Abstract
Breast cancer is a heterogenous disease that can be classified into multiple subtypes including the most aggressive basal-like and triple-negative subtypes. Understanding the heterogeneity within the normal mammary basal epithelial cells holds the key to inform us about basal-like cancer cell differentiation dynamics as well as potential cells of origin. Although it is known that the mammary basal compartment contains small pools of stem cells that fuel normal tissue morphogenesis and regeneration, a comprehensive yet focused analysis of the transcriptional makeup of the basal cells is lacking. We used single-cell RNA-sequencing and multiplexed RNA in-situ hybridization to characterize mammary basal cell heterogeneity. We used bioinformatic and computational pipelines to characterize the molecular features as well as predict differentiation dynamics and cell–cell communications of the newly identified basal cell states. We used genetic cell labeling to map the in vivo fates of cells in one of these states. We identified four major distinct transcriptional states within the mammary basal cells that exhibit gene expression signatures suggestive of different functional activity and metabolic preference. Our in vivo labeling and ex vivo organoid culture data suggest that one of these states, marked by Egr2 expression, represents a dynamic transcriptional state that all basal cells transit through during pubertal mammary morphogenesis. Our study provides a systematic approach to understanding the molecular heterogeneity of mammary basal cells and identifies previously unknown dynamics of basal cell transcriptional states.
Similar content being viewed by others
Introduction
The mammary gland contains an epithelial bilayer of basal and luminal cells that function in the production and secretion of milk from mother to offspring1,2. Multiple stem and progenitor cell populations have been identified in the basal and luminal layers, and the basal layer harbors multipotent stem cells that are capable of generating both basal and luminal progenies and reconstituting a functional mammary gland upon transplantation3,4,5. Distinct, small pools of stem cells in the basal layer have been identified by the expression of different markers such as Procr6, Bcl11b7, Lgr5/Tspan88, Cdh59, as well as by lineage-tracing Axin2-expressing cells10. However, basal cell heterogeneity in the normal mammary gland has been understudied despite knowledge of the existence of such stem cells. In particular, it is not clear how non-stem basal cells are transcriptionally organized or what their specific function and differentiation status might be.
Recent studies have utilized single-cell RNA-sequencing (scRNA-seq) to examine the normal mammary gland at single-cell resolution in both human11 and mouse9,12,13. These studies have provided foundational insights into the transcriptional landscape of the mammary epithelium. Other studies have also coupled transcriptional and epigenetic modalities at single-cell resolution to help uncover regulators of cellular identity within the mammary epithelium13,14. While these studies have advanced the understanding of the mammary epithelium, there still lacks a comprehensive and in-depth characterization of cellular states and differentiation landscape within the basal layer.
In this work, we aimed to characterize the transcriptional heterogeneity of mammary basal cells using scRNA-seq. Using differential gene expression analysis, we find four major basal cell transcriptional states. We provide a bioinformatic characterization of the molecular features of the cell states that suggest differential functional and metabolic activities. Using RNAScope, we validate the heterogeneous expression and differential enrichment of several scRNA-seq-identified basal transcriptional state markers, Acta2, Tspan8, and Egr2, in the intact mammary gland tissue. Lastly, we provide genetic cell labeling data to suggest that Egr2-expressing transcriptional state represents a dynamic one that all basal cells transit through during pubertal mammary morphogenesis. These findings regarding basal cell heterogeneity in the normal tissue lay the foundation for future work to probe the heterogeneity of their malignant counterparts in basal-like and triple negative breast cancer subtypes.
Methods
Mice
Wild-type C57BL/6 mice (Stock #000,664) and ROSA26mTmG mice (Stock #007,576) were purchased from the Jackson Laboratory. Egr2-Cre mice were reported in a previous study15, and the following primers were used for genotyping:
Forward (Egr2-Cre): CGC TTC CTC GTG CTT TAC GGT AT (480-bp product);
Forward (WT): TCA TCA GTC GGG TTA GAG CTG (312-bp product);
Reverse: GGG CTG AGG AAG ACG ACT TTA.
Egr2-Cre;ROSA26mTmG mice used for experimental analysis were generated by crossing Egr2-Cre males with ROSA26mTmG females. Crossing Egr2-Cre;ROSA26mTmG males with WT females resulted in 100% GFP expression in the mammary epithelium of female Egr2-Cre;ROSA26mTmG offspring (data not shown), suggesting germline expression of Egr2. Mice were maintained in a pathogen-free facility, following procedures that conform and have been approved by the University of California Irvine Institutional Animal Care and Use Committee.
