Abstract
The emergence and spread of resistant tuberculosis (TB) pose a threat to public health, so it is necessary to diagnose the drug-resistant forms in a clinically short time frame and closely monitor their transmission. In this study, we carried out a first whole genome sequencing (WGS)-based analysis of multidrug resistant (MDR) M. tuberculosis strains to explore the phylogenetic lineages diversity, drug resistance mechanisms, and ongoing transmission chains within the country. In total, 65 isolates phenotypically resistant to at least rifampicin and isoniazid collected in the Czech Republic in 2005–2020 were enrolled for further analysis. The agreement of the results obtained by WGS with phenotypic drug susceptibility testing (pDST) in the determination of resistance to isoniazid, rifampicin, pyrazinamide, streptomycin, second-line injectables and fluoroquinolones was more than 80%. Phylogenetic analysis of WGS data revealed that the majority of MDR M. tuberculosis isolates were the Beijing lineage 2.2.1 (n = 46/65; 70.8%), while the remaining strains belonged to Euro-American lineage. Cluster analysis with a predefined cut-off distance of less than 12 single nucleotide polymorphisms between isolates showed 19 isolates in 6 clusters (clustering rate 29.2%), located mainly in the region of the capital city of Prague. This study highlights the utility of WGS as a high-resolution approach in the diagnosis, characterization of resistance patterns, and molecular-epidemiological analysis of resistant TB in the country.
Similar content being viewed by others
Introduction
Tuberculosis (TB), an infectious disease caused by the Mycobacterium tuberculosis complex (MTBC), is still a public health problem and its treatment is challenging for clinicians. Case notifications of people newly diagnosed with TB has a declining trend, but the number of deaths rose to 1.5 million in 2020, which is probably related to the COVID-19 pandemic1. The main problem complicating the eradication and treatment of TB is the prevalence of resistant strains, predominantly multidrug resistant (MDR-TB, resistant to 2 first-line drugs: isoniazid and rifampicin) and extensively-drug resistant (XDR-TB, resistant to fluoroquinolones and injectable drugs in addition to MDR)1. In June 2021, the World Health Organization (WHO) adopted a new definition of XDR-TB as TB caused by M. tuberculosis strains that are resistant to isoniazid, rifampicin, any fluoroquinolone, and either bedaquiline or linezolid (or both)2. However, in our study, we followed the original definition of XDR, as the data is before 2021.
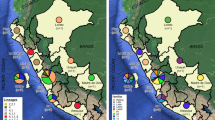
From 2005 to 2020, the incidence of TB as well as the related deaths decreased significantly in the Czech Republic, so it is currently considered a country with a low incidence of TB (Fig. 1).
Phenotypic drug susceptibility testing (pDST) is routinely performed on all TB culture-positive specimens in the country. The results from 2005 to 2020 showed a total of 480 strains of M. tuberculosis resistant to at least one antituberculosis drug. More specifically, in 2005–2020, the estimated proportion of MDR and XDR strains was 2.13% of all phenotypically tested isolates (of which 17.4% in previously treated patients)3.
Culture-based pDST on solid or liquid (Mycobacterium growth indicator tubes—MGIT) media and genotypic drug susceptibility testing (gDST) methods (such as Xpert MTB/RIF, Cepheid, Sunnyvale, California, USA and GenoType MTBDR, Hain LifeScience GmbH, Nehren, Germany) are currently preferred for the determination of M. tuberculosis resistance in clinical settings4. Limitations of pDST and gDST lead to the development and application of advanced molecular techniques such as whole genome sequencing (WGS). WGS data provide a rapid and detailed insight of mutations encoding resistance to all available antituberculosis drugs and contribute to the identification of the transmission patterns based on the genomic relatedness of M. tuberculosis strains, without the need for traditional contact tracing5,6,7,8. The molecular-epidemiological analysis of TB in the Czech Republic was conducted by so-called classical typing methods9. However, these methods interrogate only a small fraction of the M. tuberculosis genome to reconstruct the complex transmission chains and do not show sufficient discriminatory power to track the transmission links between patients10.
Despite the fact that a large proportion (more than 28%) of patients diagnosed with TB are born outside the Czech Republic, the genetic relatedness of drug-resistant M. tuberculosis strains has not yet been studied in detail11. The aim of this work was to perform the first WGS-based study using the MDR-, XDR-isolates collected in the Czech Republic during the years 2005–2020. Sequencing analysis determined the complete resistance profile and revealed the phylogenetic diversity of the strains, thus contributing to the characterization of local outbreaks and the monitoring of the dynamics of resistant strains over the years. This work will serve as a baseline to analyse the molecular epidemiology and drug-resistance of M. tuberculosis strains in the Czech Republic.
