Abstract
Microscopy by Achromatic X-rays With Emission of Laminar Light (MAXWELL) is a new X-ray/visible technique with attractive characteristics including isotropic resolution in all directions, large-volume imaging and high throughput. An ultrathin, laminar X-ray beam produced by a Wolter type I mirror irradiates the sample stimulating the emission of visible light by scintillating nanoparticles, captured by an optical system. Three-dimensional (3D) images are obtained by scanning the specimen with respect to the laminar beam. We implemented and tested the technique with a high-brightness undulator at SPring-8, demonstrating its validity for a variety of specimens. This work was performed under the Synchrotrons for Neuroscience—an Asia–Pacific Strategic Enterprise (SYNAPSE) collaboration.
Similar content being viewed by others
Introduction
We implemented and tested a new microscopy technique called MAXWELL (Microscopy by Achromatic X-rays With Emission of Laminar Light) at the SPring-8 beamline 29XU1. The parallel use of X-rays and visible light achieves nanoscale resolution in all three-dimensions—avoiding the anisotropy of other microscopies—and is primarily conceived for large-volume biology specimens, including single-molecule imaging.
High resolution in the z-direction (perpendicular to the sample surface) is specifically important when using microscopy to identify microstructures, for example the neuron connections2. In standard visible microscopy, the z-resolution is limited by the elongated point spread function. This causes the aforementioned resolution anisotropy and makes it often difficult or impossible to extract accurate information on complex microstructures. The problem is even worse when super-resolution is achieved only in the xy-directions. With MAXWELL, the resolution anisotropy and the subsequent problems are avoided.
MAXWELL is implemented by irradiating the specimen with an X-ray beam shaped by a Wolter mirror as a very thin (< 65 nm) sheet (“nanoplane”). The X-rays stimulate the emission of visible light from the irradiated volume3,4 due to the presence of implanted scintillating nanoparticles (SciNPs)5,6,7. 3D imaging is realized by scanning the specimen through the nanoplane while sequentially capturing the visible emission.
Results and discussion
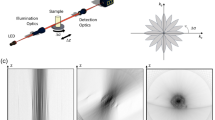
Figure 1 shows the MAXWELL instrument and the resolution estimate. The high z-resolution of MAXWELL is possible because of the short wavelengths of X-rays. However, it requires a high-performance X-ray optical device8,9,10,11,12. Much progress was made in recent years so that X-rays can now be focused below10 nm13,14,15. Among all devices, the one-dimensional type-I Wolter mirror is a clear choice for MAXWELL.
(a) Experimental setup of MAXWELL including a Wolter type-I linear focusing mirror that generates an X-ray nanoplane and a visible-light microscope system to detect the emission from X-ray-excited scintillating nanoparticles. (b) Knife edge intensity measurements performed at different positions along the optical axis. The measured thickness of the nanoplane was < 130 nm over the depth of focus of ± 175 µm. (c) A minimum FWHM size of 65 nm was observed at the focal spot.
Indeed, as other total-reflection mirrors it is almost achromatic and focuses X-rays over a relatively broad synchrotron-radiation spectral range. This corresponds to a high X-ray flux allowing high-speed imaging. The Wolter mirror produces a planar irradiation combining a nanoscale thickness (z-direction) and a large “in-focus” (xy-directions) area (> 400 µm × 400 µm). Figure 1b,c show the results of knife-edge intensity scans of the focused X-rays at different positions along the beam direction, referred to the focal point. This enables 2D full-field imaging and high-speed 3D imaging by fast scanning of large-volume (> 1 mm3) specimens. Such characteristics are difficult to achieve with other visible or X-ray focusing devices.
Furthermore, the Wolter mirror complements an elliptic-surface reflection with a hyperbolic reflection that eliminates the coma aberration16—enhancing the focusing stability, maintaining a fixed focusing size with no drifting and simplifying the alignment, which would be otherwise time consuming. To guarantee the required performances, state-of-the-art fabrication was used to produce ultrasmooth (< 0.2 nm RMS roughness and < 2 nm peak-to-valley figure error), aspherical reflecting surfaces17.