Flow cytometry
Flow cytometry was performed as described16,17. Briefly, mammary glands were isolated, minced with a razor blade, and incubated with 300 U/mL collagenase (Sigma, C9891) and 100 U/mL hyaluronidase (Sigma, H3506) for 90 min at 37 °C. The cells were treated with red blood cell lysis buffer (Sigma, R7757) for 5 min at room temperature before treating with 0.25% Trypsin (Gibco, 25,200) for 2 min at 37 °C and dispase II (2 mg/mL) (Stem Cell Technologies, 07,913) with 0.1 mg/mL DNase I (Sigma, DN25) for 2 min at 37 °C. Cells were stained for 30 min in the dark at room temperature using APC-CD45 (BD Biosciences, 559,864; 1:250), APC-CD31 (BD Biosciences, 551,262; 1:250), APC-Ter119 (BD Biosciences, 557,909; 1:250), PE/C7-Epcam (Bio Legend, 118,215; 1:250), and PerCP-Cy5.5-Cd49f (Bio Legend, 555,735; 1:250). Cells were washed and stained with Sytox blue (Invitrogen, S34857; 1:1000) before flow cytometry.
scRNA-seq analysis
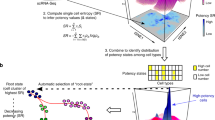
scRNA-seq experiments were performed in two separate runs using the 10X Genomics platform on FACS-sorted mammary epithelial cells from 8–9-week-old virgin females. Experimental details were as described in another study18. The sequencing data were analyzed using R version 3.6.1 and Seurat 3.1.0. Low quality cells were filtered based on mitochondrial DNA content, total number of transcripts, and total number of genes detected. Basal cells (Krt14+ cluster) were computationally isolated for further analysis. STRING version 11.0 was used to examine the protein–protein interactions using the marker genes identified by differential expression test using Seurat. Disconnected nodes were removed from the graphs, and the default interaction score (medium confidence; 0.4) was used to identify interactions. RNA Velocity analyses were performed using the R package velocyto.R (linear model) and nlvelo (non-linear model). The R package CellChat was used for analyzing the ligand-receptor communications of cells.
RNAScope
RNAScope was performed as previously described19 using ACD Bio’s reagents. Briefly, #4 mammary glands from mice were frozen in OCT and cryosectioned at 10 µm. Sectioned tissues were fixed for 1 h at room temperature with 4% PFA, and the RNAScope Multiplex Fluorescent v2 assay was run per manufacturer’s recommendations using the following probes: Acta2 (319,531-C3), Tspan8 (842,941-C1), and Egr2 (497,871-C2). Images were obtained using a Zeiss LSM700 confocal microscope and quantified using FIJI software.
Whole-mount immunofluorescence imaging
Mammary glands (#4) of 3-week-old Egr2-Cre;ROSA26mTmG and ROSA26mTmG mice were surgically isolated from adjacent tissues and spread on glass slides. Images were acquired using a Keyence BZ-X710 microscope (Keyence Corporation, Itasca, Illinois, USA).
Ethics and approval and consent to participate
All mouse experiments have been approved by and conform to the regulatory guidelines of the Institutional Animal Care and Use Committee of the University of California, Irvine. The study is reported in accordance with ARRIVE guidelines.
Results
Identification of four basal cell transcriptional states in adult virgin mouse mammary gland
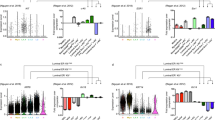
Previously, we performed scRNA-seq analysis on fluorescence-activated cell sorting (FACS)-isolated mammary epithelial cells (including both basal and luminal populations) from 8–9-week-old virgin females to characterize the heterogeneous expression of epithelial-to-mesenchymal transition (EMT)-associated genes18. However, a systematic characterization of the transcriptional diversity of basal cells has not been done in that study. To achieve this, we computationally subset the 3,651 cells that express basal cell marker Krt14 from the dataset for further analysis. Visualizing these basal cells in a UMAP projection suggests that there are no obvious batch effects (Fig. 1A). Clustering and differential gene expression analyses revealed the presence of four clusters: (1) a cluster enriched for classical myoepithelial genes (Acta2, Actg2) and thus termed “myoepithelial”; (2) a Tspan8High cluster enriched for genes the high expression of which has been previously shown to identify stem cells in the mammary basal compartment (e.g. Tspan88, Epcam20); (3) an Egr2High cluster enriched for early response- and stress-related genes (e.g. Egr2, Fos, Jun)21; and (4) a small cluster marked by proliferation-associated genes (e.g., Mki67, Pcna) (Fig. 1B–D; Supplemental Table 1). We also observed the same four cell states when each mouse was analyzed individually (Fig. S1A–D), and the marker genes used to identify each cell state in the mice showed a high degree of overlap (Fig. 1C). Importantly, the marker genes that discriminated the different transcriptional states were differentially expressed but not mutually exclusive (Fig. 1E).
