Abstract
Little is known about glandular proteins secreted from the skin- and blood-feeding ectoparasite salmon louse (Lepeophtheirus salmonis). The labial gland has ducts extending into the oral cavity of the lice, and the present study aimed to identify novel genes expressed by this gland type and to investigate their role in modulation of host parameters at the lice feeding site. Five genes associated with labial gland function were identified and named Lepeophteirus salmonis labial gland protein (LsLGP) 1–4 and 1 like (LsLGP1L). All LsLGPs were predicted to be small charged secreted proteins not encoding any known protein domains. Functional studies revealed that LsLGP1 and/or LsLGP1L regulated the expression of other labial gland genes. Immune dampening functions were indicated for LsLGP2 and 3. Whereas LsLGP2 was expressed throughout the parasitic life cycle and found to dampen inflammatory cytokines, LsLGP3 displayed an increased expression in mobile stages and appeared to dampen adaptive immune responses. Expression of LsLGP4 coincided with moulting to the mobile pre-adult I stage where hematophagous feeding is initiated, and synthetic LsLGP4 decreased the clotting time of Atlantic salmon plasma. Results from the present study confirm that the salmon louse secretes immune modulating and anti-coagulative proteins with a potential application in new immune based anti-salmon louse treatments.
Similar content being viewed by others
Introduction
Salmon louse (Lepeophtheirus salmonis) is a blood-feeding ectoparasitic copepod infecting salmonid fish species of the northern hemisphere. Salmonid aquaculture is based on rearing of high-density populations, and thus in many areas there has been a substantial increase in susceptible salmon louse hosts. The salmon louse population has therefore increased with a consequential negative impact on both farmed and wild salmonids in these areas1,2,3,4. This negative impact is a result of lice feeding on the host skin and blood, introducing erosions and light ulcers that can disturb the osmotic balance and stress the fish, eventually increasing the susceptibility to other pathogens5,6,7,8,9. High parasitic loads can be detrimental to wild post smolts migrating out the fjords10,11,12, hence farmers must keep the lice levels low to reduce infection pressure on wild salmonid populations. With the emergence of a widespread resistance towards anti-lice chemotherapeutants the use of cleaner fish has increased and new methods such as mechanical- and thermal delousing systems, have replaced medical treatments13,14. However, ethical aspects on the use of cleaner fish, and increased treatment mortality associated with the new mechanical methods have increased the need for novel control measures such as vaccines, functional feeds, or marker-assisted selection in fish breeding. Detailed knowledge of salmon louse biology and particularly the host-parasite interaction is fundamental to address this.
The life cycle of the salmon louse consists of eight developmental stages separated by moults15,16. The initial two nauplii stages are planktonic and lecithotrophic, whereas in the next copepodid stage the louse can detect a nearby swimming host and attach to its epidermis. Here, the louse passes through the parasitic phase of the copepodid stage, two attached chalimus stages and two free living pre-adult stages before it finally moults to the adult stage. During these parasitic stages, the salmon louse modulates the host immune response to avoid clearance, and when blood feeding is initiated at the mobile stages17, anti-coagulation factors are expected to be secreted. This is postulated from knowledge of other well-studied host-parasite interactions such as the ixodid tick, that also feeds on host blood for a prolonged time. Tick saliva has been found to inhibit blood clotting as well as inflammation, dendritic cell (DC) maturation and antigen presentation, T-cell proliferation and activation, neutrophil recruitment, antibody function and complement activation18. Similarly, there is some evidence indicating that the salmon louse specifically down modulates the salmonid host immune system. A significant suppression of respiratory-burst and phagocytic activity of head kidney macrophages and an increased susceptibility to viral infections have been observed in salmon louse infested Atlantic salmon (Salmo salar)9,19,20,21. In contrast to the more resistant pink (Oncorhynchus gorbuscha) and coho (O. kisutch) salmon, only a mild inflammation without a well-developed hyperplasia is seen at the juvenile lice attachment site in susceptible species such as Atlantic salmon and rainbow trout (O. mykiss)5,7,22,23,24. Even where the frontal filament penetrates the epidermis, inflammation is scarce as long as the lice are present5,25, indicating that the lice efficiently dampen immune responses in susceptible species.
The salmon louse exocrine glands are hypothesized to be important in the host-parasite interaction through the secretion of modulatory substances, as some gland types have secretory ducts ending at the host-parasite interface26,27. The labial gland, in particular, seems to be important, as it first appears at the infective copepodid stage and has secretory ducts ending distally in the oral cavity of the louse near the mandible teeth26. Labial gland products are thus likely to be deposited directly onto the lice feeding site. Nevertheless, little is known about the secretory products of the salmon louse labial glands. As parasitism has developed independently multiple times in arthropods28, modulatory proteins are not necessarily expected to be homologous even if they target similar host pathways due to convergent evolution. In the present study, we therefore aimed to identify and characterize hitherto unknown proteins expressed by the salmon louse labial gland. RNA sequencing (RNAseq) was carried out on salmon louse tissue samples with a high labial gland content, and five genes encoding proteins without predicted domains were selected and confirmed to be expressed by the labial glands by in situ hybridization. These unknown genes were characterized, and functional studies were conducted to analyse their putative modulatory effects on the Atlantic salmon immune and clotting response by in vivo gene knock down studies and by in vitro studies applying recombinant or synthetic proteins.
Results
Identification of labial gland gene candidates and localization of expression
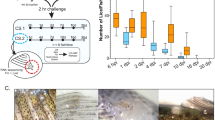
From the RNAseq data, four genes with unknown function were predicted to be expressed in the labial gland of adult lice and thus further evaluated. Transcripts for all four genes, EMLSAT00000009293, EMLSAT00000006642, EMLSAT00000005273 and EMLSAT00000000504 were localized to the labial gland by in situ hybridization, hereafter called L. salmonis labial gland protein (LsLGP) 1–4, respectively. Each L. salmonis labial gland consists of two morphologically similar secretory units that empty their content into individual reservoirs that further empty through a joint duct26. The LsLGP transcripts were only localized to the labial gland (Fig. 1), mainly to the secretory unit most distal to the duct (Fig. 1d–g). LsLGP3 was, however, also expressed by the so-called reservoir unit most proximal to the duct (Fig. 1f). The reservoir unit may thus be more involved in secrete production than first anticipated.
Localization of L. salmonis labial gland protein (LsLGP) genes by in situ analysis of a pre-adult II female salmon louse. Dark coloring indicates presence of transcripts. (a) Overview of the specimen (HE-staining) and (b) overview of the specimen hybridized with antisense LsLGP2 probe, where stars indicate tegmental type 1 glands and arrowheads indicate labial glands. Br Primitive brain, Oe Oesophagus, SE sub-epidermal tissue. A higher magnification of the labial glands treated with (c) HE, (d) antisense LsLGP1 probe, (e) antisense LsLGP2 probe, (f) antisense LsLGP3 probe (g) antisense LsLGP4 probe, (h) sense LsLGP1 probe, (i) sense LsLGP2 probe, (j) sense LsLGP3 probe, and (k) sense LsLGP4 probe.
Characterization of deduced protein sequences
To further characterize the LsLGP genes, RACE was performed for all genes and amplified products were sequenced to ensure that the transcript sequences covered the complete open reading frame (ORF) as predicted from the salmon louse genome.
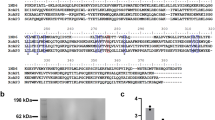
After RACE, all the LsLGP genes were found to be short proteins, with sequence lengths predicted between 101 and 195 amino acids (Fig. 2). BLAST searches did not reveal orthologous genes for any of the LsLGP genes, and no paralogous genes were identified within the salmon louse genome. Nevertheless, primers designed for LsLGP1 also amplified a gene highly similar (83%) to LsLGP1, named L. salmonis Labial Gland Protein 1 like (LsLGP1L). LsLGP1L was localized to the reverse strand around 21,000 bp up-streams of LsLGP1 in the salmon louse genome29.
