit. Iso-Seq sequencing was carried out on a Sequel II PacBio instrument using a single SMRT cell.
Iso-Seq reads were processed through the IsoSeq v3 pipeline to generate high quality polished isoforms. Briefly, raw data is converted from multiple subreads per ZMW into a single consensus CCS read (minimum read quality of 0.9), demultiplexed and clustered (minimum predicted accuracy of 0.99). The aim of the clustering step is to collapse the CCS reads derived from different isoforms of the same gene or from the same transcript isoform but which differ slightly in their 5′ and 3′ ends, typically as a result of RNA degradation.
Nuclear genome assembly and assessment
A summary of all reads obtained during this study can be found in Table 1. Jellyfish (RRID:SCR_005491)30 and genomescope2 (RRID:SCR_017014)31,32 were used to estimate the size, unique sequence content, and heterozygosity of the O. australiensis KR genome using 17-mers in the KR Illumina short read libraries. Raw MinION reads for O. australiensis KR were corrected and then de novo assembled using Canu v1.9 (RRID:SCR_015880)33 as follows: canu -p oryza -d canu genomeSize = 965 m -nanopore-raw all.fastq.gz -gridOptions = "–time = 72:00:00" -obtovlThreads = 24 -batMemory = 9. The MinION long reads and the Illumina short reads were mapped to the resulting contigs using minimap2 v2.17 (RRID:SCR_018550)34. Alignment files were used to polish the genome assembly with HyPo v1.0.335. JVarkit was used to visualise long-read coverage of the contigs36. Contigs were checked for microbial contamination by querying BLAST-generated taxon ID numbers with the lineage command of taxonkit v0.6.037. Contigs found to contain non-eukaryotic sequences were interrogated using the online genome browser IGV-Web (RRID:SCR_011793) by mapping long reads back to the assembly and checking the coverage and overlap of long reads with non-suspect sequences38. Purge Haplotigs (v1.1.2) was used to determine whether the wild rice genome assembly contained any haplotigs or junk sequences39.
The quality of the genome assembly was assessed by analysing the quality of the LTR-RT elements of the O. australiensis KR genome. LTR-RT elements are difficult to assemble. As such, LTR-RT assemblies provide a proxy for the quality of the rest of the genome. LTR regions were identified with LTR_Finder_Parallel v1.1 (RRID:SCR_018969)40 and LTR_Harvest41. The outputs were used as input for LTR_Retriever v2.8.7 (RRID:SCR_017623)—to generate LTR Assembly Index (LAI) scores42. The quality of the assembly was also assessed with BUSCO v4 (RRID:SCR_015008)43 using the Poales database (poales_odb10.2019-11-20; contains 4896 sequences) as the lineage. Contigs were mapped to the O. sativa Nipponbare reference genome (GCA_001433935.1) with minimap2 and visualised with MashMap v2.044.
Pseudomolecules for the assembly were generated using RagTag v1.0.045 using version 7 of the O. sativa Nipponbare reference genome46 as a guide for the ordering and orientation of contigs into pseudomolecules (Supplementary File S1a). Contigs that could not be assigned to a pseudomolecule were concatenated (with 100 Ns at contig boundaries) into a ‘chromosome’ called ChrUn, though we used the unassigned contigs themselves for most of the downstream analyses. To understand why the unassigned contigs could not be placed into a pseudomolecule, the identity of a random subset of the unassigned contigs was determined by BLAST searches and the average LTR content of ChrUn was determined by LTR_Retriever. Contigs flagged as repeats, haplotigs or junk by Purge Haplotigs were kept in the assembly for completeness but were not included in scaffolding (see above). JVarkit’s WGSplotter was used to visualise long-read genome coverage36. BLAST was used to determine the identity of a random subset of the unassigned contigs. Subsequent annotation of the unassigned contigs (see below) also helped in determining the identity of the sequences that could not be assembled into pseudomolecules. The quality of the scaffolded assembly was assessed again with BUSCO and LAI scores, as above. LTR_Retriever was also used to produce estimates for LTR-RT insertion times. The quality of the assembly was further checked by mapping ONT long reads to the pseudomolecules to obtain a read mapping rate and to estimate read coverage across the assembly.
Comparison of O. australiensis KR assembly to the O. sativa Nipponbare assembly
The EDTA-masked (see below) pseudomolecules were aligned to the Nipponbare genome using minimap2 with the ‘asm20’ setting. The mapping file was used as input for dotPlotly47 to generate a dot plot of the whole-genome alignment with the following settings: “-m 2000 -q 500000 -k 12 -l -p 12”. The resulting plot was used to interrogate suspected structural variations (SVs) in the wild rice genome. We used O. sativa vs O. punctata Chr2 (CM002489.2; a BB genome rice), O. sativa vs O. australiensis KR Chr2, and O. australiensis KR vs O. punctata Chr2 alignments to inspect a region on Chr2 in these species which has been reported as an inversion in non-AA rice genome1. Alignments were made as described above, except “-m 5000” was used for dotPlotly visualisation. To check whether reference bias in the O. australiensis scaffolds was affecting the Chr2 SVs, we scaffolded the KEEP contigs using the O. punctata reference genome (AVCL0200000). We aligned the O. punctata-guided O. australiensis Chr2 to O. punctata Chr2, and then to O. sativa Nipponbare Chr2 as above.
To check whether the SVs were real, contig coordinates within the scaffolds were extracted from the RagTag AGP file and these were used to build a ‘cytoband’ file for karyoploteR48. The coordinates for the SVs (± 10 kbp) identified in the O. australiensis KR and O. sativa genome alignment were added to the ‘cytoband’ file, and these were used to visualise the boundaries between contigs and SV events using karyoploteR. Further, long reads were mapped to the O. australiensis KR scaffolds to check the level of coverage across SV junctions. SV junctions with low coverage were coloured red to identify them as suspicious.