Identification of four major transcriptional states in basal cells of adult virgin mammary gland. (A) UMAP projection of mammary basal cells colored by mouse. (B) UMAP projection as in (A) colored by transcriptional state. (C) Venn diagram showing overlap in marker genes for the transcriptional states identified in both mice. Each circle represents the number of marker genes for a particular transcriptional state [same color code as in (B)] in each mouse. (D) Heatmap of top 10 marker genes for each transcriptional state identified in (B). (E) Boxplots of the indicated marker genes (marked in red in D) across all four basal transcriptional states.
To examine the robustness of these transcriptional states, we also sequenced FACS-sorted mammary epithelial cells from 8–9-week-old virgin mice deficient in EMT-inducing transcription factor Zeb118 and computationally subset basal cells for further analysis (Fig. S1E, F). In addition to a proliferating basal cell population, three other basal transcriptional states were observed in each mutant mouse analyzed (Fig. S1G–J). The top markers of each of these transcriptional states were largely similar, albeit not identical, to those in the wild-type mice. Therefore, even in Zeb1-deficient mammary glands where basal stem cell function is compromised18, similar molecular heterogeneity of bulk basal cells still exists, featuring four major transcriptional states.
We also interrogated the published scRNA-seq data on KRT14-expressing cells of the human breast from three individuals11. Generally, top marker genes for the three “non-proliferating” mouse basal cell states were detected at much lower frequencies in these human samples (Supplemental Table 2). For example, Acta2, which has been previously reported to be expressed in > 96% of mouse mammary basal cells20, was detected in 99.6% of the mouse basal cells in our scRNA-seq analysis but ACTA2 was only detected in 38.6% of the human KRT14+ cells. Moreover, Egr2 was expressed in 29.8% of mouse basal cells, but EGR2 was only detected in 5.6% of human KRT14+ cells. Nevertheless, we were still able to observe transcriptional states in the human KRT14+ population that resembled the “myoepithelial” and proliferating basal cell states in mouse (Fig. S1K–M). These data reveal both disparities and similarities in the transcriptional states of mammary basal cells between mouse and human.
Molecular features of the basal cell states
Next, we sought to characterize the molecular features of the basal cell states in mice. Intrigued by their upregulated expression of known stemness-associated genes Tspan8 and Epcam, we wondered if Tspan8High cells also display enriched expression of other genes reported to mark mammary basal/stem cells (e.g. Procr6, Bcl11b7, Cdh59, Lgr58). Using a gene scoring approach to calculate the average expression of a “stemness gene” signature (Supplementary Table 3), we found that the Egr2High basal cells had the highest average score whereas the proliferating basal cells the lowest (Fig. 2A). Overall, the discriminating power of this signature is non-remarkable.
Molecular features of the basal transcriptional states. (A–C) Boxplots displaying gene scoring of each basal transcriptional state using gene signatures for mammary stemness (A), glycolysis (B), and OxPhos (C). p values in (A-C) were generated using Mann–Whitney U tests. (D) PPIs for each cell state (myoepithelial, pink; Tspan8High, green; Egr2High, blue; proliferating, purple), where each node is protein coded by the marker gene and each edge is a predicted direct or indirect interaction. (E) Bar plots displaying the number of PPIs for each transcriptional state. (F–G) Projections of RNA Velocity fields onto UMAP from Fig. 1B using linear (F) and non-linear (G) models. (H–I) Dot plots indicating the outgoing (H) and incoming (I) signaling contributions from each transcriptional state for significant signaling pathways identified. (J) Signaling pattern of non-canonical WNT signaling.
