Abstract
The crosstalk between osteosarcoma (OS) development and abnormally expressed microRNA (miR)-601 is not explored explicitly. Here, we identified the downregulated miR-601 in osteosarcoma (OS) through a comprehensive bioinformatics analysis of GEO Datasets. The results indicated that miR-601 was downregulated in both OS cells and tissues. The OS patients with reduced expression of miR-601 displayed worse prognosis. The results of in vitro and in vivo assay revealed that elevated miR-601 inhibited the proliferative, migratory and invasive capacities in OS cells. Mechanically, miR-601 exerted its function via targeting oncogene protein kinase membrane associated tyrosine/threonine 1 (PKMYT1) at post-transcriptional level. Moreover, miR-601 was attenuated by c-Myb at transcriptional level. Taken together, our studies reveal that miR-601 is a suppressive gene negatively correlated with malignancy of OS.
Similar content being viewed by others
Introduction
Osteosarcoma (OS), the most common osteogenic malignant tumor, is most commonly seen in children and adolescents1,2. It is a heterogeneous tumor with complex pathogenesis, and the pathogenesis has not been fully clarified. Although the incidence of OS is not high, it has a high mortality rate of disability, which brings great mental and economic burden to patients, their families and society3. Pulmonary metastasis is a common cause of death in OS patients. Although surgical resection combined with neoadjuvant chemotherapy reduced mortality in OS patients, 5 year survival remained low, approximately 70% in non-metastatic patients compared with 11 to 30% in metastatic patients4. Despite numerous progress has been made, the commitments for early detection, novel therapies and lower mortality are largely futile.
The protein kinase membrane associated tyrosine/threonine 1 (PKMYT1) gene encodes a member of the serine/threonine Protein kinase family5,6. The protein of PKMYT1 is a membrane-associated kinase, which regulates cell cycle negatively via phosphorylating and extinguishing cyclin-dependent kinase 1 during G2/M phase7. Recent studies have shown that PKMYT1 is overexpressed in many malignant tumors8,9. Meanwhile, PKMYT1 has been reported to drive the progression of colorectal cancer9, prostate cancer10, gastric cancer11, non-small cell lung cancer12. A literature has reported that PKMYT1 is upregulated in OS tissues13. And high levels of PKMYT1 had lower rates of both overall survival and metastasis-free survival of OS patients than those with low levels of PKMYT113. However, the underlying mechanisms of PKMYT1 in OS remains dimly.
Recently, microRNAs (miRNAs) have been illustrated to play crucial roles in various physiological conditions, as well as pathological conditions, including OS14,15. Owing to the role in the progression, invasion and metastasis of OS cells, microRNA has been validated as a certain potential as a prognostic indicator or therapeutic target. Despite lots of miRNAs have been validated as promoters or suppressors in the generation and progression of OS, such as miR-14016, miR-328-3p17, miR-34a18, miR-2119 and miR-215-5p20, miR-601 has not been systematically researched in OS.
Here, we aimed to elucidate the specific roles miR-601 involved in OS. The microarray results indicated that miR-601 was lowly expressed in OS cells. Our results showed the same trend in OS cells and tissues. Furthermore, attenuated level of miR-601 in OS patients showed a poor prognosis. Additionally, we found that miR-601 inhibited OS progression by targeting PKMYT1. We also clarified that miR-601 was repressed by the upstream transcription factor of c-Myb.
Results
miR-601 is downregulated in OS and related to poor outcome
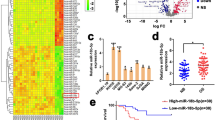
To identify the functional miRNA in OS, we conducted a bioinformatics analysis via utilizing the miRNA expression profiles of GSE: 28,423 in GEO datasets. Among the differentially expressed miRNAs in 4 normal bone cell lines and 19 OS cell lines, we found that the role and mechanism of down-regulated miR-601 in OS are rarely reported (Fig. 1a). The results of differential analysis showed that miR-601 was dramatically down-regulated in OS cells (Fig. 1b). Consistently, RT-qPCR results further supported that miR-601 was dramatically reduced in 4 OS cells lines compared with the normal bone cells (hFOB1.19) (Fig. 1c). Meanwhile, we further measured the level of miR-601 in 66 tissues and paired OS normal tissues using RT-qPCR analysis. The results suggested that miR-601 was remarkably reduced in OS tissues compared with adjacent normal tissues (Fig. 1d, e). Moreover, the association between the overall survival of OS patients and miR-601 expression was verified by Kaplan–Meier analysis. The result suggested that lower miR-601 level was associated with shorter overall survival (Fig. 1f). Based on the aforementioned information, we concluded that miR-601 was downregulated in OS, and that lower miR-601 expression was correlated with poor prognosis in OS.