To identify single nucleotide variations (SNVs) in the wild rice genome, O. australiensis KR Illumina short reads were mapped to the Nipponbare genome with minimap2. The resulting BAM file was used as input for the SAMtools (RRID:SCR_002105)49 sub-command mpileup, and BCFtools (RRID:SCR_005227) called SNV variants. VCFtools (RRID:SCR_001235)50 filtered low quality SNVs with low-read support (minQ = 30; minDP = 3). High quality SNVs were visualised using karyoploteR in RStudio with a window size of 70 kbp.
Genome repeat annotation
LTR_Retriever (see above) was used to provide an estimate of LTR-RT elements in the O. australiensis KR contigs and scaffolds. Repetitive DNA elements were annotated using the Extensive de novo TE Annotator (EDTA; Supplementary File S2b)51. A manually curated rice repeat-element library provided by the developers of EDTA was used to identify repeats in the O. australiensis KR genome. A library containing O. sativa ssp. japonica transcript sequences including untranslated regions (UTRs) was downloaded from the IOMAP Genomes Database52 and used by EDTA to mask endogenous genes from its annotation pipelines. EDTA also identifies repeat sequences in the input genome that were not part of the repeat library used for interrogation and then constructs a repeat library specific to the input assembly. Total repeat content was also estimated and annotated using the RepeatMasker-based Repeat Masking tool included in OmicsBox version 2.0.1053,54. The custom O. australiensis KR-specific repeat library generated by EDTA was used as input for the RMBlast search engine, while other settings were left as default.
Protein-coding gene annotation
Evidence-based gene-finding was carried out with the OmicsBox Gene Finding tool with O. sativa set as the most closely related species53,54. A protein evidence file containing protein sequences from 12 species/accessions of rice downloaded from the IOMAP Genomes Database52 and the Iso-Seq isoforms were used as evidence for gene finding. Genes were searched on both strands of the pseudomolecules and the unassigned contigs and alternative splice variants were allowed. The resulting gene coordinates were used as input for the built-in Functional Gene Annotation workflow. This workflow uses RepeatMasker to mask repeat elements in the genome assembly before carrying out gene prediction with Augustus. Then, both Blast2GO and InterProScan pipelines were applied to search for gene and protein matches for the identified genes. Annotations were further supplemented by searching orthologues of the identified genes/proteins using EggNOG-mapper in OmicsBox. Annotations for the contigs and scaffolds were exported as GFF3 files (Supplementary Files S3c, S4d). To check whether the GFF3 files contained TE gene annotations, we used a list of known plant TE families from the Atlas of Plant Transposable Elements55.
O. australiensis genotype diversity and genetic similarity
To identify SNVs between four genotypes of O. australiensis with demonstrated differences in salt tolerance20, the O. australiensis CH, D, and VR Illumina short reads (Table 1) were mapped to the scaffolded O. australiensis KR genome assembly using minimap2 and visualised as above. Keep River short reads were also mapped back against the Keep River genome to provide a baseline of SNVs. Mpileup was used to call variants between the O. australiensis KR genome assembly and the mapped reads. VCF files were filtered using bcftools such that only reads with a mapping quality > 30, and only SNVs with a depth of 5× were considered. The patterns of SNV density described here were visually inspected.
Genetic similarity between the O. australiensis genotypes was determined using the Illumina short reads for each genotype as input for kWIP56. kWIP uses a Weighted Inner Product of different k-mer hash values derived from khmer (RRID:SCR_001156)57 to reduce the effect that erroneous read data has on estimates of genetic similarity. The estimate of genetic similarity was also extended to the Oryza genus using publicly available Illumina short read data derived from genomic DNA samples (see Supplementary File S5e for the list of accessions).
Results
Genome assembly, making pseudomolecules and genome quality
To generate long-read sequence data for assembly, DNA from a single O. australiensis Keep River (KR) plant was extracted, purified, and sequenced on three ONT MinIOn flow cells (KR1, KR2, and KR3). Additionally, the long-read DNA extract was used to generate Illumina NovaSeq short read data for O. australiensis KR. DNA extraction and Illumina short-read sequencing was also performed on O. australiensis genotypes CH, D, and VR (Fig. 1, Table 1). We generated 49 Gbp (2.01 million reads) of ONT MinION long read data and 19 Gb (138.1 million reads) of Illumina short read data for the O. australiensis Keep River accession. Further, we generated 3.14 Gbp (1.25 million reads) of PacBio Iso-Seq data from five distinct tissue types (Table 1, Supplementary Fig. S1). Finally, we generated 20 Gbp, 12 Gbp, and 19 Gbp of Illumina short read data for three additional O. australiensis genotypes (CH, D, VR, respectively; Table 1).
K-mer analysis of O. australiensis KR short reads by jellyfish and genomescope2 estimated the genome size to be ~ 822 Mbp. About 41.9% of the k-mers were unique, which suggested that the genome contained 58.1% repeat content. Heterozygosity was estimated to be 0.001%. Based on these results, we did not expect heterozygosity to have greatly affected contig assembly.
Canu assembled a genome containing 980 Mbp across 1956 contigs (Table 2). This represents 101.5% of the expected O. australiensis genome size. The contig N50 was 1.9 Mbp, the largest contig was 24.9 Mbp in length, while the smallest contig was 1324 bp in length. After polishing with HyPo the size of the assembly increased to 996 Mbp (103.2% of the expected genome size). We achieved an average coverage of 38 × and the contigs aligned well with the O. sativa Nipponbare reference genome (Supplementary Figs. S2, S3). BUSCO and LAI scores of 91.9% (complete) and 15.2, respectively, indicate that we have assembled a highly complete contigs. We identified 93 haplotigs, 124 junk contigs, 889 repeat contigs, and 850 primary contigs using Purge Haplotigs (Table 2). Thus, after purging the assembly of all but the primary contigs, the final assembly size was 859 Mbp (cv. 965 Mbp estimated by flow cytometry15, and 822 Mbp estimated by k-mer counting—see above).