Results
Sample collection
A total of 65 samples of MDR M. tuberculosis strains were included, representing 60% of the total number of MDR-TB cases diagnosed during the study. A limited number of samples were due to the inability to reactivate some cultures in MGIT medium. Samples have been collected continuously over the years, specifically: 3 in 2005, 1 in 2006, 3 in 2008, 7 in 2009, 2 in 2010, 4 in 2014, 9 in 2015, 4 in 2016, 9 in 2017, 10 in 2018, 12 in 2019 and 1 in 2020. Subsequent isolates from the same TB patient were included in the study (only in two cases: CZ800-17/CZ541-18 and CZ628-17/CZ149-18) as long as the isolation date is more than six months after the isolation date of the previous isolate. The study included 14 women and 47 men with a mean age of 40 years (ranging from 23 to 81 years).
Sequencing data quality
The minimum criteria for sequencing data quality were: mean coverage ≥ 30 and mapped reads ≥ 90% of the reference genome covered by the sequence read. These requirements were met for all samples, as the average depth of coverage was 90× (ranging from 39× to 158×) and mapped reads 98.59% (ranging from 94.71 to 99.33%).
Drug susceptibility testing
Phenotypic testing confirmed resistance to rifampicin and isoniazid (i.e. MDR-TB) in all 65 samples. Moreover, resistance to at least one additional first-line or second-line antituberculosis drug was determined in 64 samples (98.46%, Table 1).
Sequencing data revealed a wide range of gene mutations associated with resistance to first-line and second-line antituberculosis drugs, including delamanide (Table 2). Moreover, resistance to rifampicin was primarily caused by the mutations at codon 450 (S450L) in the rpoB gene (56/65 resistant isolates)12. In addition, mutations at codon 435 (A435V; A435T; 4 patients), 445 (H445L; 2 patients) and 452 (L452P; 1 patient) were also characterized in the rpoB gene. An additional mutation in the rpoC gene (L527V; P452S; I491T) was identified in 11 samples (16.92%)12. Compensatory mutations in the rpoC gene have been primarily associated with the rpoB S450L mutation and Euro-American sublineage 4.1.2.1, which is in concordance with other studies examining the significance of these mutations13. Deletion in the rpoB gene (1302_1307delGGACCA) was detected in one phenotypic resistant isolate (CZ412-18). Isoniazid-resistant isolates most frequently showed mutations in the katG gene (S315T, 64/65). A mutation in the inhA gene (S94A) was found in one isolate. Sixteen isolates (24.62%) showed simultaneous mutations in the fabG1 gene promoter region (15C>T, 14/65; 8T>C, 2/65)14. Fifty-seven ethambutol-resistant isolates harboured mutations in the embB (primarily M306I/ M306L, 27/57; followed by G497A; A435A; T334H; T319C; G406A; S297A; P404S) and embA (including c.-16C>T, c.-12C>T; 5/57) genes15,16. Mutations encoding resistance to ethambutol were also identified in 23 phenotypically-sensitive isolates, which is the main reason for the low reliability (sensitivity and specificity 97.22% and 17.24%, respectively) of pDST for ethambutol. Compared to other antituberculosis drugs, the greatest mutational variability was observed in pncA gene encoding the resistance to pyrazinamide. Twenty-two different non-synonymous mutations and 2 novel deletion polymorphisms (287_287delA; 524_524delT) in pncA gene have been identified with 95% sensitivity and 73% specificity (Table 2, Supplementary Table 2)17. The most frequent were mutations at codons 63 (3/37) and 160 (3/37). Genotypic resistance to streptomycin was confirmed in 58/65 (87.7%) (sensitivity 93.33% and specificity 80%). Mutations in the rpsL (predominantly L43A—47/58 and L88A—7/58) and rrs (514a>c; 11/58) genes were most common18. Resistance across the second-line injections (kanamycin, amikacin, capreomycin) was induced by mutations occurring in the rrs (1401a>g), eis (10g>a; 12c>t) and tlyA (733_742delACGCAGACCG) genes and detected in 21 isolates (32.31%)19. Of these, 10 isolates (47.62%) were resistant to all second-line injectables, 10 isolates (47.62%) resistant to kanamycin and one isolate (4.61%) resistant to kanamycin simultaneously with capreomycin. Regarding fluoroquinolone resistance, the 17 isolates (26.15%) harboured various mutations only in the gyrA gene (A90V; A94G; A94A; S91P)20. In overall, the resistance to second-line injectables and fluoroquinolones was determined with a sensitivity and specificity of 80% / 93.75% and 82.22%/93.88%, respectively.
In addition, mutations associated with resistance to other second-line antituberculosis drugs have been identified, resulting in: 33 isolates (50.77%) resistant to ethionamide (mutation in ethA gene: 110_110delT; 1054_1054delC; T314I; M1A; A67P; 1010_1010delA; 341_341delT; 32_32delC; 1386_1386delT; mutation in fabG1 gene: 15C>T; 8C>T) (sensitivity 72% and specificity 60%), 3 isolates (4.61%) resistant to paraaminosalicylic acid (folC G40G; A49P), and 2 isolates (3.08%) resistant to linezolid (rplC C154A). Two isolates (3.08%) showed gene mutations in the ddn gene (T88STOP) encoding delamanide resistance21. No mutations in the genes encoding resistance to bedaquiline and clofazimine have been identified.