MAXWELL offers clear advantages with respect to other approaches for improving the z-resolution. For example, 4pi microscopy18,19and fluorescence lightsheet microscopy or selective plane illumination microscopy20,21,22,23,24 did not overcome the diffraction limit for z-resolution while adding constraints to the optical systems. Super-resolution in the z-direction was achieved using special fluorophores25,26,27 or by extracting the z-position of the fluorophores by quantifying their anisotropic aberration28. However, the long imaging time and the special specimen-labeling processes limit their use29.
On the contrary, MAXWELL achieves nanoscale z-resolution directly with X-ray focusing, without image processing. Since our SciNPs are much smaller than the X-ray beam thickness, the z-resolution is linked to the latter. Figure 1c shows a beam thickness of 65 nm. But this is not an absolute limit: considering the current resolution performance for X-ray focusing, further improvements are expected.
In addition to the thin X-ray nanoplane, small-size SciNPs are required to reach nanoscale z-resolution. For biological applications, small nanoparticles are also critical for precise administration to specific locations in tissues. And their visible emission should be ideally intense enough for single-particle detection. Tests of this capability, single particle imaging, using specimens with individual molecules are underway.
Note that the X-ray dose on the specimen when working at the highest resolution is ~ 1015 photons/mm2/s, which is quite high for biology tissues. There are margins for improving the apparatus performance and reduce this maximum dose. However, the feasibility of experiments on live specimens must still be assessed.
We extensively tested several 10 nm SciNP families, including NaGdF4:Eu and NaGdF4:Tb nanoparticles30,31 with red (~ 615 nm) or green (543 nm) emission, and CsBrPb3 nanoparticles with green emission (~ 530 nm)32. The TEM images of Fig. S1 show that the particle size is ~ 10 nm. The surface of these nanoparticles can be modified to conjugate linkers, such as streptavidin, to label specific macromolecules decorated with biotin. Such surface modifications, combined with tissue clearing33,34, allow 3D imaging deep into the specimen and single-molecule imaging for large samples.
Furthermore, with the relatively simple procedure4 of different dopings, SciNPs generate different scintillation colors (Fig. S2) and allowing multi-color imaging. We performed biocompatibility tests of these different SciNPs on live cells and found that there is only toxicity at very high concentration, 250 mg/mL (Fig. S3).
Figure 2 shows images assessing the resolution in the xy and z-directions by finding the smallest light spots from the emission of SciNPs (CsBrPb3) dispersed on a test micropatterns. The images were taken with a 50× lens, 0.7 numerical aperture (NA). We see, in particular (Fig. 2a), two spots due to different particles and the corresponding intensity scan in the xy plane. From the distance between the brightest points and from the widths of the spots, we estimate a xy-resolution of ≈400 nm, consistent with the diffraction limit for visible light and with the NA. For the z-direction, Fig. 2b,c indicate a better resolution, of the order of at least 70 nm, consistent with the X-ray sheet width.
In the present situation, isotropic resolution can be achieved by increasing the X-ray sheet width and reducing the z-resolution. However, our final objective is to obtain isotropy by improving the xy-resolution, specifically using the “super-resolution” deconvolution28,29. Tests in that direction are underway.
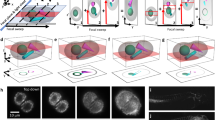
Figure 3a shows one of the first MAXWELL field tests: a reconstructed image of a thin HCT116 cell specimen. The cells were cultured on a glass substrate with SciNPs (a mixture of NaGdF4:Eu and NaGdF4:Tb) internalized via the endocytosis process; the cell culture was then washed with PBS (phosphate buffer solution) to remove SciNPs from the culture media. The MAXWELL images show that the SciNPs only reside in cells, confirming preliminary tests with ultraviolet-excited fluorescence microscopy (Fig. 3b).