miR-601 is downregulated in OS and related to poor outcome. (a) A histogram showed the 16 most downregulated miRNAs in OS cells compared with normal osteocytes. (b) The histogram showed the miR-601 expression in OS cells compared with normal osteocytes. (c) The miR-601 expression was evaluated by RT-qPCR assay in 5 OS cell lines and normal bone cells. (d, e) The expression of miR-601 was evaluated by RT-qPCR assay in 66 paired OS tissues and normal tissues (NB). (f) Kaplan–Meier curves analysis showed overall survival of 66 OS patients in miR-601 high and low groups. Patients were divided into "high" and "low" group with the 50th percentile expression of miR-601 as the critical point.
Overexpression of miR-601 promotes proliferation and metastasis of OS cells
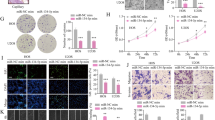
To elucidate the biological function of miR-601 in OS cells, miR-601 mimic were used to upregulate the expression of miR-601, and the efficiency was verified via RT-qPCR assay (Fig. 2a). The MTT assay results indicated that the proliferation abilities was dramatically decreased in OS cells with miR-601 overexpression (Fig. 2b). Meanwhile, the transwell assay results suggested that miR-601 upregulation remarkably impaired the migration and invasion of OS cells (Fig. 2c, d). Moreover, subcutaneous xenograft model and pulmonary metastasis model were performed to confirm the biological function of miR-601 in vivo. In accord with the in vitro results, miR-601 overexpression remarkably reduced the tumor volume and weight compared with the control group (Fig. 2e-g). Furthermore, mice injected with OS cells, transfected with LV-miR-601, showed decreased number of metastatic foci in their lungs compared with those in the mice of control group (Fig. 2h). The results of hematoxylin and eosin (H&E) staining indicated that miR-601 overexpression dramatically reduced the volume of lung metastatic lesions compared with the control group (Fig. 2i). What’s more, the mice in the miR-601 overexpression group showed higher survival rate than the control group (Fig. 2j). Collectively, the results suggested that miR-601 repressed the proliferation and metastasis of OS cells.
Overexpression of miR-601 promotes proliferation and metastasis of OS cells. (a) The miR-601 expression was evaluated by RT-qPCR assay in U2OS and 143b cells with miR-601 overexpression. (b) The proliferation of U2OS/143b cells was measures by the MTT experiment. (c, d) The invasion and migration abilities of U2OS and 143b cells was assessed by the transwell assay. (e) The nude mice (6 per group) were subcutaneously injected with U2OS cells transfected with lentivirus vector carrying the miR-601 sequence (LV-miR-601) or negative control (LV-miR-NC). (f, g) The volume and weight of xenograft tumor of nude mice. (h, i) U2OS cells, transfected with lentivirus vector carrying the miR-601 sequence (LV-miR-601) or negative control (LV-miR-NC), were injected into the nude mice (6 per group) from tail vein. The metastasis on lung was assessed via H&E staining. (j) The survival of nude mice was analyzed.
miR-601 mediates the downregulation of PKMYT1
To validate the potential target of miR-601, the databases of RNA22v2, TargetScan7.1, miRTarget and miRPathDB were utilized (Fig. 3a). Among the 16 genes screened from the above databases, PKMYT1 was reported to be implicated in various cancer types 6. Meanwhile, the putative miR-601 binding sites were predicted in PKMYT1 (Fig. 3b). The RT-qPCR results showed that both miR-601 overexpression and miR-601 depletion could not affect the mRNA level of PKMYT1 (Fig. 3c, d). Whereas, miR-601 overexpression remarkably reduced the protein level of PKMYT1 (Fig. 3e). In addition, depletion of miR-601 had the opposite effect (Fig. 3f), indicating that miR-601 regulated the expression of PKMYT1 in posttranscriptional level. Next, we ought to prove the direct interaction between miR-601 and PKMYT1 mRNA. The biotin based RNA pulldown assay indicated that there was a direct combination between miR-601 and the mRNA of PKMYT1 (Fig. 3g). Furthermore, in order to demonstrate the specificity of binding sites, Luciferase reporting experiments were performed. The wild-type and mutant PKMYT1-mRNA-3 'UTR sequences were inserted into the luciferase reporter plasmids to construct the luciferase reporter plasmids of PKMYT1-3' UTR (WT: wild-type; MUT: mutant-type). The results suggested that overexpression of miR-601 reduced the WT luciferase activity, while inhibition of miR-601 enhanced the WT luciferase activity (Fig. 3h, i). However, the change of miR-601 expression had no significant effect on the luciferase activity of the MUT. These results suggest that miR-601 regulated PKMYT1 translation via binding to the region of PKMYT1-mRNA-3'UTR. Taken together, PKMYT1 is regulated by miR-601 at posttranscriptional level and a potential target of miR-601.