In overall, 10 isolates were determined as XDR.
Phylogenetic analysis and identification of transmission cluster
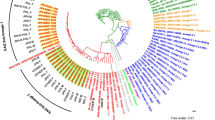
Two major lineages were observed through the WGS analysis. Phylogenetic analysis showed that most isolates (46; 70.77%) belonged to lineage 2 (specifically to 2.2.1 Beijing genotype family). Twenty isolates (30.77%) were included in lineage 4. The classification of isolates into sublineages was as follows: 9 belonged to sublineage 4.1.2.1 (Haarlem), 6 to sublineage 4.3.3 (LAM, T), 3 to sublineage 4.2.1 (Ural) and 1 to sublineage 4.3.4.2 (LAM, T). One isolate (CZ445-15) consisted of a mix of 4.3.3 and 4.1.2.1 sublineages. Additional subclassification for strains belonging to the same sublineage can be seen in the maximum likelihood phylogenetic tree (Fig. 2).
Maximum likelihood phylogenetic tree generated from 64 MDR isolates collected in the Czech Republic during the years 2005–2020 together with their lineage, cluster assignment, and genotypic resistance to first- and second-line antituberculosis drugs. The tree was annotated using iTOL v6 (https://itol.embl.de/). The first column denotes the lineages. The next 5 columns show genomic relatedness within clusters (CL12, CL5, CL2 and CL0—sets the maximum number of SNPs that differ from the genetically closest isolate). Mutations encoded resistance are represented by filled circles (presence of mutation) or empty circles (absence of mutation) icons. MDR multi-drug resistant, XDR extensively-drug resistant, drug-resistant (including mono- and poly-resistant).
Cluster analysis with a predefined maximum cut-off of 12 SNPs difference revealed 6 clusters containing 15 MDR isolates and 4 XDR isolates (n = 19; clustering rate 29.23%) with a maximum size of 2–5 isolates for each cluster (Fig. 2). Nine isolates were obtained from patients with newly diagnosed TB and 10 from patients with confirmed recurrent TB. These clusters consisted mainly of strains from Beijing sublineage 2.2.1 (73.68%) and to a lesser extent from Euro-American sublineage 4.1.2.1 (Haarlem) (26.32%). A difference of 0–2 SNPs between patients was determined in 8 cases (Fig. 2, CL0, CL2), indicating a recent transmission events and the difference 3–5 SNPs was documented in only 2 patients, indicating the direct transmission (Fig. 2, CL5). Using a threshold 6–12 SNPs to look at older transmission events, 9 patients in 4 clusters were identified (Fig. 2, CL12). The distance between the individual clusters ranged from 35 SNPs (between cluster 5 and 6) to 1066 SNPs (between cluster 3 and 1) (Fig. 3).
Minimum spanning tree based on SNP differences between the strains, including the XDR-TB and MDR-TB strains collected in Czech Republic during 2005 and 2020. Maximum distance set to 12 SNPs for linked transmission. Distant matrix generated from MTBseq (version 1.0.2, https://github.com/ngs-fzb/MTBseq_source) and a minimum spanning tree was constructed using GrapeTree software (https://achtman-lab.github.io/GrapeTree/MSTree_holder.html).
Four isolates (80%) from cluster 1 come from the capital city of Prague. Patient CZ8842 is from another region (Nový Jičín) but was first diagnosed with MDR-TB in 2005 (as well as patients CZ188 and CZ218), suggesting an association with other isolates from this cluster. Epidemiological data on patients grouped in clusters 2 and 3 also indicated the spread of a resistant strain of M. tuberculosis from Prague region. Cluster 5 was formed by three isolates, two of which related to the same accommodation facility in the town of Šenov. The remaining clusters consisted of only two isolates that came from geographically separated areas (Supplementary Table 1).
Discussion
WGS is a molecular-genetic method that is becoming increasingly preferred in clinical laboratories; however, it has not yet been performed on continuously collected MDR and XDR isolates. Therefore, we conducted the first WGS study in the country. Sequencing data in our study showed that in the last 15 years, MDR/XDR strains of M. tuberculosis belonging to the Beijing 2.2.1 lineage were the most common in the Czech Republic (70.77% of all isolates), followed by the Euro-American lineage 4 (29.23%). The results correlate with studies that have confirmed a growing incidence and transmission of MDR strains of this lineage in Eastern Europe, predominantly due to selective advantages including hypervirulence, hypermutability and increased transmissibility22,23,24,25. In comparison, the molecular-epidemiological study based on IS6110-RFLP conducted in Czech Republic in 2006 showed that only 7.1% of all isolates (11/155) belonged to the Beijing lineage, but represented up to 36% of all resistant isolates9. We assume that the prevalence of the Beijing lineage is evolutionary associated with migration events from Ukraine, which began in early 1990s. It reflects that the Ukrainian national minority is currently one of the largest membership bases in the Czech Republic26. In general, the prevalence of Beijing strains among MDR isolates is of growing concern, especially due to evolutionary success in terms of dispersion and adaptation to different human populations; therefore, appropriate measures should be taken for public health surveillance27.