HCT 116 cells cocultured with SciNPs. (a) 3D reconstructed MAXWELL image. (b) image of the same sample taken with UV irradiation. Note that the color was presented from the 3D visualization process and does not represent the emission color of the SciNPs. (c) MAXWELL image of HCT 116 cells loaded with SciNPs. The z-direction has a scale 5 times larger than the xy-directions to show the 3D structure of the SciNPs. Note that particles appear distributed in the same plane because the cultured cells spread on the glass substrate, their distribution becoming very thin in the z-direction. (d) TEM images of an HCT 116 cell revealing that many NaGdF4:Eu (left) and NaGdF4:Tb (right) nanoparticles were internalized in the cytoplasm. The cell nuclei are marked by asterisks. Scale bars: (a,b) 50 µm; (c) 10 µm; (d) 5 µm.
The bright spots in Fig. 3 are quite large. This is consistent with the cellular internalization of nanoparticles: they are aggregated by the endosomes and form large clusters of 0.1–2 µm size35,36. To show the 3D shape of these clusters, the z-scale of Fig. 3c is 5 times larger than for the x and y directions. Transmission electron microscopy (TEM) images (Fig. 3d) also confirm the nanoparticle clustering in the cell cytoplasm.
Figure 4a,b show another MAXWELL test: a 3D reconstructed image of a drosophila fly with SciNPs deposited in the tracheal and digestive systems. The specimen thickness was ~ 200 µm. The upper (close to the objective lens) part of the inset was removed to better detect SciNPs in the thorax.
MAXWELL images show a drosophila fly with SciNPs (NaGdF4:Eu) administrated to delineate the tracheal and intestine features. The 3D image, (b), was constructed by stacking single slice images such as that of (a). Note that the head part is not reconstructed in (b). The skeleton (blue) is visible due to X-ray autofluorescence. The segmentation of autofluorescence and SciNPs are based on their intensities. (c) 3D reconstructed MAXWELL image of an entire Drosophila larva. (d) Confocal laser scanning microscopy of the same Drosophila specimen exhibits high contrast but the visibility of the SciNPs is much reduced. Scale bars: (a, b) and (d) 100 µm; (c) 50 µm.
Figure 4c and the corresponding video clips (Supplementary Materials S02) show MAXWELL images of a thinner specimen, a Drosophila larva. The entire body was imaged without dissection. The X-ray-induced luminescence (blue) from the skeleton (labeled in blue Fig. 4b,c) provides a clear outline of the body37. The locations of the SciNPs (marked in green) delineate instead the tubular tracheal and digestive systems.
For comparison, Fig. 4d shows a visible-light confocal laser scanning microscopy image (FV-3000, Olympus Co., Tokyo) of the same fly specimen. This high-contrast 3D imaging could not detect the SciNPs, which were probably hidden by the much more intense fluorescence from other features.
Note that the signal-to-noise ratio (SNR) derived from MAXWELL images such as Fig. 4a reaches 11–12 decibels, much higher than the levels of confocal microscopy (Fig. 4d)—also thanks to the dark background. The high SNR would allow the detection of single SciNP, which is very stable in the X-ray irradiation without blinking or degradation of their emission.
Since the SciNPs formed in this case > 1 µm conglomerates, a focusing to a ~ 65 nm (Fig. 4c) or a relaxed ~ 170 nm (Fig. 4a,b) X-ray plane was used to irradiate the entire specimen. Note the uniform image quality over the xy planes, which is the result of the large depth of focus of the Wolter mirror. In this specific case, the X-ray nanoplane thickness of ~ 350 nm was maintained over a ~ (650 µm)2 area, sufficient for the required field of view in the xy directions. No deterioration of the resolution and contrast was detected over the entire volume. On the contrary, a visible lightsheet would produce only small-area “in-focus” images, because of the depth of focus of several µm.
Figure 5 shows a 3D reconstructed MAXWELL image of a slice of a mouse brain with glioma tumors. SciNPs were found, for example in Fig. 5b, accumulated in the tumor related microvessels via blood circulation36. A small amount of SciNPs was detected outside the microvessels, likely due to fenestration of the angiogenic microvessels.