Scaffolding resulted in the placement of 693 of the 850 primary contigs into one of 12 pseudomolecules and increased assembly size to ~ 861 Mbp. Pseudomolecules contained 81.5% of all assembled contigs (~ 812 Mbp of 996 Mbp) and long read coverage increased to 46×(Table 2, Supplementary Fig. S4, Supplementary File S1a). The remaining 157 contigs (46.9 Mbp) could not be placed into a pseudomolecule. Analysis of the unassigned contigs by LTR_Retriever showed that these sequences were largely made up of repeat sequences (~ 80% LTR-RT content, versus ~ 60% in the pseudomolecules), explaining why they could not be placed in the scaffolds. After scaffolding with KEEP contigs only, the BUSCO completeness score of the assembly was 97.5% and the LAI was 17.6. To further assess the quality of our scaffolded genome, we mapped ONT long-reads to the pseudomolecules, where 99.87% of ONT long-reads aligned to our genome with high quality (average MAPQ = 47).
After aligning our assembly long-reads (i.e., KR1, KR2, and KR3) to our assembled genome we observed several positions in the O. australiensis KR assembly that have low long-read coverage (< 20x, cv. 46× scaffold coverage; Supplementary Fig. S4). These regions predominantly occur on chromosome 4 and coincide with regions that are composed of smaller contigs (Supplementary Fig. S5). Additionally, chromosome 4 was constructed from a larger number of contigs than all other pseudomolecules (76 contigs compared to an average of 56 ± 9 contigs per pseudomolecule; Supplementary Fig. S5).
Structural variations (SVs)
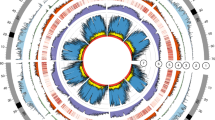
We aligned the O. australiensis KR assembly to the O. sativa Nipponbare assembly and observed a high level of synteny between the two genomes (Fig. 2). We also observed 378 instances of non-syntenic alignment (e.g., sequence that occurs on O. sativa chromosome 1 aligning to O. australiensis KR pseudomolecule 3). Thus, the genome–genome alignment indicated the presence of SVs. We investigated 21 SVs further (8 translocations, 7 inverted translocations, 5 inversions, and 1 duplication). The largest SV detected was an inversion that occurred on Chr11 and was 2.95 Mbp long within the O. sativa Nipponbare genome and 7.2 Mbp long in the O. australiensis KR genome assembly. We show that some of the SVs are unlikely to be caused by scaffolding errors (Supplementary Fig. S5). Seven of the 21 reported SVs cross contig–contig boundaries. Further, by mapping the O. australiensis KR long reads to the pseudomolecules and observing continuous mapping of long reads at a depth of 31–75× coverage across SV junctions, we show that the investigated SVs are likely not artefacts of errors in the assembly of the contigs. All reported SVs require further investigation before they can be confirmed.
Alignment of scaffolded O. australiensis KR contigs (named Chr1–12) to the O. sativa Nipponbare reference genome. The wild rice genome was aligned to the domestic rice genome using minimap2 and visualised using dotplotly. The 21 SVs that were investigated further are circled in red (some circles contain multiple SVs). Note: chromosomes do not appear in numerical order—dotPlotly orders the target sequence (O. sativa, here) by chromosome size; ChrUn is not included as it did not contain any large alignments.
Recently, Stein et al.1 showed an inversion on chromosome two of non-AA rice genomes relative to AA genome rice. This inversion was not detected in the assembly presented here (Fig. 2). We verified the inversion reported by Stein et al.1 and the lack of inversion in the O. australiensis KR assembly by aligning O. sativa Nipponbare Chr2 with O. punctata Chr2 (Supplementary Fig. S6A) and O. australiensis KR Chr2 (Supplementary Fig. S6B), respectively. When we aligned O. australiensis KR Chr2 to O. punctata Chr2 we saw an inversion similar to the O. sativa Nipponbare vs O. punctata Chr2 alignment (Supplementary Fig. S6C). To check the validity of this finding, we mapped O. australiensis KR long reads back to the assembly and observed 38–43× coverage at the alleged SV junction. Further, the coordinates of the alleged SV (Chr2:74,761,274–74,941,712) in the O. australiensis KR assembly, derived from the alignment of O. australiensis KR Chr2 to O. punctata Chr2 (Supplementary Fig. S6C), sit well within the boundaries of a contig (tig00003125 occupies Chr2: 69,779,629–76,584,758). After aligning O. punctata-guided O. australiensis Chr2 to O. punctata Chr2 we again observed an inversion at these coordinates, but no inversion when O. punctata-guided O. australiensis Chr2 and O. sativa Nipponbare Chr2 (data not shown).
Transcripts and genome annotation
Using a single PacBio Sequel II 8M SMRT Cell we were able to generate 3.3 million polymerase reads. Once these data were collapsed to Circular Consensus Sequencing (CCS) reads and demultiplexed, we had ~183–305 k CCS reads per tissue. Once the demultiplexed CCS reads were clustered we had ~21–35 k polished high quality (HQ) isoforms per tissue (Table 3). BUSCO analysis indicated that the Iso-Seq transcripts from the five selected tissues accounted for ~ 40% of the 4896 sequences in the Poales database. Only the HQ transcripts were used for annotation (low quality transcripts were discarded). A number of transcript isoforms were detected (see Supplementary Fig. S1B–D for examples of transcript isoforms for genes involved in stress response in rice).
Following the production of pseudomolecules via reference-guided scaffolding, LTR_Retriever identified 608.7 Mbp (61.6%) LTR-RT content in the assembly (assigned and unassigned contigs inclusive). EDTA identified 730.1 Mbp of repeat sequences in the pseudomolecules (Table 4, Supplementary File S2b), including 53.07% LTR-RT sequences. A peak in insertion events for both Gypsy and Copia LTR-RT elements was observed at ~ 0.5 million years ago (MYA; Supplementary Fig. S7). Gypsy repeat elements are more abundant than Copia elements in the O. australiensis KR genome. A third ‘unknown’ LTR-RT superfamily shows a slow increase in copy number over the last four million years (Supplementary Fig. S7).