miR-601 mediates the downregulation of PKMYT1. (a) Venn diagram displayed the targets of miR-601 predicted by the databases of RNA22v2, TargetScam 7.1, miRTarget and miRPathDB. (b) Schematic diagram displayed the putative binding sequence of 3’-UTR-PKMYT1- mRNA and miR-601. The mutated sites of the 3’-UTR-PKMYT1- mRNA was showed. (c, d) The expression of PKMYT1 mRNA in miR-601 upregulated or downregulated U2OS and 143b cells was measured via RT-qPCR experiment. (e, f) The protein of PKMYT1 in miR-601 upregulated or downregulated U2OS and 143b cells was measured via Western blot experiment. The original blots/gels are presented in Supplementary Dataset File. (g) The enrichment of PKMYT1 on biotinylated miR-NC and miR-601 probe was evaluated by biotin-based pulldown assay. The GAPDH mRNA served as control. (h, i) Luciferase activity of U2OS and 143b cells was accessed through luciferase reporter assay.
miR-601 exerts its tumor suppressor function in OS by targeting PKMYT1
Next, we continued to explore that miR-601 played its function of inhibiting proliferation and metastasis through regulating the level of its downstream target PKMYT1 in OS. The results indicated that upregulation of miR-601 significantly inhibited the protein level of endogenous PKMYT1 (Fig. 4a). MTT assay suggested that upregulation of PKMYT1 significantly facilitated the proliferation of OS cells, which was neutralized by overexpression of miR-601 (Fig. 4b). Similarly, it was observed that elevated expression of miR-601 dramatically attenuated the enhanced invasion and metastasis induced by augmentation of PKMYT1 (Fig. 4c, d). Moreover, we also detected the PKMYT1 protein level in OS tissues via Immunohistochemistry (IHC) assay. The results revealed that PKMYT1 expression was high in OS tissues with low miR-601-expression and low in OS tissues with high miR-601-expression (Fig. 4e), suggesting a negative correlation between miR-601 expression and PKMYT1 protein expression in OS tissues. These results above revealed that PKMYT1 functioned as a tumor-promotor in OS cells, and miR-601 exerted a tumor suppressor effect by inhibiting its expression.
miR-601 exerts its tumor suppressor function in OS by targeting PKMYT1. (a–d) U2OS/143b cells were transfected with PKMYT1 overexpression plasmid, miR-601 mimic or negative control. (a) The protein level of PKMYT1 were detected by Western blot assay in U2OS and 143b cells. The original blots/gels are presented in Supplementary Dataset File. (b–d) MTT and transwell assays assess the proliferation, invasion and migration of U2OS and 143b cells. (e) IHC assay detected the PKMYT1 protein expression in OS tissues with miR-601-low and miR-601-high. Scale bar: 100 μm.
c-Myb is involved in the downregulation of miR-601 in OS cells
To explore the underlying mechanism of miR-601 downregulation in OS, the transcription factors that may bind to the promoter region of miR-601 were analyzed. It was found that the promoter region of MIR601, host gene of miR-601, contained the binding site of the oncogene c-Myb (Fig. 5a). It has been reported that the nuclear proto-oncogene c-Myb plays a crucial carcinogenic role in the malignance of a variety of cancers21,22. Therefore, we hypothesized that c-Myb might regulate the expression of miR-601 at transcriptional level. In order to verify our conjecture, we performed a ChIP assay. The results indicated that c-Myb bound to the promoter of MIR601 (Fig. 5a). Next, three small interfering RNAs targeting c-Myb ware utilized to deplete the expression of c-Myb in OS cells. The efficiency of knockdown was evaluate via conducting RT-qPCR and Western blot experiments (Fig. 5b, c). The luciferase assay suggested depletion of c-Myb significantly promoted the activity of the MIR601 promoter (Fig. 5d). More importantly, c-Myb silencing significantly enhanced the expression of miR-601 in OS cells (Fig. 5e). Additionally, overexpression of c-Myb dramatically repressed the activity of the MIR601 promoter, as well as the expression of miR-601 (Fig. 5f–i).