(a,b) MAXWELL images of mouse brain perfused with SciNPs (NaGdF4:Eu) to delineate the microvessels induced by tumor angiogenesis. SciNPs found outside the blood vessels, such as those marked by yellow arrows and circles, are attributed to the leakage through the fenestrations on the angiogenic microvessel walls. Red arrows mark microvessels with diameter ≤ 10 µm. (c) A micrograph obtained with UV illumination shows the red light emission of SciNPs aggregated in the microvessels. (d) Low-magnification MAXWELL images showing the aggregation of SciNPs in a region of a mouse brain. Inset: the entire brain imaged after tissue clearing. Scale bars: (a) 10 µm; (b,c) 50 µm; (d) 1 mm; inset: 5 mm.
Large-volume (> 1 mm3) imaging of biology specimen with MAXWELL is influenced by the self-absorption of emitted visible light by the specimen. This problem was alleviated with the FocusClear tissue clearing agent38 originally developed for visible microscopy. Figure 5d shows a MAXWELL image of a large region of cleared mouse brain tissue, without sectioning.
As previously mentioned, the SciNP emission spectra can be modified with different dopants while maintaining a small particle size and the surface properties5,6,30. Figure 6a shows a MAXWELL image of an HCT-116 cell specimen loaded with NaGdF4:Eu (red emitting) and NaGdF4:Tb (green emitting) SciNPs. Two separate MAXWELL 3D images obtained with red and green filters were merged into this picture.
(a) Fused micrograph of two MAXWELL images taken with red or green filters, showing HCT-116 cells loaded with NaGdF4:Eu (red) and NaGdF4:Tb (green) SciNPs. Red and green circles mark those SciNPs emitting only red or green light. The yellow squares mark regions of mixed SciNPs (yellow). Zoomed-in images are shown on the right of (a). (b) Procedure to analyze and identify the color of each voxel of the merged 3D volume, a), with the emission spectra of red and green SciNPs. The voxels of strong green and red emission intensity are colored in yellow. Scale bar: (a) 50 µm; inset) 1 µm.
Figure 6b illustrates the procedure to identify the emitting specific colors of each voxel of a light emitting region of interest using the emission spectra of the SciNPs. The 3D green-filtered and red-filtered images were aligned and merged. Each voxel exhibits both green and red emission. The red/green intensity ratio of each voxel was analyzed to identify its leading color—see for example those marked with red and green circles. Voxels with high red and green emission are marked in yellow: they are probably clusters of red and green SciNPs too close to separate. Note that the very high z-resolution, evident in the perpendicular views (the inset of 6a-1), facilitates the identification of nearby SciNPs of different colors (marked by a yellow square).
The key message of this last test is that SciNPs emitting different colors can enhance the results of MAXWELL using simple color filters. Other tests with different nanoparticles are underway. But we can already conclude that the combination of MAXWELL with nanoparticle manipulation and tissue clearing offers excellent flexibility in adapting the new microscopy to the needs of different experiments.
Methods
The X-ray source for MAXWELL is the high-brightness SPring-8 BL29XU undulator equipped with a Si (111) double crystal monochromator1. A vertical slit with adjustable 10–30 µm width, at 45 m from the Wolter mirror, defines the virtual source.
The parameters of the Wolter mirror are the following: average total reflection angle of 5 mrad, half angular aperture of 1.5 × 10–3, substrate lengths of 232.2 mm and focal lengths of 229.6 mm. The mirror was fabricated with an ultra-smooth < 0.2 nm RMS surface roughness and an extremely small figure error (< 2 nm peak-to-valley)16,39 and coated with platinum. The diffraction-limited focus size at 10 keV X-ray is 41.3 nm. The angular range where coma aberration can be ignored is 80 µrad39.
The intensity distribution of the X-ray nanoplane and the corresponding focus size were measured using the dark-field knife-edge scanning method40. A platinum wire of 100 µm diameter was the knife edge. The intensity distributions along the beam direction, referred to the focal point, (e.g., Fig. 1b) were measured with 9 keV X-rays and a virtual-source size of 10 µm. The white bars indicating the FWHM (full width at half maximum) revealed a nanoplane thickness < 130 nm over a distance > 175 µm. The best focus size was 65 nm (FWHM).