In relation to resistance, XDR-TB was significantly more often detected in MDR-TB strains of the Beijing genotype (9/10; 90%) than in MDR-TB strains of non-Beijing genotypes (1/10; 10%). These data are supported by a study by Beer et al., which confirmed a significantly increased proportion of Beijing genotype (84%) among all XDR strains collected in the European Union28. In general, the most prevalent gene mutations reported in our study were rpoB S450L for rifampicin, katG S315T for isoniazid, embB M306I for ethambutol, pncA H57A for pyrazinamide, rpsL L43A for streptomycin, gyrA A94G for fluoroquinolones, rrs 1401a>g and consistent with other studies29,30. Moreover, pDST was performed on all samples; therefore, the identified gene variants can potentially contribute to increasing the sensitivity of currently used line-probe assays and developing new genotypic arrays. In comparison with the available catalogues summarizing the mutations associated with resistance, we identified two novel potential resistance-confirming indels in the pncA gene (287_287del; 524_524del)29,31. These mutations require further detailed investigation to determine their clinical relevance and impact on minimal inhibitory concentration to pyrazinamide. Additionally, we reported the first two strains of M. tuberculosis exhibiting genotypic resistance to new antituberculosis drug delamanid. The resistance was encoded by mutations in ddn gene. Interestingly, delamanid-resistant strains were isolated from patients without a prior treatment regimen containing this drug, probably due to a high rate of spontaneous mutations in the target gene32. These data are also supported by other studies that have confirmed resistance in patients without previous exposure to this drug21,33. Our results highlight the need to determine susceptibility to delamanid in clinical laboratories in order to prevent the spread and selection of delamanid-resistant strains.
Phylogenetic analysis of M. tuberculosis genomes from subsequent samples from the same patient confirmed reinfection with a different strain of M. tuberculosis (difference more than 12 SNPs) in patient 800-17/541-18 (man, 31/32 years old). The same analysis from isolates from patient 628-17/149-18 (man, 37/38 years old) showed a difference of 0 SNPs between strains. However, additional genotypic resistance to linezolid developed within 1 year (Fig. 2, Supplementary Table 2). We hypothesize that mutations associated with resistance to linezolid developed due to non-adherence to treatment regimen or incorrect combination and dosing of antituberculosis drugs34.
The overall sensitivities and specificities for WGS, as mentioned in the results (“Drug susceptibility testing” section), demonstrated more than 80% concordance of the results obtained by WGS with pDST in the determination of resistance to isoniazid, rifampicin, pyrazinamide, streptomycin, second-line injectables, and fluoroquinolones, which is consistent with other available studies. studies14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37. The lower specificity of WGS in ethambutol resistance is due to the genotypic resistance of 23 phenotypically sensitive isolates. These isolates showed mutations in the embB gene, especially at codons 306, 406 and 497. Mutations at these codons were also identified in phenotypically resistant isolates. This phenomenon is not uncommon and has been observed in other studies, as these mutations may only slightly increase the minimum inhibitory concentration and thus lead to false negative pDST results38,39,40.