c-Myb is involved in the downregulation of miR-601 in OS cells. (a) (Up) Schematic diagram displayed the putative c-Myb binding sites on the promoter of MIR601. (Down) Chip assay was performed using anti-c-Myb or anti-IgG antibodies. (b, c) The knockdown efficiency of c-Myb was evaluated by RT-qPCR assay and Western blot assay. The original blots/gels are presented in Supplementary Dataset File. (d) (Up) Schematic diagram displayed the wild type and mutant type of putative binding sites of c-Myb on the promoter of MIR601. (Down) The luciferase activity of c-Myb knockdown U2OS and 143b cells, co-transfected with luciferase reporter plasmid coding MIR601 promoter sequence or scrambled vector, was accessed by luciferase reporter assay. (e) The miR-601 expression was evaluated by RT-qPCR assay in U2OS and 143b cells with c-Myb knockdown. (f, g) The level of c-Myb mRNA and protein were detected via RT-qPCR and Western blot assay in c-Myb overexpression U2OS and 143b cells. The original blots/gels are presented in Supplementary Dataset File. (h) The luciferase activity of c-Myb overexpression U2OS and 143b cells, co-transfected with luciferase reporter plasmid coding MIR601 promoter sequence or scrambled vector, was accessed by luciferase reporter assay. (i) The miR-601 expression was evaluated by RT-qPCR assay in U2OS and 143b cells. (j) The mRNA level of c-Myb were measured via RT-qPCR assay in 66 paired OS tissues and normal tissues. (k) Pearson correlation analysis displayed the correlation between miR-601 and c-Myb in OS tissues.
To further verify the relationship between c-Myb and miR-601, we detected the level of c-Myb in the tissues of OS patients. The results suggested that the level of c-Myb in OS tissues was higher than that in paired normal tissues (Fig. 5j). Correlation analysis results indicated that the expression of c-Myb was negatively correlated with miR-601 in OS tissues (Fig. 5k). Therefore, our results supports that c-Myb is responsible for the inhibition of miR-601 in OS cells.
Discussion
miRNAs are a class of small, endogenous, evolutionarily highly conserved single-stranded non-coding RNAs that function primarily at the post-transcriptional level of target genes23,24. As we all know, the current medical level of the society cannot have a good treatment effect for patients with advanced cancer25. Therefore, the study of tumor-related mechanisms is particularly important. With the discovery and understanding of miRNAs and their target gene networks, miRNAs are crucial regulators in the proliferation, apoptosis, cell cycle progression, invasion and metastasis of cancer cells26. In this study, miR-601, which has rarely been reported in OS, was selected as the object of study. The results of in vitro and in vivo assay demonstrated that miR-601 plays a crucial role in the proliferation and metastasis of OS as a tumor suppressor. Mechanism exploration have shown that miR-601 is inhibited by c-Myb at the transcriptional level. Meanwhile, miR-601 inhibited the expression of PKMYT1 at the post-transcriptional level, thus exerting its tumor suppressive function.
It has been elucidated that miR-601 is involved in many cancers, including pancreatic cancer, breast cancer, colorectal cancer and so on27,28,29. In these cancers, miR-601 was identified as tumor suppressor. Whereas, the role of miR-601 in OS remains excusive. First of all, through analyzing the data from GEO Datasets, we discovered that miR-601 was reduced in OS cells. The RT-qPCR assay further verified that miR-601 was downregulated in both OS cells and tissues. More importantly, clinical data suggested that OS patients with downregulated miR-601 level had a shorter overall survival. Additionally, a series of functional experiments revealed that high level of miR-601 inhibited the proliferation, invasion and metastasis of OS cells. Therefore, we concluded that miR-601 also functioned as a tumor suppressor in OS.
Studies have demonstrated that PKMYT1 is overexpressed in solid tumors, and elevated PKMYT1 levels are associated with tumor progression, metastasis, and poor prognosis11,30,31. In our studies, we found that miR-601 exerted its anti-tumor function via targeting PKMYT1 at post-transcriptional level. Meanwhile, the results indicated that overexpression of PKMYT1 obviously promoted the proliferation, invasion and migration of OS cells. While, the underlying mechanism by which PKMYT1 performs its function in OS needs further verification. According to previous literature, PKMYT1 promoted tumor cell proliferation and apoptosis resistance via activating the MAPK signaling pathway in gastric cancer32. In neuroblastoma, PKMYT1 promoted cell growth and inhibited apoptosis through stabilization of the MYCN protein33. Meanwhile, PKMYT1 also promotes carcinogenesis of non-small cell lung cancer through the Notch signaling pathway12. In hepatocellular carcinoma, PKMYT1 contributes to the enhanced cell growth and motility through the activation of the beta-catenin/TCF signaling31. It has also been elucidated that PKMYT1 promote cell proliferation, invasion and migration via targeting sirtuin 3 in ovarian cancer34. In addition, studies have shown that PKMYT1 promotes tumor progression of esophageal squamous cell carcinoma by activating AKT signaling pathway35. Collectively, PKMYT1 contributes to the malignancy of OS might through a variety of signal pathway.