With a virtual source width of 30 µm rather than 10 µm, the X-ray intensity increased 3 times. Even in this case, the focus size was < 90 nm for the entire X-ray energy range 9–11 keV.
The visible light produced by the X-ray irradiation is imaged by a microscope objective and an sCMOS camera (C14440-20UP, Hamamatsu Photonics K.K.). The camera has a 2304 × 2304 array of 6.5 µm pixels, and is used with microscope objectives with 1×, 20× or 50× magnification. The sCMOS camera has low noise: read out noise is 0.7e and dark noise is 0.5 e/s, thanks to the Peltier cooling.
The visible light microscopy image of Fig. 4d was obtained with a confocal microscopy system (FV-3000, Olympus Co., Tokyo). The image was processed for deconvolution with the CellSens software (Olympus).
The specimen is mounted on a manipulation stage with 5° of motion. The X-ray nanoplane illuminates a fixed position coinciding with the focal plane of the visible objective lens. As mentioned, the 3D MAXWELL imaging is achieved by scanning the specimen in the z-direction with respect to the X-ray nanoplane.
The NaGdF4 core nanoparticles doped with Tb3+ or Eu3+ were synthesized as follows. Gd(CH3CO2)3·xH2O (0.85 mmol) and Tb(CH3CO2)3·xH2O (0.15 mmol) (or Eu(CH3CO2)3·xH2O) were dissolved in a combination of 6 ml of oleic acid and 15 ml of 1-octadecene. The mixture was heated to 130 °C under vacuum for 60 min and then cooled down to room temperature under nitrogen atmosphere. NaOH (5 mmol) and NH4F (4 mmol) were dissolved in 2.5 and 7.5 mL of methanol and stirred overnight at room temperature. The solution was then heated to 110 °C for 30 min under vacuum to remove residual methanol and moisture, purged with nitrogen three times, and finally heated to 300 °C for 120 min under nitrogen atmosphere. Finally, nanoparticles were precipitated with ethanol and washed three cycles with cyclohexane/ethanol (1:2, v/v), and the oleate-capped nanoparticles were dried.
Transmission electron microscopy (TEM, JEOL JEM-1200EX microscope, Japan) was used to analyze the nanoparticle size distribution. A drop of staining solution (2% phosphotungstic acid) was added before the SciNPs solution was deposited on copper grids or carbon films for TEM analysis.
HCT116 cells were grown in McCoy 5A (Modified) medium supplemented by 10% fetal bovine serum. The cells were incubated with 250 μg/ml of NaGdF4:Eu or NaGdF4:Tb SciNPs overnight. The cells were washed with PBS and then trypsinized and seeded with same amount of SciNPs loaded cells on coverslips to achieve 80% confluency. After 24 h incubation the cells were washed with PBS and mounted for imaging.
All procedures involving animals were approved by the Academia Sinica Institutional Animal Care and Utilization Committee (AS IACUC, approval number: 19-07-1326) and all methods were performed in accordance with the relevant guidelines and regulations. All methods are reported in accordance with ARRIVE guidelines (https://arriveguidelines.org) for the reporting of animal experiments. Eight weeks old C57BL/6JNarl mice were purchased from the National Laboratory Animal Center, Taiwan.
A total of 5 × 104 GL261-GFP glioma cells in 2 μl PBS were inoculated via a 33 gauge needle into the basal ganglia of the right brain hemisphere of the mouse. The tumor was left to grow for 14 days before MAXWELL imaging. For the whole brain imaging as Fig. 4d, the mouse brain was treated with the FocusClear (ref. 38) tissue clearing agent for 7 days and removed from the medium before the measurement.