After cluster analysis, six clusters containing a total of 19 strains were identified (clustering rate 29.23%), most of which were formed by Beijing lineage isolates. In general, the clustering rate in our study was lower compared to the continuous 20 years genotyping study of MDR-TB in Spain (clustering rate 48.4%), 4-years drug-resistant TB WGS study in Thailand (48.5%) and the 3-years study involving MDR isolates from European Union countries (clustering rate 51.6%)41,42,43. These findings confirm the discriminatory power of WGS in terms of the definition of clonal complexes and the diversity of genotypes circulating in the country and highlight the need for a public health service to control and diagnose resistant TB. Furthermore, the results suggest that we lack some isolates within the clusters, or that resistance has developed due to inappropriate TB treatment regimens, as well as patient non-compliance. The higher clustering rate exhibited by the Beijing lineage (31.81%) compared to the Euro-American lineage (25%) is consistent with previous recent studies44,45. Also, more than 50% of clustered isolates are located in the Prague region, the capital of the Czech Republic with the largest proportion of foreign population due to employment. The overall clustering rate (29.23%) was almost half lower compared to study using Mycobacterium tuberculosis-specific multiple locus variable number tandem repeat (MIRU-VNTR) method for evaluation the genetic relatedness of MDR isolates collected in the Czech Republic during the years 2003–201128. This may be due to false clustering and lower resolution of transmission using MIRU-VNTR in countries with low TB incidence46. The list of mutations associated with resistance to first-line antituberculosis drugs in clustered isolates showed that their transmission is likely to occur at the MDR-TB level. Among the first-line antituberculosis drugs, the greatest mutational variability in clustered isolates was demonstrated in the embB (encoding resistance to ethambutol) and pncA (encoding resistance to pyrazinamide) genes. In addition, mutations in the pncA gene were also unique in most of unclustered isolates. Similar diversity has been confirmed in other studies, supporting the theory that pncA mutations still remain the leading cause of pyrazinamide resistance47,48. Also, 4 isolates integrated within cluster 1 harboured an additional mutation in the rpoC gene. In contrast, a detailed look at mutations encoding resistance to second-line antituberculosis drugs (predominantly mutations in gyrA gene that confer resistance to fluoroquinolones and rrs gene that confer resistance to aminoglycosides) revealed several differences in isolates within the same cluster (cluster 1, 3, 4, 5; Supplementary Tables 1, 2). This indicates that these clustered strains evolved differently due to non-adherence to the treatment regimen, rather than by recent transmission events. Linezolid resistance (mutation rplC C154A) was confirmed in 2 isolates within the cluster 3. These findings highlight the need to control linezolid dosing in a resistant TB treatment regimen and support the theory of linezolid resistance association with the Beijing lineage49,50.
However, our study has some limitations. Several XDR/MDR cultures could not be reactivated in MGIT medium even after several attempts. In addition, WGS results could not be compared with other genotypic methods. Nevertheless, this study confirmed the utility of WGS in surveillance of drug resistant TB strains circulating in the Czech Republic. Sequencing data analysis identified complete resistance profiles of MDR and XDR strains of M. tuberculosis, suggesting that the implementation of this methodology in routine diagnostics could improve TB control at the national level.
Conclusion
This study highlights the relevance and utility of WGS as a high-resolution approach to perform genetic analysis of drug-resistant TB in the Czech Republic. WGS has detected gene mutations encoding resistance to first-line and second-line antituberculosis drugs (including delamanide and linezolid), demonstrating its clinical importance in TB therapy. In addition, our study provided a baseline genotypic characterization of the MDR- and XDR-TB strains circulating in the Czech Republic and, in combination with epidemiological information, revealed several transmission chains that will improve the surveillance activities in the country.
Materials and methods
Sample collection
The culture or sputum smear microscopy positive TB samples used in this study were collected at the National Reference Laboratory for Mycobacteria in Prague, Czech Republic. In total, 65 samples from 2005 to 2020 (representing 60% of all MDR strains) with confirmed phenotypic resistance to at least rifampicin and isoniazid (MDR-TB) were studied. The presence of M. tuberculosis in biological material was confirmed by smear positivity and culture positivity on Löwenstein-Jensen (LJ) solid medium and in BACTEC™ MGIT™ 960 System (Becton, Dickinson and Company, East Rutherford, USA). All the work related to the manipulation of live bacteria culture was performed in biosafety level 3 laboratory.
Drug susceptibility testing
Conventional phenotypic drug susceptibility testing (pDST) by proportion method with critical concentrations of antituberculosis drugs according to WHO recommendations was performed for rifampicin (40.0 μg/ml), isoniazid (0.2 μg/ml), ethambutol (2.0 μg/ml), pyrazinamide (400.0 μg/ml), streptomycin (4.0 μg/ml), kanamycin (30.0 μg/ml), ethionamide (40.0 μg/ml), cycloserine (40.0 μg/ml), paraminosalycylic acid (1.0 μg/ml) and moxifloxacin (1.0 μg/ml) and used as a golden standard to define XDR-TB and MDR-TB51. The results were obtained after 3–6 weeks of incubation at 37 °C.
Whole genome sequencing
1 ml of early positive MGIT cultures were inactivated at 95 °C for 30 min in the biosafety level 3 laboratory. The genomic DNA from inactivated bacterial cultures was extracted according to the manufacturer’s protocol using QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany). DNA concentration was quantified by Qubit 4 technology using Qubit dsDNA HS Assay kit (Thermo Fisher Scientific, Waltham, USA). A concentration of genomic DNA in the range of 0.2–0.4 ng was used to prepare sequencing libraries using the Illumina Nextera XT library preparation kit (Illumina, San Diego, USA). Whole genome sequencing was executed on the Illumina MiSeq platform (Illumina, San Diego, USA) producing paired-end FASTQ files.