Using the HQ Iso-Seq transcripts and protein sequences from the Oryza genus as evidence, OmicsBox identified 52,090 genes and 60,325 transcripts in the assembly (Supplementary Files S3c, S4d). OmicsBox functionally annotated 35,620 genes and 43,362 transcripts. To check whether our annotation included TE genes, we searched the gene transcripts for known plant TE families55. Of our annotated genes 1033 sequences (829 Gypsy LTR-RTs, 81 Copia LTR-RTs, 4 unknown LINEs, 105 CACTA TIRs, 4 MuDR TIRs, 8 Pong TIRs, and 2 unknown non-LTR-RT repeat elements) were found to be TE genes and removed from the O. australiensis gene annotation. After accounting for the presence of TE genes we were left with 51,057 predicted genes, 34,587 functionally annotated genes, and 42,329 functionally annotated transcripts.
Genotypic diversity and genetic similarity across four O. australiensis accessions and within the genus Oryza
After mapping the O. australiensis KR short reads to the O. australiensis KR assembly, we detected ~ 2 million SNVs. In comparison, we detected ~ 2.9 million, ~ 5 million, and ~ 6.4 million SNVs when mapping the CH, D, and VR accession short reads to the O. australiensis KR assembly. In the samples we analysed, genome wide SNV number did not increase with increasing geographic distance (Fig. 1). On the other hand, Fig. 3 shows that the CH accession is most closely related to KR, followed by VR, and then D—that is, genetic distance (as determined by kWIP) increases with increasing geographic distance (Figs. 1, 3).
Genetic distance between Oryza species/accessions derived from Illumina short read libraries by kWIP. Samples are coloured based on the Oryza genome clade that they occupy (see inset Legend). kWIP clustered samples into the canonical O. sativa (AA genome) and O. officinalis (BB, CC, BBCC, CCDD, EE genomes) complexes. The O. australiensis lineage is divergent from the rest of the O. officinalis genome complex, suggesting it harbours lineage specific adaptations that could be explored for stress tolerance. Within the O. australiensis lineage, the four genotype samples re-sequenced in the present study (KR, CH, VR, and D) show genetic distances that are correlated with geographic distance. The sample divergence between KR, CH, VR, and D, as well as the other O. australiensis accessions shown here suggests that there may be within-species genomic variation that can also be explored for tolerance to stresses (e.g., genotype-specific tolerance to salt stress). Samples that appear with red borders are suspicious as they did not cluster with their corresponding genome clade. This may be due to errors in the sequencing files that were not corrected prior to running kWIP or may reflect human error during sample preparation for sequencing.
Genetic distance between 15 species/accessions (representing the AA, BB, CC, BBCC, CCDD, EE, FF, GG, HHJJ, and HHKK genomes) within the Oryza genus was estimated with kWIP. A full list of the short read accessions used can be found in Supplementary File S5e. The genetic distances estimated by kWIP allowed for the reconstruction of the Oryza phylogeny except for two ‘misplaced’ samples (10. O. ridleyi, and 13. O. australiensis; Fig. 3). We were able to re-construct the canonical O. sativa (AA) and the O. officinalis (BB, CC, CCDD, EE) genome complexes, and the remaining Oryza genomes (FF, GG, HHJJ, HHKK) form a third complex. The O. australiensis lineage diverges from the rest of the O. officinalis complex members (Fig. 3).
Mapping the O. australiensis KR short reads to the O. sativa Nipponbare reference genome revealed ~ 2.9 million SNVs between O. australiensis KR and O. sativa Nipponbare (Supplementary Fig. S8). There are regions on all O. sativa chromosomes that, from visual inspection, have low SNV density. Also, the density of SNVs appears to increase at both terminal ends of the O. sativa chromosomes, though this may be due to uneven short read mapping density across the O. sativa Nipponbare genome (Supplementary Fig. S9).
Discussion
In this study we report on the first long-read assembly of an O. australiensis nuclear genome. Our results indicate that the assembled genome is of high quality and completeness. The assembled pseudomolecules (excluding ChrUn) are, on average, 2.2 times larger than the O. sativa Nipponbare reference chromosomes, which is in line with previous observations that O. australiensis chromosomes are larger than other Oryza species when seen during mitosis58. Our scaffolded O. australiensis genome assembly (858 Mbp) is smaller than estimated by flow cytometry (965 Mbp16,58), however our k-mer based estimation of genome size using O. australiensis short reads predicted a genome of 822 Mbp. The discrepancy in size between the genome reported here and the estimated genome size for O. australiensis15,16 may come about due to irresolvable LTR-RT elements.
After aligning the ONT long reads used to assemble the O. australiensis KR contigs to the reference-guided pseudomolecules, we observed unequal coverage distribution on some regions of the pseudomolecules (Supplementary Fig. S4). Most often, coverage was halved in these regions (for example, see Chr4). Having purged the assembly of haplotigs it is unlikely that low coverage results from the presence of homologous maternal and paternal sequences being present in the assembly. Instead, it appears that the low coverage came about due to the relatively high number of small contigs that make up some parts of the pseudomolecules. For example, Chr4, which has the lowest median long read coverage, is composed of the most contigs compared to all other pseudomolecules (apart from ChrUn; Supplementary Fig. S5). The fragmented nature of the contigs that have been used to build these regions of Chr4, and the other pseudomolecules, is characteristic of repetitive genome regions that are typically difficult to assemble. To overcome the limitations imposed by highly repetitive genome regions and, thus, to resolve the sequences therein, it is recommended that future studies employ long reads sufficient for a higher genome coverage.