c-Myb, one of the MYB family members, has been considered as an oncogenic transcription factor involved in various cancers22,36. Here we find a potential binding site for c-Myb on the promoter of the miR-601. The ChIP assay results supported that c-Myb could bind to the region of miR-601 promoter. The luciferase assay validated that the activity of miR-601 was enhanced in c-Myb depleted cells and attenuated in c-Myb elevated cells. What’s more, the same trend was observed in the expression of miR-601. The clinical data further supported that miR-601 was negatively related to c-Myb in OS tissues. The above results indicated that c-Myb inhibited the expression of miR-601 via serving as a suppressive transcription factor.
In summary, our results depicted that miR-601 served as an anti-oncogene to suppress cell proliferation and metastasis through inhibition of oncogene PKMYT1. The oncogene c-Myb medicated the inhibition of miR-601 in OS cells. These observations illustrates that miR-601 is a crucial molecule and may provide a novel rationale for the treatment of OS.
Methods
Human tissue specimens
All the pairs of normal and OS tissues from a cohort of 66 patients were gotten from Huazhong University of Science and Technology Union Shenzhen Hospital. All OS patients wrote the informed consent. All experiments were approved by medical ethics committee affiliated with Huazhong University of Science and Technology Union Shenzhen Hospital and were carried out in accordance with relevant guidelines and regulations.
Cell culture
The Cell Bank of China Academy of Sciences (Shanghai, China) provides all the cell lines. α-MEM medium (HyClone, USA) containing 10% FBS (Gibco, USA) was utilized in the use of culturing U2OS, MG63 and MNNG cells. RPMI-1640 medium (Gibco, USA) containing 15% FBS was used to culture Saos-2 cells. DMEM medium (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% FBS was used to culture hFOB1.19 and 143b cells. 100 U/mL penicillin and 100 mg/mL streptomycin were supplemented in all the culture medium. The OS cell lines were cultured under the condition of 37 °C with 5% CO2 in an incubator. hFOB 1.19 cell line was cultured under the condition of 33.5 °C with 5% CO2.
Western blot
The cells were first lysed with a RIPA lysate and then the concentration of proteins was evaluated via a BCA protein assay kit (Thermo Fisher Scientific). Denaturation of protein by boiling and protein loading buffer. 10% SDS-PAGE was utilized to separate the proteins, which was then transferred to a polyvinylidene fluoride (PVDF, Millipore, Billerica, MA, USA) membranes. After incubated with 5% skimmed milk at room temperature for 1 h, the membranes were incubated with the primary antibodies against c-Myb (1:100, Abcam, ab169111) and PKMYT1 (1:100, Abcam, ab200387) at 4 °C overnight. Then the membranes were incubated with secondary antibody (1:50,000, Abcam, ab6728 or ab6721) conjugated with horseradish peroxidase at room temperature for 1 h. Finally, the Enhanced Chemiluminescence Detection System (Thermo Fisher Scientific) was applied to detect the protein signals. GAPDH served as internal control.
Cell proliferation assay
Logarithmic phase cells were collected and 2000 cells were added to each well in a 96-well plate with a volume of 100ul. After 1,2,3,4 and 5 days, 20 ul MTT solution (5 mg/ml) was added to each well and continue to be cultured in dark for 4 h. Replaced the liquid in the well with 150 ul dimethyl sulfoxide to each well to dissolve the crystal completely. The absorbance of each well was detected by ELISA reader (Thermo Fisher Scientific) at 490 nm.
Transwell
The ability of migration and invasion in OS cells was assessed via transwell assay. Place the transwell chambers (8 μm), either uncoated or coated with Matrigel (Sigma), into a 24-well plate. Logarithmic phase cells were collected, diluted with 200ul FBS-free medium, and added into the upper chamber (5 × 104 per well). 700 ul 1640 medium, supplemented with 10% FBS, was added into the lower chamber of the 24-well plate. After 24 h, the liquid in the wells was discarded. Incubate the wells with formaldehyde for 20 min. Subsequently, 0.1% crystal violet was added in to the wells for 15 min, washed with clear water, and gently wiped off the upper cells with cotton swabs. Cells were observed and counted in random 5 fields under the microscope.