Bioinformatics analysis
Paried-end Illumina read data was first prepared for analysis in the following manner: we used trimmomatic (v0.39) to remove any adapter fragments and low-quality tail-ends with quality phred-scores ≤ Q352. All reads shorter than 35 bp in length were discarded. We then used Kraken2 (v2.2.1) to screen libraries for any contaminating reads that were assigned to taxa outside of the Mycobacterium tuberculosis complex using a custom database53. The remaining reads were quality-assessed using seqkit3 (v0.15)54. To assign isolates to MTBC lineages, and identify antibiotic resistance markers (SNPs, indels and frameshifts), we used TB-profiler (v2.8.6; using the M. tuberculosis H37Rv genome as the mapping reference)29. We identified one isolate (CZ445-15) that consisted of a mix of two lineages, which was excluded from the rest of the analyses.
For phylogenetic and cluster analysis we identified SNPs in the following manner: Reads were mapped against the hypothetical MTBC ancestor reference genome, described previously, using BWA (v0.7.17), and de-duplicated, to remove optical PCR duplicate reads, using picard tools (v2.12; https://broadinstitute.github.io/picard/)55. We then used samtools to identify fixed variants with a frequency of at least 70% and removed any variant identified within repetitive genomic regions (PE/PPE), insertion sequences, as well as prophages. A concatenated alignment of 3657 identified SNP positions was then generated from the resulting vcf-files which was used to generate a distance matrix of SNP distances using the dna.dist function implemented in the R package ape56. We used IQ-TREE (v1.6) to construct a maximum likelihood phylogeny using an GTR + F + ASC evolutionary model. The IQ-tree software searches through several evolutionary models and selects for a “best-fit model” using the Bayesian Information Criterion (BIC) score assigned to each tested model. The GTR + F + ASC model consistently scored the highest.
We then used the iTol web-based phylogeny tool (v5) to visualize the tree57,58. A distant matrix was generated from MTBseq and a minimum spanning tree was constructed using GrapeTree software (https://github.com/achtman-lab/GrapeTree) with the maximum distance threshold of 12 SNPs (due to reporting an analysis of strains collected in 15 years timeframe) for linked transmission59,60.
Ethics statement
This study was approved by the Ethics committee of Jessenius Faculty of Medicine in Martin (EK 72/2018), Comenius University in Bratislava, Slovakia. All methods were carried out in accordance with relevant guidelines and regulations. Informed consent was obtained from all participants or, if participants are under 18, from a parent and/or legal guardian.
Data availability
WGS raw reads were submitted to the European Nucleotide Archive as FASTQ files under study Accession No. PRJEB48710.
References
World Health Organization. Global Tuberculosis Report. Blood (World Health Organization, 2015).
Viney, K. et al. New definitions of pre-extensively and extensively drug-resistant tuberculosis: Update from the World Health Organization. Eur. Respir. J. 57, 4 (2021).
Registr tuberkulózy. https://tbc.uzis.cz/. Accessed 9 Jan 2022.
WHO. Rapid Communication: Molecular Assays as Initial Tests for the Diagnosis of Tuberculosis and Rifampicin Resistance 1–8 (World Health Organization, 2020).
Faksri, K. et al. Comparisons of whole-genome sequencing and phenotypic drug susceptibility testing for Mycobacterium tuberculosis causing MDR-TB and XDR-TB in Thailand. Int. J. Antimicrob. Agents 54, 109–116 (2019).
van Beek, J., Haanperä, M., Smit, P. W., Mentula, S. & Soini, H. Evaluation of whole genome sequencing and software tools for drug susceptibility testing of Mycobacterium tuberculosis. Clin. Microbiol. Infect. 25, 82–86 (2019).
Walker, T. M. et al. A cluster of multidrug-resistant Mycobacterium tuberculosis among patients arriving in Europe from the Horn of Africa: A molecular epidemiological study. Lancet. Infect. Dis 18, 431–440 (2018).
Jabbar, A. et al. Whole genome sequencing of drug resistant Mycobacterium tuberculosis isolates from a high burden tuberculosis region of North West Pakistan. Sci. Rep. 9, 1–9 (2019).
Prodinger, W. M. et al. Molecular epidemiology of tuberculosis in the Czech Republic, 2004: Analysis of M. tuberculosis complex isolates originating from the City of Prague, South Moravia and the Moravian-Silesian region. Central Eur. J. Public Health 14, 168–174 (2006).
Kamerbeek, J. et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35, 907–914 (1997).
Země narození | Registr tuberkulózy. https://tbc.uzis.cz/cs/browser/birth-country?type=czechiaVis&yearOfDisease=2020 (2020).
Sheed Khan, A. et al. Characterization of rifampicin-resistant Mycobacterium tuberculosis in Khyber Pakhtunkhwa, Pakistan. Sci. Rep. 11, 14194 (2021).
De Vos, M. et al. Putative compensatory mutations in the rpoc gene of rifampin-resistant Mycobacterium tuberculosis are associated with ongoing transmission. Antimicrob. Agents Chemother. 57, 827–832 (2013).