We found that ~ 74% of the assembly is repeat sequence, of which ~ 55.5–61.6% was identified as LTR-RTs. These estimates are in line with previous estimates (~ 65%) of O. australiensis genome repeat content15. Stein et al.1 report a maximum total repeat content of 49.6% for O. punctata (BB genome), and a minimum of 27.2% for O. meridionalis (AA genome), thus O. australiensis has the highest total repeat content among the few Oryza species sequenced to date. We show that there was a relatively recent (~ 0.5 MYA) increase in LTR-RT (Gypsy and Copia) composition of the O. australiensis KR genome, in line with previous observations of O. australiensis Bacterial Artificial Chromosome (BAC) library sequences15. Similar trends of expansion for these LTR-RT types have been documented for both O. sativa indica and O. sativa ssp. japonica varieties1. However, along with a recent expansion of these LTR-RTs, the O. sativa genomes have undergone subsequent substantial gene loss, which has played a role in determining the genome size59. We also report on the expansion of an unknown LTR-RT family that appears to be in progress in modern-day O. australiensis plants, similar to the two O. sativa subspecies mentioned above and other O. australiensis accessions15. Piegu et al.15 showed that the LTR-RT element called ‘Kangourou’, a low copy number LTR-RT relative to Gypsy or Copia, had a peak burst time of ~ 2.3 MYA. The magnitude of the copy number burst in the O. australiensis genome of Kangourou in previous studies using BAC libraries15 matches that of the ‘unknown’ LTR-RT element we report here, however our results suggest a much more recent (~ 0.2 MYA) but slower increase in copy number. Other species of wild rice appear to have more ancient LTR-RT family (Gypsy, Copia, and unknown) expansions. For example, the Gypsy, Copia, and unknown LTR-RT families of O. brachyantha peaked in abundance around 1.8, 2, and 2.5 MYA, respectively1. Shenton et al.13 show a burst in the Mutator MuDR2 family composition of the O. officinalis genome complex. Further, O. meridionalis seems to have more dated (~ 1–1.5 MYA) expansions and then contractions of the same LTR-RTs relative to O. australiensis KR1. However, given the differences in the assembly methods and qualities of the various Oryza genomes, it is difficult to make direct comparisons of LTR-RT family ages and so these speculations should be viewed with caution.
After accounting for TE genes in the annotation file, we predicted 51,057 genes and functionally annotated 34,587 of those genes. This is in line with previously reported Oryza genome annotations. For example, Stein et al.1 report a maximum of 38,550 annotated loci for O. sativa ssp. japonica, and a minimum of 24,208 for O. brachyantha. In comparison, Release 7 of the O. sativa Nipponbare genome contains 39,045 non-TE annotated genes and 49,066 non-TE gene models46. Thus, the number of genes/transcripts annotated in this version of the O. australiensis KR genome assembly is comparable to that of the O. sativa Nipponbare version 7 genome. This is consistent with the hypothesis that the size of the O. australiensis genome is the result of the expansion of LTR-RT elements, rather than gene duplication events15. However, similar numbers of coding genes invite speculation about a functional role for expansion of LTR-RT elements. For example, it is conceivable that the especially adverse environments where O. australiensis evolved in northern Australia have selected for expansion of particular gene families involved in tolerance to stress.
Comparing the O. australiensis KR genome assembly to the O. sativa Nipponbare assembly, we identified a number of large structural variations. Recently, Stein et al.1 showed an inversion on chromosome two of non-AA rice genomes relative to AA genome rice. The same inversion appears in Leersia perrieri and Brachypodium distachyon and so it may be a conserved feature in grass genomes, except in rice with the AA genome1. This inversion was not detected by reference-guided construction of the O. australiensis assembly (Fig. 2), suggesting a need for deeper coverage of this region in future studies. However, we can report 36–45× coverage at the putative inversion junction in the O. australiensis KR assembly, and we show that the putative SV sits well within the boundaries of a single contig. Hence, we suggest that this region of the O. australiensis KR assembly is not the result of misassembly. The possibility remains that it is an artefact of reference-biased scaffolding, however we found that the inversion is missing in the O. australiensis assembly even after using a BB rice genome (O. punctata) to order and orient the contigs (data not shown), so this is unlikely. The SV reported by Stein et al.1 in non-AA rice genomes may truly be absent from the O. australiensis KR genome. One hypothesis for its absence is hybridisation between the co-occurring AA-genome species (O. meridionalis and O. rufipogon) and O. australiensis, thus eliminating the Chr2 SV in the O. australiensis KR genome. However, given the difficulty in producing viable progeny from artificial crosses between the AA genome species and O. australiensis60, we find this to be an improbable explanation for the observed results. Thus, in the absence of further evidence, all structural variations should be viewed with caution.
Given that the 21 large SVs we observed do not appear to be the result of either scaffolding or assembly artefacts, if these SVs prove to be real, they highlight considerable differences in the structure of the O. australiensis KR assembly and O. sativa. These SVs, and the substantial repeat content of the wild rice genome, would offer insight into why crossing the AA genome O. sativa and the EE genome O. australiensis is difficult and why successful crosses have abnormal chromosomal arrangements and the resulting progeny are sterile61,62. To validate the putative SVs, future scaffolding attempts could make use of other Oryza species for reference-guided scaffolding (e.g., the O. officinalis assembly may be a better candidate for scaffolding than the O. sativa Nipponbare assembly as O. australiensis is a member of the O. officinalis complex). However, this would still present biases in contig ordering and orientation. Therefore, we recommend that future scaffolding attempts either make use of pre-existing O. australiensis BAC libraries and FingerPrinted Contigs (FPC) to order and orient the contigs1,13 or use Hi-C technology to generate new data for O. australiensis. Given the still experimental nature of Hi-C and difficulties in obtaining high coverage of linked reads these efforts may require significant investment.
Along with structural variations in the O. australiensis KR genome relative to O. sativa Nipponbare, we also uncovered many SNVs (Supplementary Fig. S8). Interestingly, there were more SNVs between some O. australiensis genotypes and O. australiensis KR than there were between O. sativa and O. australiensis KR based on short read mapping. The density of SNVs increases at the terminal regions of the O. sativa chromosomes. Conversely, there is a region on each chromosome where the density of SNVs is low. These regions may represent the centromeric repeat elements of the chromosome (Supplementary Fig. S8). While centromeric regions have a common functional role in organising cell division, their sequences can be diverse between species. However, in the case of rice and other cereals, they are highly conserved63,64. For example, Gao et al.65 showed that O. australiensis and O. sativa share similar centromeric retrotransposon repeat (CRR) sequences even though other rice species such as O. brachyantha have unique CRR profiles. This helps explain why sequence similarity between O. australiensis KR and O. sativa at centromeric regions is higher than at other regions of the genome.