Transfection
The miR-601 mimic, miR-601 inhibitor, short interference RNAs targeting c-Myb and corresponding negative control were purchased from RiboBio (Guangzhou, China). Lipofectamine 2000 (Invitrogen) was utilized for the cell transfection based on the given instructions. Lentivirus vector carrying the miR-601 sequence (LV-miR-601) and negative control (LV-miR-NC) were brought from GeneChem (Shanghai, China). RT-qPCR assay was conducted to evaluate the efficiency of transfection. GAPDH and U6 were served as loading control. The sequence of siRNAs were shown in Table 1.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
TRIzol Reagent from Invitrogen was applied to extract total RNA from the OS tissues and cultured cells. The cDNA was synthesized via utilizing the PrimeScriptTM RT reagent Kit (Takara, Dalian, China) and miRNA First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA) based on the given instructions. SYBR Premix Ex TaqTM (TaKaRa) was applied to perform the qPCR based on the given protocols. U6 snRNA served as control to normalize the amplified transcript level of miRNA. β-actin served as control for the amplified transcript level of mRNA. The primers were shown in Table 2.
Luciferase assay
The procedure was same as described previously37. To identify the binding between miR-601 and 3’UTR of PKMYT1 mRNA, 3’UTR of PKMYT1 mRNA sequence containing putative binding site of miR-601 (wild type, WT; mutant type, MUT) was constructed into luciferase reporter plasmid. To assess the promoter activity of MIR601, MIR601 promoter sequence containing putative binding site of c-Myb (WT or MUT) was constructed into luciferase reporter plasmid. The U2OS and 143b cells were transfected with luciferase reporter plasmid and pRL-TK plasmid. 48 h later, the luciferase activity was detected via a luciferase activity kit (Promega, USA) according to the directions. Renilla luciferase served as a normalizing control.
Chromatin immunoprecipitation (ChIP) assay
The procedure was same as described previously38. ChIP assay was conducted using a ChIP assay kit (Millipore, New Bedford, MA) according to the given instructions. The cells were grown in a 10 cm petri dish. The formaldehyde was used to crosslink the proteins and DNA. The DNA is interrupted by ultrasound to an average size of 300–500 bp. Take 10 ul of lysate as Input. The lysate was divided into two parts, anti-c-Myb (cell signaling technology, #12,319) and anti-IgG (cell signaling technology, #2729) antibodies were added respectively, and agarose beads were added to precipitate DNA and protein complex. The DNA and protein complex were purified, crosslinking was terminated with 5 M NaCl, and the purified DNA was analyzed by qPCR experiment.
Biotin miRNA pull-down assay
The single-stranded biotinylated miR-601 or miR-NC probe was brought by the GeneChem (Shanghai, China). Briefly, 4 × 106 U2OS and 143b cells were firstly transfected with 50 μM biotinylated miR-601 or miR-NC for 36 h. Then the cells were lysed followed by the cells lysate were incubated with the biotinylated probe at 4 °C overnight. Then add the streptavidin coated agarose beads (Invitrogen, USA) into the mixture medium that was further incubated at 4 °C for 4 h. Washed and separate the precipitates. Extract total RNA from the precipitates and analyzed the purified RNA via RT-qPCR analysis.
IHC assay
Firstly, the paraffin sections were dewaxed in xylene and gradient ethanol (100%, 95%, 80%, 70%). After washing with water and PBS, paraffin sections were treated with Triton X-100 and 30% H2O2 for permeability and sealing. After washing again, the slides were placed in a sodium citrate buffer (pH = 6.0, 0.01 M), heated and boiled in the microwave for 10 min. Then 10% goat serum was added for sealing at 37℃ for 30 min. Then the diluted primary antibody against PKMYT1 was added and incubated overnight in the refrigerator at 4 °C. The next day, after washing with PBS, the diluted secondary antibody was added and incubated in an incubator at 37 °C for 1 h. DAB solution was added, and hematoxylin was redyed. Then dehydration was performed, xylene was used to make slices transparent, and neutral resin was added to seal the slices. Finally, the stained sections were imaged at 200X under an inverted microscope.