Wu, X. et al. Use of whole-genome sequencing to predict Mycobacterium tuberculosis drug resistance in Shanghai, China. Int. J. Infect. Dis. 96, 48–53 (2020).
Plinke, C. et al. embCAB sequence variation among ethambutol-resistant Mycobacterium tuberculosis isolates without embB306 mutation. J. Antimicrob. Chemother. 65, 1359–1367 (2010).
Ramaswamy, S. V. et al. Molecular genetic analysis of nucleotide polymorphisms associated with ethambutol resistance in human isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44, 326–336 (2000).
World Health Organization. Catalogue of Mutations in Mycobacterium tuberculosis Complex and Their Association with Drug Resistance (World Health Organization, 2021).
Jagielski, T. et al. Screening for streptomycin resistance-conferring mutations in Mycobacterium tuberculosis clinical isolates from Poland. PLoS ONE 9, e100078 (2014).
Ali, A. et al. Whole genome sequencing based characterization of extensively drug-resistant Mycobacterium tuberculosis isolates from pakistan. PLoS ONE 10, e0117771 (2015).
von Groll, A. et al. Fluoroquinolone resistance in Mycobacterium tuberculosis and mutations in gyrA and gyrB. Antimicrob. Agents Chemother. 53, 4498–4500 (2009).
Schena, E. et al. Delamanid susceptibility testing of Mycobacterium tuberculosis using the resazurin microtitre assay and the BACTEC™ MGIT™ 960 system. J. Antimicrob. Chemother. 71, 1532–1539 (2016).
Casali, N. et al. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res. 22, 735–745 (2012).
Ford, C. B. et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat. Genet. 45, 784–790 (2013).
Merker, M. et al. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat. Genet. 47, 242–249 (2015).
Merker, M. et al. Multidrug- and extensively drug-resistant Mycobacterium tuberculosis Beijing clades, Ukraine, 2015—Volume 26, Number 3–March 2020—Emerging Infectious Diseases journal—CDC. Emerg. Infect. Dis. 26, 481–490 (2020).
Leontiyeva, Y. Ukrainians in the Czech Republic: On the pathway from temporary foreign workers to one of the largest minority groups. In Ukrainian Migration to the European Union (eds Fedyuk, O. & Kindler, M.) 133–149 (Springer, 2016).
Wiens, K. E. et al. Global variation in bacterial strains that cause tuberculosis disease: A systematic review and meta-analysis. BMC Med. 16, 1–13 (2018).
De Beer, J. L., Ködmön, C., van der Werf, M. J., van Ingen, J. & van Soolingen, D. Molecular surveillance of multi- and extensively drug-resistant tuberculosis transmission in the European Union from 2003 to 2011. Eurosurveillance 19, 20742 (2014).
Phelan, J. E. et al. Integrating informatics tools and portable sequencing technology for rapid detection of resistance to anti-tuberculous drugs. Genome Med. 11, 1–7 (2019).
Coll, F. et al. Genome-wide analysis of multi- and extensively drug-resistant Mycobacterium tuberculosis. Nat. Genet. 50, 307–316 (2018).
World Health Organization. Drug Resistance Profiles of Mycobacterium tuberculosis Complex and Factors Associated with Drug Resi (World Health Organization, 2021).
Fujiwara, M., Kawasaki, M., Hariguchi, N., Liu, Y. & Matsumoto, M. Mechanisms of resistance to delamanid, a drug for Mycobacterium tuberculosis. Tuberculosis 108, 186–194 (2018).
Pang, Y. et al. In vitro drug susceptibility of bedaquiline, delamanid, linezolid, clofazimine, moxifloxacin, and gatifloxacin against extensively drug-resistant tuberculosis in Beijing, China. Antimicrob. Agents Chemother. https://doi.org/10.1128/AAC.00900-17 (2017).
Nambiar, R. et al. Linezolid resistance in Mycobacterium tuberculosis isolates at a tertiary care centre in Mumbai, India. Indian J. Med. Res. 154, 85 (2021).
The CRyPTIC Consortium and the 100,000 Genomes Project. Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N. Engl. J. Med. 379, 1403–1415 (2018).
Coll, F. et al. Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med. 7, 1–10 (2015).
Chen, X. et al. Evaluation of whole-genome sequence method to diagnose resistance of 13 anti-tuberculosis drugs and characterize resistance genes in clinical multi-drug resistance Mycobacterium tuberculosis isolates from China. Front. Microbiol. 10, 1741 (2019).
Cheng, S., Cui, Z., Li, Y. & Hu, Z. Diagnostic accuracy of a molecular drug susceptibility testing method for the antituberculosis drug ethambutol: A systematic review and meta-analysis. J. Clin. Microbiol. 52, 2913–2924 (2014).
Engström, A., Morcillo, N., Imperiale, B., Hoffner, S. E. & Juréen, P. Detection of first- and second-line drug resistance in Mycobacterium tuberculosis clinical isolates by pyrosequencing. J. Clin. Microbiol. 50, 2026–2033 (2012).