Overview of the predicted L. salmonis labial gland protein (LsLGP) 1–4. Upper panel (table): Both the length of the complete ORF and the length of mature sequences (in parentheses) is given, while the putative mass in kDa is given for the mature proteins only. Aa amino acids, pI theoretical isoelectric point, #C number of cysteines, PRR proline rich region. Lower panel: Predicted leader sequences are indicated with a grey background, while predicted N-linked glycosylation site are indicated with a blue background. Positively and negatively charged amino acids are shown in blue and red, respectively, while cysteines are depicted in green. PRR’s are underlined, while the two amino acid deletions identified in LsLGP4 is marked with squares. LsLGP1 and 1L are aligned to show identical (*) and similar (: or.) amino acids.
Scanning the InterPro database with the putative LsLGP protein sequences revealed no homology to any known protein domains, though all proteins were predicted to contain a leader sequence (Fig. 2). Moreover, all sequences were found to have a high content of charged amino acids (aa’s), where LsLGP1 and LsLGP1L were predicted to be acidic, and LsLGP2–4 predicted to be basic proteins (Fig. 2). In addition to the many negatively charged residues encoded by LsLGP1L and LsLGP1, both proteins were found to contain a high percentage of asparagine and the small hydrophobic residues leucine and valine. Moreover, one N-linked glycosylation site was predicted in both sequences. LsLGP2 was found to contain six cysteines N-terminally possibly forming disulphide bridges, followed by a rather long proline rich region (PRR), APPP(AP)20, and a short C-terminus with mainly positively charged amino acids (KGGRRRRDVPIVFDH). LsLGP3 is the largest of the LsLGP proteins, containing no cysteines or other identifiable patterns, though with an overweight of charged amino acids scattered throughout the protein sequence. In LsLGP4, the smallest of the sequenced LsLGPs, a PRR was also found, R(RP)3. A total number of 28 clones were sequenced, and of these were four found to have a 6 bp deletion in the PRR region, shortening it to two RP repeats (R(RP)2). Another 6 bp deletion was seen in seven other clones in connection to an exon–intron boundary excluding a glycine and an isoleucine. As no paralogue LsLGP4 genes were identified in the salmon louse genome, these transcripts may be considered as allelic variants.
Ontogenetic expression
Generally, the labial gland genes were found to be marginally expressed in eggs and planktonic stages (Fig. 3), though significantly increased transcript levels of LsLGP1L, 1, and 2 were observed in planktonic copepodids (Fig. 3a–c). The levels of these three transcripts further increased rapidly after attachment to the host, with the highest expression detected in 4 dpi copepodids when compared to all other analysed timepoints. During the chalimus II and pre-adult stages, the levels of LsLGP1L, 1, and 2 were significantly lower than during the initial establishing phase of the copepodid, but still significantly higher than in the planktonic nauplius stages. A significant increase was again detected in adult lice, either for both females and males (LsLGP1L and 2) or males only (LsLGP1). A steady increase of LsLGP3 was found during the parasitic stages, with the highest mRNA level detected in adult lice, both females and males (Fig. 3d). LsLGP4 was found to be the last transcript to increase its expression above that of planktonic stages, with a significant increase detected first when the lice had moulted into the mobile pre-adult I stage (Fig. 3e). The LsLGP4 transcript level was also found to be at its highest in adult lice, with the highest expression detected in males, likely to reflect the higher labial gland to body size ratio in males. In fact, for all LsLGPs besides LsLGP1L, a higher average transcript level was observed in adult males compared to adult females.
Level of L. salmonis labial gland protein (LsLGP) transcripts during development and starvation related to the LsEF1α and LsADT3 reference genes (N = 5). (a–e) Developmental expression in fertilized egg sacs (egg), nauplius (nau) I and II, planktonic copepodids (c free) and copepodids 2 days (c 2d) and 4 days (c 4d) post infestation, chalimus (chal) I and II, pre-adult (pad) I and II and adult lice (ad). For female adult lice both young (Y) and mature (M) lice were analyzed. mRNA level was related to that of fertilized egg sacs. Statistically significant differences are denoted with different letters (p ≥ 0.05). (f–j) Expression in adult females at 0, 24 and 48 h (h) after removal from salmon. mRNA levels were related to that of the lice taken directly off the host (0 h).
Expression in starved lice
As a rapid down regulation of some intestinal transcripts involved in blood-feeding has been detected in starved adult lice30,31, we analyzed the expression of the labial gland genes during starvation. The analysis was performed to explore whether the gene products may be involved in digestive processes rather than, or in addition to, host modulation. None of the genes were, however, found to be modulated when lice were incubated off the host for 48 h (Fig. 3f–j).
Functional studies
LsLGP1/1L KD copepodids showed decreased levels of all known labial gland genes
To investigate whether the LsLGP proteins manage to dampen the Atlantic salmon immune response in vivo, double stranded RNA (dsRNA) induced KD of LsLGP transcripts was performed in lice followed by subsequent in vivo challenge studies. LsLGP1 and LsLGP1L are highly similar genes, hence, dsLGP1 targeted both genes simultaneously. A KD efficiency of 97.7 and 95.7 were detected for LsLGP1 and LsLGP1L, respectively (Fig. 4a). Interestingly, a significant decreased transcript level of LsLGP2 was also detected in LsLGP1/1L KD lice, with an efficiency of 60%. We therefore analyzed the expression of four additional genes confirmed to be expressed in the lice labial gland (unpublished data), to further explore the regulative effect of LsLGP1/1L. These transcripts were also found to be significantly decreased in KD animals with KD efficiencies between 49 and 62% (Fig. 4a). Therefore, it was not possible to analyze the specific modulatory role of LsLGP1 and 1L by this approach, as any differences detected could be attributed to any of the other labial gland genes.
Expression of L. salmonis labial gland protein (LsLGP) genes after dsRNA treatment. (a) Expression of LsLGP genes in dsLsLGP1/1L treated copepodids 3 days post infestation related to the LsEF1α and LsADT3 reference genes (N = 8). In addition to KD of the targeted genes, KD of LsLGP2 and four additional, yet unpublished, genes confirmed to be expressed in the lice labial gland called L. salmonis labial gland astacin (LsLGA) 2–4 and L. salmonis labial gland chymotrypsin 1 (LsLGCT1), were analyzed. (b) Expression of LsLGP2 in dsLGP2 treated copepodids 3 days post infestation related to the LsEF1α and LsADT3 reference genes (N = 8). Statistical significance was teste with Student’s t-test (p ≥ 0.05) between control and dsRNA treated group and is denoted with an asterisk.
Infestations with LsLGP2 KD lice induced an increase in immune gene transcripts
LsLGP2 was the only labial gland transcript shown to be downmodulated by dsLGP2. The first attempt for LsLGP2 KD applying normal concentration of dsLGP232 did, however, only result in a transient suboptimal knock down (Fig. 4b). This was also seen when the concentration of dsLGP2 was slightly increased (15 ng/μl), and a doubling of the concentration was required to obtain KD efficiencies above 90%. But, as KD was transient, the immune response against KD lice were only evaluated early when the LsLGP2 expression was at its highest and KD still sufficient. Expression of immune genes, selected to indicate the involvement of LsLGP2 in inflammation, complement activation and activation of innate and adaptive immune cells, was analyzed in LsLGP2 KD and control lice infested fish. Immune gene transcripts were analyzed from skin both directly underneath lice (infested site) and in nearby unaffected skin sites at three days post infestation with copepodids. Generally, when comparing immune gene expression in fish infested with control lice to that infested with KD lice, a higher average expression in KD infested fish were seen for all genes analyzed (Fig. 5). The mRNA level of IL1β, IL6, IL8, MMP13, NCCRP1, and IL4/13a were significantly higher in skin directly underneath the lice in both control and KD infested fish, while the TNFα mRNA level were only significantly elevated to that of unaffected sites in KD infested fish. Moreover, a significant higher (p ≥ 0.5) expression was detected for the key pro-inflammatory cytokines IL1β, IL6, and IL8, the anti-inflammatory cytokine IL10, and the T helper 2 related cytokine IL4/13a in KD infested fish when compared to skin underneath control lice at the site of infestation. While the T-cell response did not seem to be significantly affected by the LsLGP2 KD, the opposite was indicated for B-cell responses. All immunoglobulin transcript levels were elevated at both unaffected and infested sites in KD infested fish, with a significant increase in KD infested fish seen at both unaffected an infested skin for IgT and at unaffected skin only for IgD. At the time of sampling, no lice had as expected molted into the chalimus I stage, and similar lice numbers were seen between the control (9.2 copepodids/fish ± 3.9) and KD infested groups (10.8 copepodids/fish ± 2.8).