In vivo assay
Four-week-old BALB/c nude mice, brought from Shanghai Laboratory Animal Research Center, China, were used for the xenograft model and metastasis model. 2 × 106 LV-miR-601 or LV-miR-NC transfected U2OS cells were subcutaneously injected into the mice (6 per group) in the right hip to construct the xenograft model. Tumor volumes were recorded every 3 days. The mice were sacrificed at 27 days. Solid tumors were removed and weighted. The equation V = 0.5 × L (length) × W2 (width) was applied to evaluate the tumor volume. For the metastasis model, 2 × 106 LV-miR-601 or LV-miR-NC transfected U2OS cells were injected into the mice (6 per group) from the tail vein. Lung metastasis was confirmed by H&E staining. The animal experiments were reviewed and approved by the Animal Care and Use Committee affiliated with Huazhong University of Science and Technology Union Shenzhen Hospital and were carried out in accordance with relevant guidelines and regulations in compliance with the ARRIVE guidelines.
Statistical analysis
All experiments were conducted in triplicate and displayed with one representative experimental result. In addition, the statistics were analyzed by GraphPad Prim version 8.0.0 (GraphPad Software, San Diego, California USA, www.graphpad.com) and SPSS 18.0 software (SPSSlnc., Chicago, IL, USA, www.spss.com). The results are shown as the mean value ± standard deviation. Student’s t test and ANOVA were employed to statist the comparisons between groups. Kaplan–Meier survival curves was established with the log-rank test. P < 0.05 was considered significant (*P < 0.05; **P < 0.01; ***P < 0.001, NS indicating no significant).
References
Lilienthal, I. & Herold, N. Targeting molecular mechanisms underlying treatment efficacy and resistance in osteosarcoma: a review of current and future strategies. Int. J. Mol. Sci. 21, 6885 (2020).
Ritter, J. & Bielack, S. S. Osteosarcoma. Ann. Oncol. 21, i320–i325 (2010).
Harrison, D. J. et al. Current and future therapeutic approaches for osteosarcoma. Expert Rev. Anticancer Ther. 18, 39–50 (2018).
Makielski, K. M. et al. Risk factors for development of canine and human osteosarcoma: A comparative review. Vet. Sci. 6, 48 (2019).
Fattaey, A. & Booher, R. N. Myt1: A Wee1-type kinase that phosphorylates Cdc2 on residue Thr14. Prog. Cell Cycle Res. 3, 233–240 (1997).
Asquith, C., Laitinen, T. & East, M. P. PKMYT1: A forgotten member of the WEE1 family. Nat. Rev. Drug Discov. 19, 157 (2020).
Schmidt, M. et al. Regulation of G2/M transition by inhibition of WEE1 and PKMYT1 kinases. Molecules. 22, 2045 (2017).
Lewis, C. W. et al. Upregulation of Myt1 promotes acquired resistance of cancer cells to Wee1 inhibition. Cancer Res. 79, 5971–5985 (2019).
Jeong, D. et al. Protein kinase, membrane associated tyrosine/threonine 1 is associated with the progression of colorectal cancer. Oncol. Rep. 39, 2829–2836 (2018).
Wang, J. et al. PKMYT1 is associated with prostate cancer malignancy and may serve as a therapeutic target. Gene 744, 144608 (2020).
Wang, X. M. et al. Effects of MCRS1 on proliferation, migration, invasion, and epithelial mesenchymal transition of gastric cancer cells by interacting with Pkmyt1 protein kinase. Cell Signal. 59, 171–181 (2019).
Sun, Q. S. et al. Overexpression of PKMYT1 indicates the poor prognosis and enhances proliferation and tumorigenesis in non-small cell lung cancer via activation of Notch signal pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 4210–4219 (2019).
Wu, Y. et al. Systematic screening for potential therapeutic targets in osteosarcoma through a kinome-wide CRISPR-Cas9 library. Cancer Boil. Med. 17, 782–794 (2020).
Lee, Y. S. & Dutta, A. MicroRNAs in cancer. Annu. Rev. Pathol. 4, 199–227 (2009).
Wang, J. et al. The role of miRNA in the diagnosis, prognosis, and treatment of osteosarcoma. Cancer Biother. Radiopharm. 34, 605–613 (2019).
Song, B. et al. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene 28, 4065–4074 (2009).
Yang, Z. et al. miR-3283-p enhances the radiosensitivity of osteosarcoma and regulates apoptosis and cell viability via H2AX. Oncol. Rep. 39, 545–553 (2018).
Li, Q. C. et al. miR-34a increases cisplatin sensitivity of osteosarcoma cells in vitro through up-regulation of c-Myc and Bim signal. Cancer Biomark. 21, 135–144 (2017).