Interestingly, when comparing four genotypes of O. australiensis that had been documented for their contrasting salt tolerance20, we found that SNV density was not well correlated with the geographical distance between each of the populations (Fig. 1). For example, VR had at least 1.5 million more SNVs than the other two genotypes (when compared with KR), yet it was collected from a site far closer to Keep River than the D genotype. We speculate that SNV densities may be more related to the diversity of alleles required for adaptation to stresses than physical separation between populations. For example ‘relative salt tolerance’20 (Fig. 1) appeared to correlate better with the abundance of SNV abundance than simple distance between populations.
Contrary to the SNV data, the genetic distances estimated by kWIP do reflect the geographic distance between the O. australiensis genotypes (Fig. 3). That is, populations of O. australiensis genotypes that occur closer to each other in space are more genetically similar than genotypes that occur further apart. This is expected as O. australiensis is a mostly inbreeding species (~ 3% outcrossing), so gene flow and introgressions between genotypes should be low. Thus, we assume that these populations of O. australiensis are reproductively isolated and, as such, their genomes are evolving independently. If this is true, we expect to see local adaptation in these genotypes66, a hypothesis consistent with the findings of Yichie et al.20.
The genetic-distance tree generated by kWIP follows previously published Oryza phylogenies13. Importantly, the O. australiensis lineage diverges from the rest of the O. officinalis complex members (the B, C, and D genomes) suggesting that we can expect to see lineage specific adaptation in the O. australiensis genome. Given that this species occupies a diverse yet adverse suite of environmental niches, the genomic variation that underpins the divergence of the O. australiensis lineage may be associated with tolerance to multiple abiotic and biotic stresses. The functionally annotated genome that we present for O. australiensis KR here is therefore a resource that can be used to explore this genomic variation to uncover genes involved in stress tolerance.
Conclusions
Here we present the first reference-quality genome of O. australiensis KR—a wild species of rice native to the northern regions of Australia. While O. australiensis co-occurs with other Oryza species, it is genetically and phenotypically unique among the Oryza genus and is the sole member of the EE genome clade67. It demonstrates unrivalled tolerance to extreme environmental conditions, including heat, drought and soil salinity3,18,21,68. The leaf anatomy and associated photosynthetic efficiency of O. australiensis are also unique traits22 and its grain has distinct pigmentation and starch composition23,24. Such traits make it an important genetic resource in the Oryza genus, as recognised formally by the OMAP and IOMAP11,12. However, no group has successfully assembled the nuclear genome for this species until now. It is expected that this genomic resource will enable phenomics in wild cereal relatives and lead to the selection of useful traits for the improvement of domestic rice cultivars, and in the breeding and domestication of O. australiensis itself. For example, given that we now know the positions of genes in the O. australiensis KR assembly, one can use the assembly alongside short reads from diverse O. australiensis accessions to identify SNVs in genes associated with tolerance in accessions from adverse environments. Similarly, with further Iso-Seq data one could identify novel transcript isoforms across different O. australiensis accessions under multiple stresses. Furthermore, given the role that gene regulation plays in response to stress, the O. australiensis KR assembly will uncover promoters of genes of interest. This work could extend to the use of long read data (either PacBio or ONT reads) to investigate the response of the epigenome to abiotic stresses. Finally, an expanded collection of O. australiensis accessions with contrasting phenotypes under extreme conditions will facilitate the identification of markers for stress tolerance via strategic inclusion of tolerant accessions to make mapping populations.
Data availability
This Whole Genome project (i.e., the Canu-generated purged contigs reported here) has been deposited at DDBJ/ENA/GenBank under the accession JAIFGZ000000000. The version described in this paper is version JAIFGZ010000000. All long- and short-read data associated with the reported genome assembly have been deposited at the Sequence Read Archive and GenBank in NCBI under BioProject PRJNA743927. The AGP file required to construct the pseudomolecules from the Canu contigs, the annotation for protein-coding genes in the contigs, the annotation for repeat elements in the scaffolds, and the annotation for protein-coding genes in the scaffolds (all GFF3 files) have been provided as Supplementary Files and can be accessed via figshare: https://doi.org/10.6084/m9.figshare.c.5875592.v2.
Abbreviations
- AGP:
-
A golden path
- ANU:
-
Australian National University
- BAM:
-
Binary Alignment Map
- BCF:
-
Binary variant call format
- BLAST:
-
Basic local alignment search tool
- bp:
-
Base pairs
- BUSCO:
-
Benchmarking universal single-copy orthologs
- Chr:
-
Chromosome
- ChrUn:
-
Chromosome unassigned
- DVC(R):
-
Deputy Vice Chancellor (Research)
- EDTA:
-
Ethylenediaminetetraacetic acid
- EDTA:
-
Extensive de novo TE annotator
- GFF3:
-
General Feature Format 3
- GO:
-
Gene Ontology
- HPC:
-
High performance computer
- HQ:
-
High quality
- IGV:
-
Integrative genomics viewer
- IOMAP:
-
International Oryza Map Alignment Project
- Iso-Seq:
-
Isoform sequencing
- Kbp:
-
Kilo base pairs
- LAI:
-
LTR assembly index
- LQ:
-
Low quality
- LTR:
-
Long terminal repeat
- RT:
-
Retrotransposon
- Mbp:
-
Mega base pairs
- MYA:
-
Million years ago
- NCBI:
-
National Center for Biotechnology Information
- NSW:
-
New South Wales
- OMAP:
-
Oryza Map Alignment Project
- ONT:
-
Oxford Nanopore Technologies
- PAF:
-
Pairwise mApping format
- PEG:
-
Polyethylene glycol
- PIF:
-
P Instability factor
- PVP:
-
Polyvinylpyrrolidone
- RIN:
-
RNA integrity number
- RMBlast:
-
RepeatMasker compatible NCBI BLAST suite
- SAM:
-
Sequence Alignment Map
- SNV:
-
Single nucleotide variation
- SV:
-
Structural variation
- TIR:
-
Terminal inverted repeat
- VCF:
-
Variant call format
References
Stein, J. C. et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 50, 285–296 (2018).
Brar, D. S. & Khush, G. S. Wild relatives of rice: A valuable genetic resource for genomics and breeding research. In The Wild Oryza Genomes (eds Mondal, T. K. & Henry, R. J.) 1–25 (Springer, 2018).