Relative transcript level (2−∆∆Ct ± SD) of selected Atlantic salmon immune gene transcripts in response to control (Ctr) and LsLGP2 knock-down (KD) copepodids 3 days post infestation. The expression in unaffected and affected sites on infested fish (N = 8) were related to elongation factor 1 alpha (elf1α) and tripartite motif (trim) genes (∆Ct) and to non-infested control fish (∆∆Ct). *Indicates significant differences (p ≥ 0.05) when compared to skin from non-infested fish. #Indicates significant differences (p ≥ 0.05) between unaffected and affected skin of infested fish. Significant difference (p ≥ 0.05) between skin sites infested with ctr and KD lice are also denoted with an asterisk (*).
Recombinant LsLGP3 decreased the expression of B- and T-cell markers in primary head kidney leucocytes
As LsLGP3 and 4 were only expressed in mobile lice stages, adult lice were used for KD studies of these two genes. No differences in the immune response between skin sites infested with KD and control animals were detected in in vivo experiments. This may be due to the mobility of the lice at this lice stage and thus difficulties obtaining samples where mobile lice have been feeding for a prolonged period. Consequently, as an alternative, recombinant LsLGP3 (recLGP3) and synthetic LsLGP4 (synLGP4) was administered to primary head kidney leucocytes in vitro to analyze their immune modulative potential.
As around 0.3 μg/ml LPS was co-purified together with recLGP3, LPS was also included in the assay both as control and in combination with recLGP3. Analysis of selected Atlantic salmon immune genes in recLGP3 stimulated leucocytes revealed a significant up-regulation of IL8 and IL4/13a, and down modulation of IFNγ, CD8α, IgD and IgT (Fig. 6). This down-modulation was not seen in response to LPS alone. No regulation was detected in synLGP4 stimulated leucocytes (Supplementary Fig. S1).
Relative transcript level (2−∆∆Ct ± SD) of selected Atlantic salmon immune gene transcripts in head kidney leucocytes after a 4-h stimulation with recombinant LsLGP3 (recLGP3) (N = 3). Three quantities of recLGP3 were applied, 150, 112.5 and 75 μg/well. The immune gene expression in control (ctr), LPS, and recLGP3 treated leucocytes were related to elongation factor 1 alpha (elf1α) (∆Ct), using the expression in control cells as calibrator (∆∆Ct). Significant difference (p ≥ 0.05) between ctr and recLGP3 treated leucocytes are denoted with an asterisk (*).
Synthetic LsLGP4 inhibit blood coagulation
As both LsLGP3 and 4 were found to be highly expressed in the mobile stages, their possible inhibition of blood clotting and hence importance in blood feeding were analysed. The prothrombin time test was used to measure blood coagulation, where the clotting time was determined by plasma gel formation in recLGP3/synLGP4 treated plasma compared to the control. SynLGP4 treated plasma demonstrated a significant increase in clotting time compared to the relatively short clotting time in control and recLGP3 treated plasma (Fig. 7a). We therefore conducted a KD experiment injecting pre-adult II female lice and allowing them to develop into adult egg producing lice on fish, to analyse if a reduced LsLGP4 level would affect blood feeding or fecundity at 32 dpi (Fig. 7b–e). An LsLGP4 knock down of 98% did, however, not alter the number of animals with blood in gut. Knock down did also not seem to inhibit anti-coagulation of blood within the gut, as peristaltic movement with liquid blood could be seen to the same degree as in control lice. Moreover, KD did not affect the size of the lice body, though, a small but still significant decrease in the egg string length were detected in the dsLGP4 treated lice.
Analysis of LsLGP3 and 4’s role in blood coagulation. (a) Coagulation efficiency in control (CTR), recLGP3 (75 μg) and synLGP4 (50 and 75 μg) treated plasma measured as time to plasma gel formation after thromboplastin addition (minutes ± SD; N = 3). Some synLGP4 treated samples did not coagulate during the experiment and was set to 45 min. (b–e) Knock down (KD) study of LsLGP4 during the mobile salmon louse stages to analyse the importance of LsLGP4 in blood feeding. (b) Relative transcript level (2−∆∆Ct ± SD) of LsLGP4 in control (dsCTR) and dsLGP4 treated lice at 32 dpi, related to the LsEF1α and LsADT3 reference genes (N = 5). (c) Average number of recovered lice per fish (± SD) in fish (N = 3) infested with 10 control or dsLGP4 treated lice/fish. (d) Percentage of lice with visible blood in intestine at sampling in control and dsLGP4 treated lice (dsCTR N = 16, dsLGP4 N = 18). (e) The body length (BL) and egg string length (EL) of control and dsLGP4 treated lice (mm ± SD, dsCTR N = 16, dsLGP4 N = 18). Significant difference between control and treatment group are denoted with one asterisk (*) if p ≥ 0.05, two asterisk if p ≥ 0.01 and three asterisk if p ≥ 0.001.
Discussion
Blood feeding parasites are expected to secrete factors with immune modulative and anti-coagulative functions, but the presence of such factors have not been confirmed in glandular secretions of the salmon louse. The present study report genes expressed by the labial gland, a salmon louse gland type that is likely to deposit its content directly onto the lice feeding site. These LsLGP genes were not found to encode any known protein domains that could help elucidate their function. Signal peptides were, however, identified, indicating that the LsLGPs are secreted. All five labial gland proteins were found to be relatively short, with several charged residues that may act in ligand binding.
The highly similar LsLGP1 and LsLGP1L were both predicted to be acidic proteins at physiological conditions, with a potential role in binding positively charged ligands. As LsLGP1L expression was elevated already in planktonic copepodids, it is likely that LsLPG1L is necessary shortly after the copepodids settles on a suitable host. At this stage, the salmon lice attachment and feeding activity inflict tissue damage to the host skin with a low induction of inflammation in susceptible salmonids5,6,7,22,25. In higher vertebrates, extracellular calcium represents one of the earliest signals after tissue damage, functioning as an immune cell attractant and supporting the wound healing response33,34. To interfere with this part of the host response, ticks secrete a calreticulin-like protein that also has regions dominated by acidic residues representing putative calcium-binding regions35. As Ca2+ tend to be bound by oxygen atoms36, it is tempting to speculate that the many negatively charged oxygen atoms within the multiple Asp and Glu side chains of LsLGP1/1L may bind calcium ions. Extracellular calcium is also important for keratinocyte differentiation and proliferation, where a low concentration induces proliferation and higher concentrations induces differentiation in higher vertebrates37. To regenerate the epidermis, wound healing includes both these processes, where calcium seems to be necessary for normal regenerative skin responses38. In the resistant coho salmon, salmon louse induces epidermal hyperplasia that buries and kills the lice23, while in the susceptible rainbow trout a milder hyperplastic response is induced underneath mobile lice7. Thus, it may be vital for the salmon louse to control the concentration of extracellular calcium to regulate epidermal epithelial cell proliferation. Wound healing is in general a conserved process39,40, but little is known about piscine calcium-sensing receptors, and how extracellular calcium affects the fish skin during wound healing and inflammatory responses. Further studies confirming the ligand of LsLGP1/1L is needed, and additional positively charged molecules should also be evaluated. As the KD studies indicated that LsLGP1 and/or 1L are likely key regulators for labial gland secretion, further structural and functional characterization of LsLGP1/1L might provide interesting targets for lice control. Though, if LsLGP1/1L is glycosylated as predicted, eucaryotic expression systems would be necessary to produce functional proteins.