Sekar, D. et al. Dissecting the functional role of microRNA 21 in osteosarcoma. Cancer Gene Ther. 26, 179–182 (2019).
Monterde-Cruz, L. et al. Circulating miR-215-5p and miR-642a-5p as potential biomarker for diagnosis of osteosarcoma in Mexican population. Hum Cell. 31, 292–299 (2018).
Mitra, P. Transcription regulation of MYB: a potential and novel therapeutic target in cancer. Ann. Transl. Med. 6, 443 (2018).
Ramsay, R. G. & Gonda, T. J. MYB function in normal and cancer cells. Nat. Rev. Cancer. 8, 523–534 (2008).
Rupaimoole, R. & Slack, F. J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 16, 203–222 (2017).
Ali, S. Z. et al. Regulatory mechanism of MicroRNA expression in cancer. Int. J. Mol. Sci. 21, 1723 (2020).
Isakoff, M. S. et al. Osteosarcoma: Current treatment and a collaborative pathway to success. J. Clin. Oncol. 33, 3029–3035 (2015).
Hata, A. & Kashima, R. Dysregulation of microRNA biogenesis machinery in cancer. Crit. Rev. Biochem. Mol. Biol. 51, 121–134 (2016).
Hu, J. Y. et al. miR-601 is a prognostic marker and suppresses cell growth and invasion by targeting PTP4A1 in breast cancer. Biomed. Pharmacother. 79, 247–253 (2016).
Cao, W. et al. Identification of miR-601 as a novel regulator in the development of pancreatic cancer. Biochem. Biophys. Res. Commun. 483, 638–644 (2017).
Wang, Q. et al. Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer. Plos One. 7, e44398 (2012).
Ghelli, L. D. R. A. et al. A WEE1 family business: regulation of mitosis, cancer progression, and therapeutic target. J. Hematol. Oncol. 13, 126 (2020).
Liu, L. et al. PKMYT1 promoted the growth and motility of hepatocellular carcinoma cells by activating beta-catenin/TCF signaling. Exp. Cell Res. 358, 209–216 (2017).
Zhang, Q. Y., Chen, X. Q., Liu, X. C. & Wu, D. M. PKMYT1 promotes gastric cancer cell proliferation and apoptosis resistance. Onco. Targets Ther. 13, 7747–7757 (2020).
Chayka, O., D’Acunto, C. W., Middleton, O., Arab, M. & Sala, A. Identification and pharmacological inactivation of the MYCN gene network as a therapeutic strategy for neuroblastic tumor cells. J. Biol. Chem. 290, 2198–2212 (2015).
Xuan, Z. H. et al. PKMYT1 aggravates the progression of ovarian cancer by targeting SIRT3. Eur. Rev. Med. Pharmacol. Sci. 24, 5259–5266 (2020).
Zhang, Q. et al. Overexpressed PKMYT1 promotes tumor progression and associates with poor survival in esophageal squamous cell carcinoma. Cancer Manag. Res. 11, 7813–7824 (2019).
Fry, E. A. & Inoue, K. c-MYB and DMTF1 in cancer. Cancer Invest. 37, 46–65 (2019).
Zeng, Z. et al. LncRNA-MTA2TR functions as a promoter in pancreatic cancer via driving deacetylation-dependent accumulation of HIF-1α. Theranostics. 9, 5298–5314 (2019).
Deng, S. et al. Nutrient stress-dysregulated antisense lncRNA GLS-AS impairs GLS-mediated metabolism and represses pancreatic cancer progression. Cancer Res. 79, 1398–1412 (2019).
Acknowledgements
The research was funded by the Science and Technology Plan of Nanshan District of Shenzhen City, China (Grant number: No. 2020113 (Normal)), the project of Huazhong University of Science and Technology Union Shenzhen Hospital (NY2021024) and the project of Shenzhen Nanshan District Science and Technology (NS2021145).
Author information
Authors and Affiliations
Contributions
Designed and supervised the research: C.J.; Performed cellular experiments: P.L., J.F.; Conducted animal experiments: H.C., F.H; Provided the clinical data: P.L., J.F., S.X., H.L., S.Z., J.L.; Analyzed the data: P.L., J.F.; Wrote the manuscript: P.L.. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Luo, P., Fang, J., Chen, H. et al. c-Myb-mediated inhibition of miR-601 in facilitating malignance of osteosarcoma via augmentation of PKMYT1. Sci Rep 12, 6692 (2022). https://doi.org/10.1038/s41598-022-10684-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-10684-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.