Atwell, B. J., Wang, H. & Scafaro, A. P. Could abiotic stress tolerance in wild relatives of rice be used to improve Oryza sativa? Plant Sci. 215–216, 48–58 (2014).
Bisht, D. S., Solanke, A. U. & Mondal, T. K. Informatics of wild relatives of rice. In The Wild Oryza Genomes (eds Mondal, T. K. & Henry, R. J.) 27–40 (Springer, 2018).
Huang, X. et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42, 961–967 (2010).
Bolser, D., Staines, D. M., Pritchard, E. & Kersey, P. Ensembl plants: Integrating tools for visualizing, mining, and analyzing plant genomics data. Methods Mol. Biol. 1374, 115–140 (2016).
Henry, R. J. et al. Australian Oryza: Utility and conservation. Rice 3, 235–241 (2010).
Hiromi, K.-K. et al. OryzaGenome2.1: Database of diverse genotypes in wild Oryza species. Rice 14, 00468 (2021).
Yu, H. et al. A route to de novo domestication of wild allotetraploid rice. Cell 184, 1156–1170 (2021).
Song, S., Tian, D., Zhang, Z., Hu, S. & Yu, J. Rice genomics: Over the past two decades and into the future. Genomics Proteomics Bioinform. 16, 397–404 (2018).
Wing, R. A. et al. The Oryza map alignment project: The golden path to unlocking the genetic potential of wild rice species. Plant Mol. Biol. 59, 53–62 (2005).
Jacquemin, J., Bhatia, D., Singh, K. & Wing, R. A. The International Oryza Map Alignment Project: Development of a genus-wide comparative genomics platform to help solve the 9 billion-people question. Curr. Opin. Plant Biol. 16, 147–156 (2013).
Shenton, M. et al. Evolution and diversity of the wild rice Oryza officinalis complex, across continents, genome types, and ploidy levels. Genome Biol. Evol. 12, 413–428 (2020).
Shi, C. et al. The draft genome sequence of an upland wild rice species, Oryza granulata. Sci. Data 7, 131 (2020).
Piegu, B. et al. Doubling genome size without polyploidization: Dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 16, 1262–1269 (2006).
Ammiraju, J. S. S. et al. The Oryza bacterial artificial chromosome library resource: Construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res. 16, 140–147 (2006).
Zhang, Q. J. & Gao, L. Z. Rapid and recent evolution of LTR retrotransposons drives rice genome evolution during the speciation of AA-genome Oryza species. G3 Genes Genomes Genet. 7, 1875–1885 (2017).
Phillips, A. L., Scafaro, A. P. & Atwell, B. J. Photosynthetic traits of Australian wild rice (Oryza australiensis) confer tolerance to extreme daytime temperatures. Plant Mol. Biol. https://doi.org/10.1007/s11103-021-01210-3 (2022).
Scafaro, A. P. et al. A thermotolerant variant of Rubisco activase from a wild relative improves growth and seed yield in rice under heat stress. Front. Plant Sci. 9, 1–11 (2018).
Yichie, Y., Brien, C., Berger, B., Roberts, T. H. & Atwell, B. J. Salinity tolerance in Australian wild Oryza species varies widely and matches that observed in O. sativa. Rice 11, 1–14 (2018).
Hamzelou, S. et al. Wild and cultivated species of rice have distinctive proteomic responses to drought. Int. J. Mol. Sci. 21, 5980 (2020).
Mathan, J., Singh, A., Jathar, V. & Ranjan, A. High photosynthesis rate in two wild rice species is driven by leaf anatomy mediating high Rubisco activity and electron transport rate. J. Exp. Bot. https://doi.org/10.1093/jxb/erab313 (2021).
Tikapunya, T., Fox, G., Furtado, A. & Henry, R. Grain physical characteristic of the Australian wild rices. Plant Genet. Resour. 15, 409–420 (2016).
Tikapunya, T. et al. Molecular structures and properties of starches of Australian wild rice. Carbohydr. Polym. 172, 213–222 (2017).
Joly-Lopez, Z. et al. An inferred fitness consequence map of the rice genome. Nat. Plants 6, 119–130 (2020).
Michael, T. P. & VanBuren, R. Building near-complete plant genomes. Curr. Opin. Plant Biol. 54, 26–33 (2020).
Jones, A. et al. High-molecular weight DNA extraction, clean-up and size selection for long-read sequencing. PLoS ONE 16, e0253830 (2021).
Jones, A., Borevitz, J., Warthmann, N. & Jones, A. Cost-conscious generation of multiplexed short-read DNA libraries for whole genome sequencing. Protocols.io. https://doi.org/10.17504/protocols.io.unbevan (2018).
Betts, N. S. et al. Isolation of tissues and preservation of RNA from intact, germinated barley grain. Plant J. 91, 754–765 (2017).
Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011).
Vurture, G. W. et al. GenomeScope: Fast reference-free genome profiling from short reads. Bioinformatics 33, 2202–2204 (2017).
Ranallo-Benavidez, T. R., Jaron, K. S. & Schatz, M. C. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 11, 1432 (2020).
Koren, S. et al. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017).
Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Kundu, R., Casey, J. & Sung, W.-K. Hypo: Super fast & accurate polisher for long read genome assemblies. BioRxiv. https://doi.org/10.1101/2019.12.19.882506 (2019).
Lindenbaum, P. JVarkit: Java-Based Utilities for Bioinformatics. https://doi.org/10.6084/m9.figshare.1425030.v1 (2015).
Shen, W. & Xiong, J. TaxonKit: A cross-platform and efficient NCBI taxonomy toolkit. BioRxiv. https://doi.org/10.1101/513523 (2019).
Thorvaldsdóttir, H., Robinson, J. T. & Mesirov, J. P. Integrative genomics viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2012).
Roach, M. J., Schmidt, S. A. & Borneman, A. R. Purge Haplotigs: Allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinform. 19, 460 (2018).
Ou, S. & Jiang, N. LTR_FINDER_parallel: Parallelization of LTR_FINDER enabling rapid identification of long terminal repeat retrotransposons. Mob. DNA 10, 48 (2019).