LsLGP2-4 were, in contrast to LsLGP1 and 1L, predicted to be basic proteins at physiological conditions. Additionally, the putative protein sequence of LsLGP2 contains a rather long PRR. As the cyclic five-membered ring of proline is constraining the rotation of its backbone41, this long PRR may cause LsLGP2 to be a conformationally loose protein. Proline is a good hydrogen-bond acceptor due to a more accessible and electron-rich carbonyl group than those of other natural amino acids42,43. Therefore, (XP)n PRRs are believed to function as rigid and sticky arms that can bind rapidly and reversibly to other proteins41, and thus the long PRR of LsLGP2 may work together with the positively charged C-terminus in ligand binding. LsLGP2 KD lice, presumably secreting a lower concentration of LsLGP2, induced a significant increase of the pro-inflammatory salmon transcripts IL1β, IL6, IL8 and TNFα compared to control lice. This indicates that LsLGP2 inhibits the action of an immune component or cell directly, or indirectly by binding something that induces inflammation. Moreover, LsLGP2 KD lice induced a significantly higher local expression level of IL4/13a, and an increased average expression of IgM, IgT and IgD was seen at both unaffected and affected skin mainly underneath KD lice. In zebrafish, recombinant IL4/13A promotes Th2-type immune responses by increasing B-cell proliferation and antibody production44, while recombinant rainbow trout IL4/13A augments B-cell IgM secretion45. It is, however, not known how teleost IL4/13A influences IgT and IgD expression, although zebrafish IL4 increases the number of IgZ-2 positive (IgZ-2+) B-cells46. While these cells are IgM+IgZ-2+ cells, rainbow trout IgT+ cells do not express IgM or IgD on their surface46,47. Moreover, rainbow trout have three B-cell types expressing IgM and/or IgD; one double positive line expressing both IgM and IgD and two single positive IgM+ or IgD+ B-cell types48,49. IgD single positive B cells have, as well as IgT+ B-cells, been suggested to be important in mucosal immunity, and IgT has been identified as a limiting factor for protozoan parasites infecting mucosal surfaces47,48,49,50. Further study is needed to resolve if the reported increase is of biological relevance caused by an increased expression level of resident B-cells or increased influx of IgT+ and IgD+ B-cells.
LsLGP3 also seems to modulate the expression of IgD and IgT, as recLGP3 induced a down modulation of these two transcripts in addition to CD8α and IFNγ in head kidney leucocytes. A significant reduction of adaptive immune responses has also been indicated in vivo when fish are infested with mobile lice. A down modulation of adaptive immune transcripts in both rainbow trout and Atlantic salmon infested with pre-adult and/or adult lice, and an increased susceptibility to infectious salmon anaemia virus in Atlantic salmon infested with mobile lice have been observed7,9,51. Interestingly, the expression of LsLGP3 reaches its highest level in pre-adult and particularly the adult stages. At the adult stage, the lice increase in size, particularly the females that are typically 10–12 mm compared to the 6–7 mm males. Even though adult lice are mobile, the females often gather in groups where they reside at specific locations on the fish typically behind the adipose and anal fins52,53. Moreover, the feeding activity of the lice intensify, and it is now breaking through the basement membrane to reach the dermal blood supply inducing an increase in the influx of immune cells to the site of infestation6,7. This relatively more intense feeding activity may increase the need to dampen host responses, and the present study indicate that salmon louse labial gland proteins have the ability for such immune suppression. However, LPS contamination represents a possible source of error when studying the immune response towards bacterial derived recombinant proteins, as LPS is a potent immune stimulator54. And as a low concentration of LPS was detected in the recLGP3 solution, this could potentially have affected the results in the present study. LPS removal from recombinant proteins requires a rather harsh treatment of the protein with a high potential of altering its function. Therefore, instead of removing LPS from the recLGP3 solution, LPS was included as a control when stimulating the leucocytes, where similar concentration of LPS as what was detected in the recLGP3 solution did not induce any significant regulation of the selected immune genes. This indicates that the co-purified LPS did not induce the down streams effect seen in the recLGP3 treated leucocytes, nevertheless, one should keep this possible source of error in mind when concluding on the present results.
Besides immune responses, coagulation is another important host pathway to suppress for blood feeding parasites. As LsLGP4 is not expressed before the lice reaches the stage where blood-feeding commonly is initiated, an anti-coagulant role for LsLGP4 could be suggested. This was further supported in an in vitro coagulation test, where a highly significant delay of coagulation was observed in response to synLGP4. The LsLGP4 knock down studies suggested that this protein did not affect the viscosity of the blood within the lice gut, nor the development and growth of the lice. This could indicate that functional amounts of LsLGP4 was not ingested together with the blood during feeding. However, additional gland- or gut derived anti-coagulants are also likely to be secreted by the lice as seen in other blood-feeding parasites such as ticks18, and could have masked in vivo effects of LsLGP4 KD. The secretory ducts of the labial glands are found distally in the mouth tube near the mandible teeth26, and it is thus likely that the labial gland secrete has its main effect directly at the lice feeding site rather than within the lice gut. This is further supported by the constitutive expression level of the LsLGPs observed in starved lice, in contrast to the decrease typically seen for genes involved in digestion30,31. The minor yet significant reduction in egg string length of dsLGP4-treated animals implies, however, that even though LsLGP4 is not essential for a normal feeding activity, it affects the nutritional status of the lice to some degree. Potentially, LsLGP4 KD could complicate the initiation of a blood-meal intake if LsLGP4 is involved in decreasing blood-clot formation at the site of feeding.
Previous studies of salmon louse secretions have focused on secretory/excretory products (SEPs) extracted both from dopamine treated and untreated salmon louse55,56,57. Many components such as astacins, serine proteases, protease inhibitors, calreticulin, and prostaglandin E2 (PGE2) have been identified in these products, and functional studies have revealed an immune dampening and anti-chemotactic effect of SEPs56,58. However, many of the proteins identified in SEPs are proteins expressed by tegumental glands secreting the mucus covering the lice, intracellular proteins or proteins involved in nutritional uptake or digestion, while the LsLGPs identified in the present study could not be identified in the SEP data sets26,29,55,56,57. Probably, the LsLGP’s concentration is low compared to gut and tegumental gland derived proteins, or the release of labial gland secretion is either not constant or not regulated by dopamine but requires host factors. Therefore, this indicate that using salmon louse SEPs in in vitro studies of immune modulation is likely to introduce experimental errors, as digestion enzymes have a low specificity and will non-specifically degrade host immune cells and immune active factors. It is also unlikely that such enzymes are deposited on the host skin during infestation given the low severity of lice induced lesions6,7,23,25. Furthermore, it has not been possible to experimentally confirm that lice derived PGE2 is in fact taking part in the host-parasite interaction59,60, emphasizing that more studies like this, directly addressing the regulation and mode of action of glandular proteins, are needed to analyse the interaction taking place at the site of infestation. New knowledge on this topic may represent a promising fundament for the development of new immune based anti-salmon louse treatments. The present study has identified salmon louse immune modulatory and anti-coagulative proteins and represents an important initial step towards such developments.
Materials and methods
Ethical statement
All experiments were carried out in strict accordance with relevant guidelines and regulations. The study was carried out with approval granted from the Ethic committee of Norwegian Food Safety Authority (approval numbers 8589) following ARRIVE guidelines.
Source of L. salmonis
A laboratory strain of L. salmonis salmonis (LsGulen) was maintained on farmed Atlantic salmon according to Hamre et al.61. Salmon were hand fed on a commercial diet (Skretting Nutra Olympic 4.0 mm) and reared in sea water with a salinity of 34.5 ppt and a temperature of 9 ± 0.5 °C if not otherwise mentioned. Eggs, nauplii, copepodids and adult lice maintained off the host were kept in incubators with continuous flow-through of seawater from the same supply as the fish tanks61.
All salmon louse stages were collected for ontogenetic analysis in pentadruplicate samples (biological replicates). For the earliest life stages, multiple animals were pooled to yield sufficient total RNA for further analysis as follows: Eggs: 1 eggsac (string) containing approximately 200 developing embryos. Nauplius I, nauplius II and copepodids (free-living): approximately 100 larvae. Copepodids 2 days post infestation (dpi) and 4 dpi: 60 larvae. Chalimus I: 30 animals, chalimus II: 20 animals, female and male pre-adult I and II, and adult stages: single animals. Additionally, both young and mature adult females were collected. Young females were sampled prior to enlargement of the genital segment and the extrusion of the first egg string, while mature females were sampled when bearing external egg strings.