Before perfusion with SciNPs, the mouse was sacrificed by an overdose of Zoletil 50 (Virbac Laboratories, Carros, France, 1 mg/20 g bodyweight) administered by intraperitoneal injection. A catheter (PE-08, BB31695, ID 0.2 mm, OD 0.36 mm, Scientific Commodities, Inc.) was inserted into the right common carotid artery of the mouse and vessel is fixed with silk sutures. The tubing was secured with two knots around the CCA and then perfused with 2 ml heparinized (500 U/mL) NaGdF4:Eu SciNPs (10 mg/mL in 5% glucose buffer). The brain tissue specimens were immersed in 4% paraformaldehyde for 24 h. After fixation, the tissues were washed by PBS three times and sliced to 100 μm thickness by a vibratome (VT1200s, Leica Biosystems, Buffalo Grove, IL, USA). The slices were embedded in EverBrite™ Hardset Mounting Medium with DAPI (Biotium, Fremont, CA, USA).
References
Tamasaku, K. et al. SPring-8 RIKEN beamline III for coherent X-ray optics. Nucl. Instr. Methods A 467–468, 686. https://doi.org/10.1016/S0168-9002(01)00446-6 (2001).
Chin, A. L. et al. A synchrotron X-ray imaging strategy to map large animal brains. Chin. J. Phys. 65, 24–32. https://doi.org/10.1016/j.cjph.2020.01.010 (2020).
Dieguez, E., Arizmendi, L. & Cabrera, J. M. X-ray induced luminescence, photoluminescence and thermoluminescence of Bi4Ge3O12. J. Phys. C Solid State Phys. 18, 4777–4783. https://doi.org/10.1088/0022-3719/18/24/021 (1985).
Jacobsen, C., Lindaas, S., Williams, S. & Zhang, X. Scanning luminescence X-ray microscopy: Imaging fluorescence dyes at suboptical resolution. J. Microsc. 172, 121–129. https://doi.org/10.1111/j.1365-2818.1993.tb03403.x (1993).
Wang, F., Deng, R. & Liu, X. Preparation of core-shell NaGdF4 nanoparticles doped with luminescent lanthanide ions to be used as upconversion-based probes. Nat. Protoc. 9, 1634–1644. https://doi.org/10.1038/nprot.2014.111 (2014).
Kamkaew, A., Chen, F., Zhan, Y., Majewski, R. L. & Cai, W. Scintillating nanoparticles as energy mediators for enhanced photodynamic therapy. ACS Nano 10, 3918–3935. https://doi.org/10.1021/acsnano.6b01401 (2016).
Sudheendra, L. et al. NaGdF4:Eu3+ nanoparticles for enhanced X-ray excited optical imaging. Chem. Mater. 26, 1881–1888. https://doi.org/10.1021/cm404044n (2014).
Karvinen, P. et al. Kinoform diffractive lenses for efficient nano-focusing of hard X-rays. Opt. Express 22, 16676–16685. https://doi.org/10.1364/OE.22.016676 (2014).
Chu, Y. S. et al. Hard-X-ray microscopy with Fresnel zone plates reaches 40 nm Rayleigh resolution. Appl. Phys. Lett. 92, 103119. https://doi.org/10.1063/1.2857476 (2008).
Murray, K. T. et al. Multilayer Laue lenses at high X-ray energies: Performance and applications. Opt. Exp. 27, 7120–7138. https://doi.org/10.1364/OE.27.007120 (2019).
Wu, S. R., Hwu, Y. & Margaritondo, G. Hard-X-ray zone plates: Recent progress. Materials 5, 1752–1773. https://doi.org/10.3390/ma5101752 (2012).
Kang, H. C. et al. Focusing of hard X-rays to 16 nanometers with a multilayer Laue lens. Appl. Phys. Lett. 92, 221114. https://doi.org/10.1063/1.2912503 (2008).
Mimura, H. et al. Breaking the 10 nm barrier in hard-X-ray focusing. Nat. Phys. 122, 25. https://doi.org/10.1038/nphys1457 (2010).
Huang, X. et al. 11 nm hard X-ray focus from a large-aperture multilayer Laue lens. Sci. Rep. 3, 3562. https://doi.org/10.1038/srep03562 (2013).