Plinke, C., Walter, K., Aly, S., Ehlers, S. & Niemann, S. Mycobacterium tuberculosis embB codon 306 mutations confer moderately increased resistance to ethambutol in vitro and in vivo. Antimicrob. Agents Chemother. 55, 2891–2896 (2011).
Iglesias, M. J. et al. The value of the continuous genotyping of multi-drug resistant tuberculosis over 20 years in Spain. Sci. Rep. 10, 1–12 (2020).
Tagliani, E. et al. Use of a whole genome sequencing-based approach for Mycobacterium tuberculosis surveillance in Europe in 2017–2019: An ECDC pilot study. Eur. Respir. J. 57, 2002272 (2021).
Nonghanphithak, D. et al. Clusters of drug-resistant Mycobacterium tuberculosis detected by whole-genome sequence analysis of nationwide sample, Thailand, 2014–2017. Emerg. Infect. Dis. 27, 813–822 (2021).
Liu, Y. et al. The study on the association between Beijing genotype family and drug susceptibility phenotypes of Mycobacterium tuberculosis in Beijing. Sci. Rep. 7, 1–7 (2017).
Sun, L. L. et al. Insight into multidrug-resistant Beijing genotype Mycobacterium tuberculosis isolates in Myanmar. Int. J. Infect. Dis. 76, 109–119 (2018).
Alaridah, N. et al. Transmission dynamics study of tuberculosis isolates with whole genome sequencing in southern Sweden. Sci. Rep. 9, 1–9 (2019).
Li, K. et al. Characterization of pncA mutations and prediction of PZA resistance in Mycobacterium tuberculosis clinical isolates from Chongqing, China. Front. Microbiol. 11, 3252 (2021).
Allana, S. et al. PncA gene mutations associated with pyrazinamide resistance in drug-resistant Tuberculosis, South Africa and Georgia. Emerg. Infect. Dis. 23, 491–495 (2017).
Zhang, Z., Pang, Y., Wang, Y., Liu, C. & Zhao, Y. Beijing genotype of Mycobacterium tuberculosis is significantly associated with linezolid resistance in multidrug-resistant and extensively drug-resistant tuberculosis in China. Int. J. Antimicrob. Agents 43, 231–235 (2014).
Wasserman, S., Meintjes, G. & Maartens, G. Linezolid in the treatment of drug-resistant tuberculosis: The challenge of its narrow therapeutic index. Expert Rev. Anti Infect. Ther. 14, 901–915 (2016).
Technical Report on Critical Concentrations for Drug Susceptibility Testing of Medicines Used in the Treatment of Drug-Resistant Tuberculosis. https://www.who.int/publications/i/item/WHO-CDS-TB-2018.5. Accessed 9 Jan 2022.
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Wood, D. E., Lu, J. & Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 20, 1–13 (2019).
Shen, W., Le, S., Li, Y. & Hu, F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 11, e0163962 (2016).
Cancino-Muñoz, I. et al. Cryptic resistance mutations associated with misdiagnoses of multidrug-resistant tuberculosis. J. Infect. Dis. 220, 316–320 (2019).
Paradis, E., Claude, J. & Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A. & Minh, B. Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Zhou, Z. et al. Grapetree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 28, 1395–1404 (2018).
Walker, T. M. et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: A retrospective observational study. Lancet. Infect. Dis. 13, 137–146 (2013).
Funding
This research was funded by Grant APVV-18-0084, Grant VEGA-1/0093/22, and Grant of Ministry of Health, Czech Republic—conceptual development of research organization (“The National Institute of Public Health—NIPH, 75010330”).
Author information
Authors and Affiliations
Contributions
M.D. wrote the first version of manuscript. V.D. and M.Šp. undertook sample collection and phenotypic drug susceptibility testing. A.S. and A.N. performed the bioinformatics analysis. M.D. and M.Šk. performed WGS. E.M.R., A.M.C., I.P. contributed in review and editing of the manuscript. I.S., D.M.C. and J.M. contributed in formal analysis. J.M. directed the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dohál, M., Dvořáková, V., Šperková, M. et al. Whole genome sequencing of multidrug-resistant Mycobacterium tuberculosis isolates collected in the Czech Republic, 2005–2020. Sci Rep 12, 7149 (2022). https://doi.org/10.1038/s41598-022-11287-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-11287-5
This article is cited by
-
Genotypic and phenotypic comparison of drug resistance profiles of clinical multidrug-resistant Mycobacterium tuberculosis isolates using whole genome sequencing in Latvia
BMC Infectious Diseases (2023)
-
Tuberculosis in Ukrainian War Refugees and Migrants in the Czech Republic and Slovakia: A Molecular Epidemiological Study
Journal of Epidemiology and Global Health (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.