To analyse the expression in starved lice, 15 adult female lice were collected from Atlantic salmon kept at 10 °C. Five lice were analysed from each time point (biological replicates), sampled directly from the host, or after being kept for 24- and 48-h off-host in running seawater.
Total RNA purification and cDNA synthesis
All tissue/lice samples for RNA isolation were collected in RNA later (LifeTechnologies), kept overnight at 4 ○C and further stored at − 20 ○C. Total RNA was isolated with a combined Tri reagent (Sigma Aldrich) and RNeasy (Qiagen) method, as previously described62, with DNase treatment performed on column. Total RNA from adult female lice were purified with Tri reagent combined with the RNeasy mini kit (Qiagen), while all other samples were purified with Tri reagent in combination with the RNeasy micro kit (Qiagen). Extracted RNA was either kept at − 80 °C until use, or cDNA synthesis was performed directly.
For PCR, cDNA synthesis was carried out using the qScript cDNA SuperMix (Quanta Bioscience), applying 1 µg lice total RNA. For real time RT-PCR, the AffinityScript qPCR cDNA Synthesis Kit (Stratagene) was used according to supplier’s recommendations in a 10 µl reaction, using 200 ng lice total RNA, 1000 ng salmon skin total RNA or 475 ng salmon leucocyte total RNA. Samples were diluted 10 or 5 times for lice and salmon samples, respectively, before storage at − 20 °C.
RNA sequencing
The region holding the labial gland was dissected out from 20 adult female lice after an over night incubation at 4 °C in RNA later (LifeTechnologies). Total RNA was purified as described in “Source of L. salmonis” section. Further library preparation, RNAseq at the Norwegian sequencing Center (Oslo) and data processing were performed as previously described by Eichner et al.63. To identify potential labial gland genes, the labial gland expression was compared to RNAseq data obtained for the different developmental stages of the lice, in addition to the gut, gonad and thoracic feet 1 and 2 expression29. If expression was high in the labial gland sample, and low or not detected in planktonic stages as well as gonads and gut, the expression were further compared to the expression in the thoracic feet where all types of tegumental glands are found26. As tegumental type 1 and 2 glands are also present in the labial gland sample, subtracting genes expressed in thoracic feet was expected to facilitate identification of unique genes expressed solely in the labial glands.
In situ hybridization
In situ hybridization of the candidate genes identified in the RNAseq data was done to confirm that they were expressed by the labial gland and not in surrounding tissues. Predicted sequences from the candidate genes were retrieved at ENSEMBLE Metazoa (https://metazoa.ensembl.org/Lepeophtheirus_salmonis/Info/Index), and gene specific primers were designed based on these sequences and BLASTED in the salmon louse genome to ensure specificity. Single stranded digoxigenin (DIG) labelled antisense and sense RNA probes of the selected genes were prepared by in vitro transcription using the DIG RNA Labelling Kit (Roche), with purified PCR products that included T7 promoters (TAATACGACTCACTATAGGGAGA) as templates (Primers listed in Table 1). PCR was performed as previously described using Q5® High-Fidelity DNA Polymerase, and the products were purified with the GenElute (™) PCR Clean-Up Kit (Sigma-Aldrich).
A pre-adult II female L. salmonis were fixed in phosphate-buffered 4% paraformaldehyde (pH 7.4) for 24 h at 4 °C. Subsequently, specimens were processed with the Histokinette 2000 (Reichert-Jung) where they were washed in PBS, dehydrated through a graded ethanol series, and embedded in paraffin wax. Horizontal sections, 4.0 μm thick, were cut with a Leica RM 225 microtome (Leica Microsystems). In situ hybridization was performed according to Dalvin et al.64, with some modifications as described earlier65. Additionally, the proteinase K treatment was prolonged to 18 min. Antisense and sense probe were applied to adjacent sections at each round of hybridization, to control for unspecific hybridization. Adjacent sections were also stained with hematoxylin (Shandon Instant Hematoxylin, Thermo Scientific) for 2.5 min and 1.5 min with 1% erythrosine (Certistain, Merck). HE stained sections were mounted in Histomount (Invitrogen).
Cloning and sequencing
Once genes were confirmed to be expressed in the labial glands, primers for rapid amplification of cDNA ends (RACE) were designed from the predicted sequences (Table 1). RACE (5ʹ and 3ʹ) was performed for all genes using the SMARTer™ RACE cDNA amplification kit according to suppliers’ instructions (Clontech). RACE PCR products were cloned using the TOPO TA Cloning® Kit for sequencing with One Shot™ TOP10 Chemically Competent E. coli (Thermo Fisher). Colonies were used as templates for PCR reaction using the Q5® High-Fidelity DNA Polymerase kit according to suppliers’ recommendation (BioLabs), applying M13 forward and reverse primers (Cycles were: initial denaturation 98 °C for 30 s, 30 cycles of 98 °C for 10 s, 55 °C for 30 s and 72 °C for 30 s/kb, and a final extension for 2 min at 72 °C). PCR products were further purified by ExoSAP-it (Affymetrix) and sequenced using the BigDye Terminator v3.1 Cycle Sequencing kit from Applied Biosystems. Sequencing was completed on an ABI prism 7700 automated sequencing apparatus at the University of Bergen sequencing facility. The open reading frame (ORF) was further confirmed by one directly sequenced PCR as described above for colonies in 25 μl reactions, applying gene specific primers and 1 μl cDNA from copepodid or adult lice.
Sequences were assembled and translated using Vector NTI Advance 10 software (Invitrogen). Sequence similarity search of ORFs against GenBank was done using NCBI BLAST in tblastn (https://blast.ncbi.nlm.nih.gov/Blast.cgi). ORFs were aligned in Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). Location of domains was predicted by InterProScan (http://www.ebi.ac.uk/interpro/search/sequence/), and signal peptides in the SignalP-5.0 server (https://services.healthtech.dtu.dk/service.php?SignalP-5.0). ProtParam was used to compute theoretical pI and molecular weights (https://web.expasy.org/cgi-bin/protparam/protparam), and glycosylation sites were predicted in the NetNGlyc 1.0 server (https://services.healthtech.dtu.dk/service.php?NetNGlyc-1.0).
Real time RT-PCR
Real time RT-PCR was performed with 1× PowerUp™ SYBR Green Master Mix (Thermo Fisher Scientific), 500 nM forward and reverse primers (Tables 1 or 2) and 2 µl diluted cDNA in 10 µl reactions. Samples were always run in duplicate (technical replicates) on the Applied Biosystems 7500 Real-Time PCR System under standard conditions (50 °C for 2 min, 95 °C for 2 min, 40 cycles of 95 °C for 15 s and 60 °C for 1 min, followed by a melt curve analysis at 60–95 °C). A five-point standard curve of fourfold dilutions was made for each assay to calculate PCR efficiencies, given by the equation E% = (101/slope − 1) × 10066. Salmon louse cDNA were further diluted 1:10, while cDNA from salmon skin or primary leucocytes were diluted 1:5. The relative differences in threshold cycle between the target gene and the geometric mean of the reference genes (ΔCT) and expression relative to a calibrator (ΔΔCT) were calculated, transformed by the equation 2−ΔΔCT67. The reference gene expression was evaluated in all datasets prior to calculation to ensure a stable expression (Supplementary Fig. S2).
RNA interference and infection studies
RNA interference (RNAi) followed by infestation studies were performed to analyse the functional role of the labial gland proteins in vivo. Double stranded RNA (dsRNA) was produced using MEGAscript® RNAi Kit (Ambion) according to supplier’s instructions with primers for the LsLGPs listed in Table 1 and for the control fragment, CYP, see Dalvin et al.68. RNAi on salmon louse nauplii was performed by soaking as previously described by Eichner et al.32. Around 60–100 nauplius I larvae, all from the same egg string, were incubated overnight (17 h) in 150 µl of seawater containing 20 ng/µ1 control or labial gland gene dsRNA (LsLGP1–3). Thereafter, all animals within a treatment group were pooled and kept in flow through incubators until at least one day after the copepodid stage was reached. For pre-adult (LsLGP5) and adult lice (LsLGP4 and 5), 50 μl of LsLGP or control dsRNA solution was stained with 1 μl Bromphenol blue to enable us to see that approximately 600 ng dsRNA were injected dorsally into the haemocoel of the cephalothorax. Pre-adult lice were incubated off-host for 4 h prior to infestation, while adult lice were incubated 4 days off-host to allow knock down (KD) of target mRNAs prior to infestation as the sampling and immune analysis were to be conducted 3 days post infestation.