Yan, H. et al. Multimodal hard X-ray imaging with resolution approaching 10 nm for studies in material science. Nano Futures 2, 011001. https://doi.org/10.1088/2399-1984/aab25d (2018).
Matsuyama, S. et al. 50-nm-resolution full-field X-ray microscope without chromatic aberration using total-reflection imaging mirrors. Sci. Rep. 7, 46358. https://doi.org/10.1038/srep46358 (2017).
Egawa, S. et al. An optical design of twin Wolter mirrors for focusing and imaging experiments with soft X-ray free electron lasers: Advances in X-ray/EUV optics and components XIV. Proc. SPIE 11108, 1110804. https://doi.org/10.1117/12.2529039 (2019).
Hell, S. & Stelzer, E. H. K. Properties of a 4Pi confocal fluorescence microscope. J. Opt. Soc. Am. A9, 2159–2166. https://doi.org/10.1364/JOSAA.9.002159 (1992).
Li, Y. et al. Accurate 4Pi single-molecule localization using an experimental PSF model. Opt. Lett. 45, 3765–3768. https://doi.org/10.1364/OL.397754 (2020).
Huisken, J. et al. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305(5686), 1007–1009. https://doi.org/10.1126/science.1100035 (2004).
Fuchs, E., Jaffe, J. S., Long, R. A. & Azam, F. Thin laser light sheet microscope for microbial oceanography. Opt. Exp. 10, 145. https://doi.org/10.1364/OE.10.000145 (2002).
Chu, L. A. et al. Rapid single-wavelength lightsheet localization microscopy for clarified tissue. Nat. Commun. 10, 4762. https://doi.org/10.1038/s41467-019-12715-3 (2019).
Planchon, T. A. et al. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Methods. 8, 417–423. https://doi.org/10.1038/nmeth.1586 (2011).
Chen, B. C. et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998. https://doi.org/10.1126/science.1257998 (2014).
Lu, C. H. et al. Lightsheet localization microscopy enables fast, large-scale, and three-dimensional super-resolution imaging. Commun. Biol. 2, 177. https://doi.org/10.1038/s42003-019-0403-9 (2019).
Uno, S. N. et al. A spontaneously blinking fluorophore based on intramolecular spirocyclization for live-cell super-resolution imaging. Nat. Chem. 6, 681–689. https://doi.org/10.1038/nchem.2002 (2014).
Velasco, M. G. M. et al. 3D super-resolution deep-tissue imaging in living mice. Optica 8, 442–450. https://doi.org/10.1364/OPTICA.416841 (2021).
Huang, B., Babcock, H. & Zhuang, X. Breaking the diffraction barrier: Super-resolution imaging of cells. Cell 143, 1047–1058. https://doi.org/10.1016/j.cell.2010.12.002 (2010).
An, S. et al. Axial plane single-molecule super-resolution microscopy of whole cells. Biomed. Opt. Exp. 11, 461–479. https://doi.org/10.1364/BOE.377890 (2020).
Ranasinghe, M. et al. X-ray excited luminescence spectroscopy and imaging with NaGdF4:Eu and Tb. Anal. Chem. https://doi.org/10.26434/chemrxiv.13175807.v1 (2020).
Jang, H. S., Woo. K., & Lim, K. Bright dual-mode green emission from selective set of dopant ions in β-Na(Y,Gd)F4:Yb, Er/β-NaGdF4:Ce, Tb core/shell nanocrystals, Opt. Exp. 20, 17107–17118. https://doi.org/10.1364/OE.20.017107 (2012).
Chen, Q. et al. All-inorganic perovskite nanocrystal scintillators. Nature 561, 88–93. https://doi.org/10.1038/s41586-018-0451-1 (2018).
Chiang, A. S. et al. Three-dimensional mapping of brain neuropils in the cockroach, Diploptera punctata. J. Comp. Neurol. 440, 1–11. https://doi.org/10.1002/cne.1365 (2001).