Ellinghaus, D., Kurtz, S. & Willhoeft, U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinform. 9, 18 (2008).
Ou, S., Chen, J. & Jiang, N. Assessing genome assembly quality using the LTR Assembly Index (LAI). Nucleic Acids Res. 46, e126 (2018).
Seppey, M., Manni, M. & Zdobnov, E. M. BUSCO: Assessing genome assembly and annotation completeness. In Gene Prediction (ed. Kollmar, M.) 227–245 (Springer, 2019).
Jain, C., Koren, S., Dilthey, A., Phillippy, A. M. & Aluru, S. A fast adaptive algorithm for computing whole-genome homology maps. Bioinformatics 34, i748–i756 (2018).
Alonge, M. et al. RaGOO: Fast and accurate reference-guided scaffolding of draft genomes. Genome Biol. 20, 224 (2019).
Kawahara, Y. et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4 (2013).
Poorten, T. dotPlotly (2018).
Gel, B. & Serra, E. karyoploteR: An R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics 33, 3088–3090 (2017).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Ou, S. et al. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 20, 275 (2019).
Wing, R. International Oryza Map Alignment Project. https://doi.org/10.7946/P2FC9Z (2017).
Götz, S. et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435 (2008).
BioBam. OmicsBox—Bioinformatics Made Easy (2019).
Pedro, D. L. F. et al. An atlas of plant transposable elements [version 1; peer review: 2 approved]. F1000Research 10, 1194 (2021).
Murray, K. D., Webers, C., Ong, C. S., Borevitz, J. & Warthmann, N. kWIP: The k-mer weighted inner product, a de novo estimator of genetic similarity. PLoS Comput. Biol. 13, e1005727 (2017).
Crusoe, M. R. et al. The khmer software package: Enabling efficient nucleotide sequence analysis [version 1; peer review: 2 approved, 1 approved with reservations]. F1000Research 4, 900 (2015).
Uozu, S. et al. Repetitive sequences: cause for variation in genome size and chromosome morphology in the genus Oryza. Plant Mol. Biol. 35, 791–799 (1997).
Vitte, C., Panaud, O. & Quesneville, H. LTR retrotransposons in rice (Oryza sativa L.): Recent burst amplifications followed by rapid DNA loss. BMC Genomics 8, 218 (2007).
Multani, D. S. et al. Development of monosomic alien addition lines and introgression of genes from Oryza australiensis Domin. to cultivated rice O. sativa L.. Theor. Appl. Genet. 88, 102–109 (1994).
Abbasi, F. M., Ahmad, H., Sajid, M., Inamullah, & Brar, D. S. Detecting nature of chromosome pairing in A and E genomes of Oryza. Cytologia (Tokyo) 74, 223–227 (2009).
Abbasi, F. M. et al. Assessment of genomic relationship between Oryza sativa and Oryza australinesis. Afr. J. Biotechnol. 9, 1312–1316 (2010).
Aragón-Alcaide, L., Miller, T., Schwarzacher, T., Reader, S. & Moore, G. A cereal centromeric sequence. Chromosoma 105, 261–268 (1996).
Jiang, J. et al. A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc. Natl. Acad. Sci. 93, 14210–14213 (1996).
Gao, D. et al. A lineage-specific centromere retrotransposon in Oryza brachyantha. Plant J. 60, 820–831 (2009).
Lascoux, M., Glémin, S. & Savolainen, O. Local adaptation in plants. eLS. https://doi.org/10.1002/9780470015902.a0025270 (2016).
Henry, R. J. Oryza australiensis Domin. In The Wild Oryza Genomes (eds Mondal, T. K. & Henry, R. J.) (Springer, 2018).
Yichie, Y. et al. Salt-treated roots of Oryza australiensis seedlings are enriched with proteins involved in energetics and transport. Proteomics 19, 1900175 (2019).
Acknowledgements
The authors would like to acknowledge Ming-Dao Chia for providing initial bioinformatics support. We thank Scott Ferguson, Lina Herliana, Chelsea Matthews, and Farhad Masoomi-Aladizgeh for the discussions about bioinformatics analyses. We thank Dr Sally Norton from the Australian Grains Genebank in Horsham Victoria, Australia for providing the seed a decade ago that led to the present study. We thank Dr. James Cowley (Founder of ‘The Figure Legend’) for his advice on designing figures. We would like to deeply thank the reviewers, whose thoughtful comments helped substantially improve the manuscript.
Funding
This work was funded by the Macquarie University DVC(R) Fund with contributions from the Department of Biological Sciences. This project was supported by the ARC Centre of Excellence in Plant Energy Biology (CE140100008). AP acknowledges support from an Australian Research Training Program Scholarship and the FJ Sandoz Scholarship from the University of Adelaide. The funding was also provided by Deputy Vice-Chancellor (Research) Discretionary Fund (51277/20).
Author information
Authors and Affiliations
Contributions
A.P., S.F., N.W.H., A.J., J.B., R.B., and B.A. contributed to technical planning after B.A. initiated the project with a successful application for a Macquarie University DVC(R) Discretionary grant. J.B. and A.J. provided technical expertise for execution of the sequencing. A.P. and A.J. performed all DNA and RNA preparations and performed all ONT MinION sequencing steps. A.J. performed Illumina sequencing steps, gave initial bioinformatics support, and supplied FASTQ files. N.W.H. performed Iso-Seq analysis and provided bioinformatics support. A.P. and S.F. performed all data analysis, genome assembly, and subsequent analyses. N.W.H. and R.B. provided access to the computing resources required. N.W.H., R.B., and B.A. provided guidance throughout. B.A. grew the plants for sequencing prior to extractions in Canberra. A.P. created all figures and wrote the initial draft of the manuscript. All authors contributed to the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Phillips, A.L., Ferguson, S., Watson-Haigh, N.S. et al. The first long-read nuclear genome assembly of Oryza australiensis, a wild rice from northern Australia. Sci Rep 12, 10823 (2022). https://doi.org/10.1038/s41598-022-14893-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-14893-5
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.