For immune studies, Atlantic salmon (average weight around 150 g) were kept individually in single tanks at 12 °C69. In each infestation experiment, three groups of fish were included (N = 6–8): an untreated group of fish, fish infested with control lice and fish infested with KD animals. Around 80 copepodids/fish or 10 adult females were added, and scaled skin samples were taken at and away from the attachment site of the lice 3 days post infestation (dpi). Immune responses were measured analyzing the transcript level of Atlantic salmon immune genes listed in Table 2. The lice number was recorded, and KD in lice transcript level was also measured and confirmed to be below 90% before subsequent analysis.
Injection of pre-adult lice II were conducted to analyse the effect of LsLGP4 KD on coagulation, feeding and development. The pre-adult lice (10/fish) were held on fish (N = 3) for 32 days, to allow for the second egg string to be extruded. After 32 days, the lice were counted and carefully removed from the fish to analyse KD of LsLGP4 transcript level. Pictures was taken to allow for size measurements in image J and visual inspection of gut blood content. All KD experiments were repeated twice with different batches of fish.
Heterologous expression and synthesis of labial gland proteins
The open reading frame (ORF) of LsLGP3 was used as templates for gene synthesis, and the gene was codon optimized to maximize expression in E. coli (GenScript). The gene was flanked by SapI restriction sites and delivered in a customized SapI-free plasmid with kanamycin selection marker (GenScript). Gene fragments excluding the signal peptides were sub-cloned into expression vectors, p1 and p12, as previously described by Bjerga et al.70, introducing an N-terminal or a C-terminal hexahistidin-tag, respectively. Before heterologous expression in E. coli MC1061, the inserts were sequenced to verify the ORF. Protein expression was induced after 2.5 h at 37 °C with 0.1% l-arabinose (Sigma) and produced over-night at 20 °C. Recombinant soluble LsLGP3 (recLGP3) was purified with the Ni–NTA Fast Start Kit (Qiagen). The buffer was changed to 1× PBS with a Pierce™ Protein concentrator, 10K MWCO (Thermo Scientific). The purity of recLGP3 was analyzed by Coomassie staining of SDS-PAGE gels (Supplementary Fig. S3), and the concentration measured on a Nanodrop Spectrophotometer (absorbance at 280 nm). Purified recLGP3 was stored at − 80 °C, and any precipitation removed by centrifugation (13,000×g, 10 min at 4 °C) before use. Moreover, the concentration of LPS-contamination was analyzed with the ToxinSensor™ Chromogenic LAL Endotoxin Assay Kit (GenScript®) according to the suppliers protocol. For LsLGP4, a synthetic LsLGP4 (synLGP4) was purchased (LeifTein). The purity of synLGP4 was analyzed with high pressure liquid chromatography to be 99.31%, and the peptide sequence was confirmed by Mass Spectrometry by the supplier.
In vitro stimulation of primary head kidney leucocytes
Atlantic salmon primary head kidney leucocytes were stimulated with recLGP3 and synLGP4 to analyze their involvement in immune modulation. Each experiment was repeated twice for each protein to be tested, as follows.
The average fish weight was 520 g for recLGP3 experiments and around 600 g for synLGP4 experiments. The leucocytes were isolated from three fish in each experiment, on a discontinuous Percoll gradient (Fisher) as previously described71, using L-15 medium (Gibco) supplemented with 10% Fetal bovine serum (Merck), heparin (Leo Pharma) and 1% penicillin (Merck). The leucocytes were counted manually with a hemocytometer, and cell viability evaluated using 0.04% Trypan Blue Stain (Gibco). Cells from each fish were plated into 96-well culture plates (Falcon), with a density of approximately 1 × 106 cells/well in 100 μl supplemented L15-medium. For recLGP3-stimulation, eight wells per fish where plated, given 1 × PBS (control), 1 μM/well LPS (Merck) or three different concentrations of protein (150, 112.5 and 75 μg protein/well). RecLGP3 was diluted to different concentrations in PBS to a final volume of 100 μl and added to the cells, whereas 100 μl PBS with or without LPS was used for control cells. For synLGP4-stimulation, four wells/fish was plated given control and the three different concentrations of the proteins as for recLGP3. SynLGP4 was diluted to different concentrations in ddH2O to a final volume of 30 μl and supplemented with L-15 medium to a final volume of 100 μl. The control cells received 30 μl ddH2O in 70 μl L-15 medium.
The cells were incubated at 16 °C. After four and 20 h, the L15-medium was removed and centrifuged for 2 min at 460×g, 4 °C, to harvest the non-adherent cells. The supernatant was removed, and the pellet was dissolved in 300 µl TRI reagent (Sigma). TRI reagent, 200 µl, was added to the adherent cells immediately after removal of the L15-medium and transferred to the non-adherent cells after repetitive mixing and scraping with the pipette. The cells were frozen in Tri reagent at − 80 °C until total RNA isolation as described in “Source of L. salmonis” section, after addition of 500 μl TRI reagent to the homogenized cells. Immune responses were measured analyzing the transcript level of Atlantic salmon immune genes listed in Table 2.
Blood coagulation measurement
To determine the anti-coagulation ability of recLGP3 and synLGP4, prothrombin time (PT) was determined by a modified Quick one-stage method72,73. Blood was collected from three Atlantic salmon (between 900 and 1400 g) into a syringe pre-filled with 10 mM sodium citrate (Merck) to a final ratio of one volume sodium citrate to nine volumes blood. The blood was centrifuged for 15 min at 180×g, 4 °C to collect the plasma. For each fish, 100 µl plasma was mixed with 100 µl 25 mM CaCl2 (Merch) and 100 µl rabbit Thromboplastin (Sigma Aldrich, 100 µg/µl in 10 mM CaCl2), and aliquoted into eight tubes at room temperature. Either 50 or 75 µg recLGP3 or 75 µg synLGP4 dissolved in 15 µl ddH2O was added to the plasma mixture, or 15 µl ddH2O for control plasma, all in duplicates. This setup gave two technical replicates per fish per treatment. The time until clot formation was measured while the tubes were continuously tilted back and forth 90 degrees but stopped after 45 min if no clotting was visible (only synLGP4 treated group). This experiment was repeated three times for LsLGP4 and twice for LsLGP3.
Statistical analysis
All statistical analysis were performed in GraphPad prism 8.0.1 (GraphPad Software). A student’s t-tests were used to test for differences when only two groups are compared (control versus treatment, α = 0.05). A one-way ANOVA was used to test for differences when more than two groups are compared (α = 0.05), using a Tukey’s multiple comparisons post hoc test.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Forseth, T. et al. The major threats to Atlantic salmon in Norway. Ices J. Mar. Sci. 74, 1496–1513 (2017).
Halttunen, E. et al. Migration of Atlantic salmon post-smolts in a fjord with high infestation pressure of salmon lice. Mar. Ecol. Prog. Ser. 592, 243–256 (2018).
Halttunen, E. et al. Sea trout adapt their migratory behaviour in response to high salmon lice concentrations. J. Fish Dis. 41, 953–967 (2018).
Vollset, K. W. et al. Disentangling the role of sea lice on the marine survival of Atlantic salmon. Ices J. Mar. Sci. 75, 50–60 (2018).
Jones, M. W., Sommerville, C. & Bron, J. The histopathology associated with the juvenile stages of Lepeophtheirus salmonis on the Atlantic salmon, Salmo salar L.. J. Fish Dis. 13, 303–310 (1990).