Richardson, D. S. & Lichtman, J. W. Clarifying tissue clearing. Cell 162, 246–257. https://doi.org/10.1016/j.cell.2015.06.067 (2015).
Grant, B. & Donaldson. J. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell. Biol. 10, 597–608. https://doi.org/10.1038/nrm2755 (2009).
Chien, C. C. et al. Gold nanoparticles as high-resolution X-ray imaging contrast agents for the analysis of tumor-related micro-vasculature. J. Nanobiotechnol. 10, 10. https://doi.org/10.1186/1477-3155-10-10 (2012).
Rezende, M. V. S. et al. Sustainable preparation of ixora flower-like shaped luminescent powder by recycling crab shell biowaste. Optik 235, 166636. https://doi.org/10.1016/j.ijleo.2021.166636 (2021).
Liu, Y. C. & Chiang, A. S. High-resolution confocal imaging and three-dimensional rendering. Methods 30, 86–93. https://doi.org/10.1016/S1046-2023(03)00010-0 (2003).
Yamada, J. et al. Hard X-ray nanoprobe scanner. IUCrJ 8. https://doi.org/10.1107/S2052252521007004 (2021) (in press).
Suzuki, Y., Takeuchi, A., Takano, H. & Takenaka, H. Performance test of Fresnel zone plate with 50 nm outermost zone width in hard X-ray region. Jpn. J. Appl. Phys. 44, 1994. https://doi.org/10.1143/JJAP.44.1994 (2005).
Acknowledgements
This work is supported by Ministry of Science and Technology (MoST, Taiwan.107-2119-M-001-047; 109-2123-M-001-001, 108-2923-M-001 -008 -MY2, 107-2923-M-001-012 -MY3); National University of Singapore (R-184-000-289-133, R-184-000-261-101, and R-184-000-310-733 to Chian-Ming Low and Eng Soon Tok; National University of Singapore—National Supercomputing Center (N-184-000-042-001 to SYNAPSE Project); Asian Office of Aerospace Research and Development (AOARD, 19IOA16). We thank SYNAPSE Project for the technical support. YH thanks Fu-Ren Kao brought the z-resolution issue in visible light microscopy to our attention and J-H Lim of PLS-II (Pohang, Republic of Korea) for his help in analyze the data. The use of the BL29XUL beamline at SPring-8 was supported by RIKEN. This work was partially supported by the Brain Research Center under the Higher Education Sprout Project, co-funded by the Ministry of Education and the Ministry of Science and Technology in Taiwan. The BIB beamline of Taiwan Photon Source of National Synchrotron Radiation Research Center (NSRRC) is supported by SYNAPSE project and MoST (Taiwan).
Author information
Authors and Affiliations
Contributions
Y.K., Y.H., G.M., and T.I. conceived the experiment and write the paper. S.M.Y., H.H.C., H.T., Y.H. performed synchrotron experiments and collected data. S.M.Y., H.H.C., H.T., Y.H., Y.K., T.T.L. processed and analyzed the image data. S.M.Y., H.H.C., C.R.C., Y.S.W., C.P., Y.Y.C., D.Y.H., C.L.Y., A.L.C., C.M.L., F.C.K.T., A.T., E.S.T., and A.S.C. prepared the biology samples for measurements. H.T., C.Y.C., K.E.Y., S.J.C., J.Y., S.M. designed, implemented, and tested the x-ray focusing optics and the visible light imaging system. Y.T.C., H.M.H., T.L.S., C.P., P.C., E.S.T., X.X.C., H.M.L., J.B., A.P.S., prepared, tested and characterized the scintillating nanoparticles and the conjugation. Y.K., S.M.Y., H.H.S., H.T., C.Y.C., K.E.Y., G.M. and Y.H. prepared the figures. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Video 1.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kohmura, Y., Yang, SM., Chen, HH. et al. The new X-ray/visible microscopy MAXWELL technique for fast three-dimensional nanoimaging with isotropic resolution. Sci Rep 12, 9668 (2022). https://doi.org/10.1038/s41598-022-13377-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-13377-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.