Jonsdottir, H., Bron, J. E., Wootten, R. & Turnbull, J. F. The histopathology associated with the preadult and adult stages of Lepeophtheirus salmonis on the Atlantic Salmon, Salmo salar L.. J. Fish Dis. 15, 521–527 (1992).
Dalvin, S. et al. Rainbow trout Oncorhynchus mykiss skin responses to salmon louse Lepeophtheirus salmonis: From copepodid to adult stage. Fish Shellfish Immunol. 103, 200–210 (2020).
Fast, M. D., Muise, D. M., Easy, R. E., Ross, N. W. & Johnson, S. C. The effects of Lepeophtheirus salmonis infections on the stress response and immunological status of Atlantic salmon (Salmo salar). Fish Shellfish Immunol. 21, 228–241 (2006).
Barker, S. E. et al. Sea lice, Lepeophtheirus salmonis (Krøyer 1837), infected Atlantic salmon (Salmo salar L.) are more susceptible to infectious salmon anemia virus. PLoS ONE 14, e0209178 (2019).
Grimnes, A. & Jakobsen, P. J. The physiological effects of salmon lice infection on post-smolt of Atlantic salmon. J. Fish Biol. 48, 1179–1194 (1996).
Tort, L. Stress and immune modulation in fish. Dev. Comp. Immunol. 35, 1366–1375 (2011).
Finstad, B., Bjørn, P. A., Grimnes, A. & Hvidsten, N. A. Laboratory and field investigations of salmon lice [Lepeophtheirus salmonis (Krøyer)] infestation on Atlantic salmon (Salmo salar L.) post-smolts. Aquac. Res. 31, 795–803 (2000).
Aaen, S. M., Helgesen, K. O., Bakke, M. J., Kaur, K. & Horsberg, T. E. Drug resistance in sea lice: A threat to salmonid aquaculture. Trends Parasitol. 31, 72–81 (2015).
Hjeltnes, B., Bang Jensen, B., Bornø, G., Haukaas, A. & Walde, C. S. Fiskehelserapporten 2018. In Veterinærinstituttet 2019 (2018).
Hamre, L. A. et al. The salmon louse Lepeophtheirus salmonis (Copepoda: Caligidae) life cycle has only two chalimus stages. PLoS ONE 8, e73539 (2013).
Johnson, S. C. & Albright, L. J. The developmental stages of Lepeophtheirus salmonis (Krøyer, 1837) (Copepoda, Caligidae). Can. J. Zool. 69, 929–950 (1991).
Heggland, E. I., Dondrup, M., Nilsen, F. & Eichner, C. Host gill attachment enables blood-feeding by the salmon louse (Lepeophtheirus salmonis) chalimus larvae and alters parasite development and transcriptome. Parasit. Vectors. https://doi.org/10.1186/s13071-020-04096-0 (2020).
Nuttall, P. A. Wonders of tick saliva. Ticks Tick Borne Dis. 10, 470–481 (2019).
Fast, M. D. et al. Susceptibility of rainbow trout Oncorhynchus mykiss, Atlantic salmon Salmo salar and coho salmon Oncorhynchus kisutch to experimental infection with sea lice Lepeophtheirus salmonis. Dis. Aquat. Organ. 52, 57–68 (2002).
Mustafa, A. et al. Effects of sea lice (Lepeophtheirus salmonis Kroyer, 1837) infestation on macrophage functions in Atlantic salmon (Salmo salar L.). Fish Shellfish Immunol. 10, 47–59 (2000).
Gamil, A. A. A., Gadan, K., Gislefoss, E. & Evensen, Ø. Sea lice (Lepeophtheirus salmonis) infestation reduces the ability of peripheral blood monocytic cells (PBMCs) to respond to and control replication of salmonid alphavirus in Atlantic salmon (Salmo salar L.). Viruses 12, 1450 (2020).
Øvergård, A. C., Hamre, L. A., Grotmol, S. & Nilsen, F. Salmon louse rhabdoviruses: Impact on louse development and transcription of selected Atlantic salmon immune genes. Dev. Comp. Immunol. 86, 86–95 (2018).
Johnson, S. C. & Albright, L. J. Comparative susceptibility and histopathology of the response of naive Atlantic, Chinook and Coho salmon to experimental infection with Lepeophtheirus salmonis (Copepoda, Caligidae). Dis. Aquat. Organ. 14, 179–193 (1992).
Braden, L. M., Barker, D. E., Koop, B. F. & Jones, S. R. Comparative defense-associated responses in salmon skin elicited by the ectoparasite Lepeophtheirus salmonis. Comp. Biochem. Physiol. D Genomics Proteomics 7, 100–109 (2012).
Bron, J. E., Sommerville, C., Jones, M. & Rae, G. H. The settlement and attachment of early stages of the salmon louse, Lepeophtheirus salmonis (Copepoda, Caligidae) on the salmon host, Salmo salar. J. Zool. 224, 201–212 (1991).
Øvergård, A. C. et al. Exocrine glands of Lepeophtheirus salmonis (Copepoda: Caligidae): Distribution, developmental appearance, and site of secretion. J. Morphol. 277, 1616–1630 (2016).
Bell, S., Bron, J. E. & Sommerville, C. The distribution of exocrine glands in Lepeophtheirus salmonis and Caligus elongatus (Copepoda: Caligidae). Contrib. Zool. 69, 9–20 (2000).
Poulin, R. & Randhawa, H. S. Evolution of parasitism along convergent lines: From ecology to genomics. Parasitology 142, S6–S15 (2015).
Skern-Mauritzen, R. et al. The salmon louse genome: Copepod features and parasitic adaptations. Genomics 113, 3666–3680 (2021).
Heggland, E. I. et al. A scavenger receptor B (CD36)-like protein is a potential mediator of intestinal heme absorption in the hematophagous ectoparasite Lepeophtheirus salmonis. Sci. Rep. 9, 4218 (2019).
Heggland, E. I., Trosse, C., Eichner, C. & Nilsen, F. Heavy and light chain homologs of ferritin are essential for blood-feeding and egg production of the ectoparasitic copepod Lepeophtheirus salmonis. Mol. Biochem. Parasitol. 232, 111197 (2019).
Eichner, C., Nilsen, F., Grotmol, S. & Dalvin, S. A method for stable gene knock-down by RNA interference in larvae of the salmon louse (Lepeophtheirus salmonis). Exp. Parasitol. 140, 44–51 (2014).
Breitwieser, G. E. Extracellular calcium as an integrator of tissue function. Int. J. Biochem. Cell Biol. 40, 1467–1480 (2008).
Olszak, I. T. et al. Extracellular calcium elicits a chemokinetic response from monocytes in vitro and in vivo. J. Clin. Investig. 105, 1299–1305 (2000).
Jaworski, D. C. et al. A secreted calreticulin protein in Ixodid tick (Amblyomma americanum) saliva. J. Insect Physiol. 41, 369–375 (1995).
Strynadka, N. C. J. & James, M. N. G. Crystal-structures of the helix-loop-helix calcium-binding proteins. Annu. Rev. Biochem. 58, 951–998 (1989).
Elsholz, F., Harteneck, C., Muller, W. & Friedland, K. Calcium—A central regulator of keratinocyte differentiation in health and disease. Eur. J. Dermatol. 24, 650–661 (2014).
Oda, Y. et al. Combined deletion of the vitamin D receptor and calcium-sensing receptor delays wound re-epithelialization. Endocrinology 158, 1929–1938 (2017).
Richardson, R. et al. Adult zebrafish as a model system for cutaneous wound-healing research. J. Investig. Dermatol. 133, 1655–1665 (2013).
Sveen, L. R. et al. Wound healing in post-smolt Atlantic salmon (Salmo salar L.). Sci. Rep. https://doi.org/10.1038/s41598-019-39080-x (2019).
Williamson, M. P. The structure and function of proline-rich regions in proteins. Biochem. J. 297, 249–260 (1994).
Veis, A. & Nawrot, C. F. Basicity differences among peptide bonds. J. Am. Chem. Soc. 92, 3910–3914 (1970).
Ball, L. J., Kuhne, R., Schneider-Mergener, J. & Oschkinat, H. Recognition of proline-rich motifs by protein-protein-interaction domains. Angew. Chem. Int. Ed. Engl. 44, 2852–2869 (2005).
Zhu, L. Y., Pan, P. P., Fang, W., Shao, J. Z. & Xiang, L. X. Essential role of IL-4 and IL-4Ralpha interaction in adaptive immunity of zebrafish: insight into the origin of Th2-like regulatory mechanism in ancient vertebrates. J. Immunol. 188, 5571–5584 (2012).
Wang, T. et al. First in-depth analysis of the novel Th2-type cytokines in salmonid fish reveals distinct patterns of expression and modulation but overlapping bioactivities. Oncotarget 7, 10917–10946 (2016).
Hu, Y. L., Xiang, L. X. & Shao, J. Z. Identification and characterization of a novel immunoglobulin Z isotype in zebrafish: Implications for a distinct B cell receptor in lower vertebrates. Mol. Immunol. 47, 738–746 (2010).
Zhang, Y. A. et al. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat. Immunol. 11, 827-U82 (2010).
Castro, R. et al. CCR7 is mainly expressed in teleost gills, where it defines an IgD+IgM- B lymphocyte subset. J. Immunol. 192, 1257–1266 (2014).
Perdiguero, P. et al. Teleost IgD+IgM- B cells mount clonally expanded and mildly mutated intestinal IgD responses in the absence of lymphoid follicles. Cell Rep. 29, 4223 (2019).
Xu, Z. et al. Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. PNAS 110, 13097–13102 (2013).
Braden, L. M., Koop, B. F. & Jones, S. R. Signatures of resistance to Lepeophtheirus salmonis include a TH2-type response at the louse-salmon interface. Dev. Comp. Immunol. 48, 178–91 (2015).
Todd, C. D. et al. Infestations of wild adult Atlantic salmon (Salmo salar L.) by the ectoparasitic copepod sea louse Lepeophtheirus salmonis Krøyer: Prevalence, intensity and the spatial distribution of males and females on the host fish. Hydrobiologia 429, 181–196 (2000).
Bui, S., Oppedal, F., Nola, V. & Barrett, L. T. Where art thou louse? A snapshot of attachment location preferences in salmon lice on Atlantic salmon hosts in sea cages. J. Fish Dis. 43, 697–706 (2020).
Wakelin, S. J. et al. “Dirty little secrets”—Endotoxin contamination of recombinant proteins. Immunol. Lett. 106, 1–7 (2006).
Hamilton, S. et al. Characterisation of proteins in excretory/secretory products collected from salmon lice, Lepeophtheirus salmonis. Parasit. Vectors 11, 294 (2018).
Fast, M. D., Johnson, S. C., Eddy, T. D., Pinto, D. & Ross, N. W. Lepeophtheirus salmonis secretory/excretory products and their effects on Atlantic salmon immune gene regulation. Parasite Immunol. 29, 179–89 (2007).
Fast, M. D. et al. Lepeophtheirus salmonis: Characterization of prostaglandin E2 in secretory products of the salmon louse by RP-HPLC and mass spectrometry. Exp. Parasitol. 107, 5–13 (2004).
Piesz, J. L., Barker, S. E. & Bricknell, I. R. Anti-chemotactic activity in the secretory/excretory products of Lepeophtheirus salmonis. Fish Shellfish Immunol. 98, 296–300 (2020).
Dalvin, S., Eichner, C., Dondrup, M. & Øvergård, A. C. Roles of three putative salmon louse (Lepeophtheirus salmonis) prostaglandin E2 synthases in physiology and host-parasite interactions. Parasit. Vectors 14, 206 (2021).
Eichner, C., Øvergård, A. C., Nilsen, F. & Dalvin, S. Molecular characterization and knock-down of salmon louse (Lepeophtheirus salmonis) prostaglandin E synthase. Exp. Parasitol. 159, 79–93 (2015).
Hamre, L. A., Glover, K. A. & Nilsen, F. Establishment and characterisation of salmon louse (Lepeophtheirus salmonis (Krøyer 1837)) laboratory strains. Parasitol. Int. 58, 451–60 (2009).
Øvergård, A. C., Nerland, A. H. & Patel, S. Evaluation of potential reference genes for real time RT-PCR studies in Atlantic halibut (Hippoglossus Hippoglossus L.); during development, in tissues of healthy and NNV-injected fish, and in anterior kidney leucocytes. BMC Mol. Biol. 11, 36 (2010).
Eichner, C., Dondrup, M. & Nilsen, F. RNA sequencing reveals distinct gene expression patterns during the development of parasitic larval stages of the salmon louse (Lepeophtheirus salmonis). J. Fish Dis. 41, 1005–1029 (2018).
Dalvin, S., Nilsen, F. & Skern-Mauritzen, R. Localization and transcription patterns of LsVasa, a molecular marker of germ cells in Lepeophtheirus salmonis (Krøyer). J. Nat. Hist. 47, 889–900 (2013).
Trosse, C., Nilsen, F. & Dalvin, S. RNA interference mediated knockdown of the KDEL receptor and COPB2 inhibits digestion and reproduction in the parasitic copepod Lepeophtheirus salmonis. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 170, 1–9 (2014).
Bustin, S. A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25, 169–93 (2000).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, 45 (2001).
Dalvin, S. et al. Functional characterisation of the maternal yolk-associated protein (LsYAP) utilising systemic RNA interference in the salmon louse (Lepeophtheirus salmonis) (Crustacea: Copepoda). Int. J. Parasitol. 39, 1407–1415 (2009).
Hamre, L. A. & Nilsen, F. Individual fish tank arrays in studies of Lepeophtheirus salmonis and lice loss variability. Dis. Aquat. Organ. 97, 47–56 (2011).
Bjerga, G. E., Arsin, H., Larsen, O., Puntervoll, P. & Kleivdal, H. T. A rapid solubility-optimized screening procedure for recombinant subtilisins in E. coli. J. Biotechnol. 222, 38–46 (2016).
Øverland, H. S., Pettersen, E. F., Rønneseth, A. & Wergeland, H. I. Phagocytosis by B-cells and neutrophils in Atlantic salmon (Salmo salar L.) and Atlantic cod (Gadus morhua L.). Fish Shellfish Immunol. 28, 193–204 (2010).
Quick, A. J. The Hemorrhagic Diseases & the Physiology of Hemostasis 1st edn. (Charles C. Thomas, 1942).
Pavlidis, M., Berry, M., Kokkari, C. & Kentouri, M. Prothrombin time, activated partial thromboplastin time and fibrinogen values in Mediterranean marine teleosts. Fish Physiol. Biochem. 21, 335–343 (1999).
Acknowledgements
This research was funded by the Research Council Norway, SFI-Sea Lice Research Centre, Grant Number 203513/O30, the Norwegian Seafood Research Fund (FHF), Grant Number 901564 and the European Union Horizon 2020 project ParaFishControl, Grant Number 634429. This output reflects only the authors’ views, and the European Union cannot be held responsible for any use that may be made of the information contained herein. Additional funding was received from NORCE Norwegian Research Centre. We thank Heidi Kongshaug, Per Gunnar Espedal, Liv Sandlund and Noelia Nuñez-Ortiz for technical assistance. We thank Sussie Dalvin for discussions.
Author information
Authors and Affiliations
Contributions
A.C.Ø.: Acquisition, analysis and interpretation of data, Writing—original draft. H.M.D.M.: Coagulation assay, Leucocyte stimulation, Writing—review and editing. L.A.H.: Fish experiments, Conceptualization, Writing—review and editing. M.D.: Analysis of transcriptomics data, Writing—review and editing. G.E.K.B. and Ø.L.: Production of recombinant protein. Writing—review and editing. J.K.C. and K.B.: Leucocyte stimulation, Writing—review and editing. F.N.: Interpretation of data, Writing—review and editing. S.G.: Conceptualization, Writing—review and editing. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Øvergård, AC., Midtbø, H.M.D., Hamre, L.A. et al. Small, charged proteins in salmon louse (Lepeophtheirus salmonis) secretions modulate Atlantic salmon (Salmo salar) immune responses and coagulation. Sci Rep 12, 7995 (2022). https://doi.org/10.1038/s41598-022-11773-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-11773-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.