Differential metabolic preference of mammary basal and luminal cells has been suggested, such that basal cells may prefer a glycolytic metabolism while luminal cells display increased oxidative phosphorylation22,23. This metabolic paradigm is of interest, given that cancerous cells have increased glycolytic activity24. Using a set of hallmark gene sets defined by the Molecular Signatures Database to score for glycolysis and oxidative phosphorylation (OxPhos) (Supplementary Table 3), we found that the proliferating basal cells have the highest glycolytic and OxPhos signatures (Fig. 2B,C), suggesting increased requirement for energy to support cellular growth and division. Of the “non-proliferating” basal cell states, the Egr2High cells scored the highest for the glycolytic signature, and the “myoepithelial” cells scored the highest for OxPhos. Interestingly, the Tspan8High cells scored the lowest for both signatures. These data reveal previously unrecognized metabolic heterogeneity within the mammary basal layer.
To gain further insights into the potential activity of each basal transcriptional state, we examined the protein–protein interaction (PPI) networks of all of the marker genes that define each state. We utilized STRING25 to derive direct (physical) and indirect (functional) associations between the marker genes. Interestingly, the number of PPIs were vastly different across the cell states, with proliferating basal cells exhibiting the highest number of PPIs and the highest number of marker genes (Fig. 2D,E; Fig. S2A), which is consistent with their highly specialized cellular activity. Although the difference in the number of marker genes for each “non-proliferating” cell state was small, the difference in the number of PPIs was more pronounced (Fig. 2E; Fig. S2A). For example, the numbers of PPIs in the Tspan8High and “myoepithelial” cells were 15 (lowest of all four states) and 113 (second highest), respectively, whereas their numbers of unique marker genes were 31 (lowest) and 39 (second lowest). This data suggest that the marker genes used to define the “myoepithelial” cells have a higher degree of physical and functional associations and may point to a functional consequence. Consistently, a gene ontology (GO) term analysis using Enrichr26,27 to probe the GO Biological Processes 2018 library revealed terms related to muscle contraction for the “myoepithelial” marker genes (Fig. S2B).
Next, we performed RNA Velocity28, a computational method that calculates the relative abundances of spliced and unspliced RNA to infer the future states of single cells. Based on the directions of the vectors arrows, which are known to associate with possible state transitions, we did not observe a clear differentiation trajectory across the different basal cell states regardless of whether we used a linear28 (Fig. 2F) or non-linear model19 (Fig. 2G) of RNA Velocity. However, we found consistent differences in the length of the vector arrows that suggest differential RNA dynamics among the different states. The “myoepithelial” and Tspan8High cells showed small RNA velocities (short or no arrows), known to associate with either quiescent or terminally differentiated cells29,30. The Egr2High cells exhibited large RNA velocities in both linear and non-linear models (Fig. 2F,G), suggesting that these cells may be in a more active and transitional cellular state compared to the others. The proliferating cells exhibited large RNA velocities in the linear but not in the perhaps more realistic non-linear model19, and in both cases, the arrows pointed away from but not back to the “non-proliferating” cells (Fig. 2F,G), raising the possibility that they may not be able to readily switch back to a “non-proliferating” state19.
Lastly, we used CellChat31 to explore ligand-receptor pairs and infer potential signaling cross-talks within the basal cell layer. CellChat identified ten signaling pathways that were significantly enriched within basal cells, and outgoing and incoming signals were largely heterogeneous across the cell states (Fig. 2H,I). It appeared that the Tspan8High cells send the most outgoing signals, and the “myoepithelial” cells receive the most incoming signals. Interestingly, non-canonical WNT (ncWNT) signaling surfaced as the most prominent in “myoepithelial” cells, which appeared to be the primary cells responding to WNT signals from the Tspan8High cells (i.e., paracrine) and, to a lesser extent, from the “myoepithelial” cells themselves (i.e., possibly autocrine) (Fig. 2J).
Collectively, our findings suggest that the Tspan8High transcriptional state associates with low number of marker genes, low PPIs, low glycolysis, slow RNA dynamics, but can potentially serve as a signaling niche32, whereas the Egr2High transcriptional state is the most dynamic of all states and the “myoepithelial” transcriptional state represents the most specialized state associated with a “mature” myoepithelial fate.
Validating basal transcriptional heterogeneity and detecting dynamic Egr2 expression in the intact mammary tissue
We next sought to examine the molecular heterogeneity in bulk basal cells in the intact mammary tissue by using RNAScope to validate the differential expression of a top marker gene from each “non-proliferating” basal transcriptional state. Specifically, we probed for the expression of Acta2, Tspan8, and Egr2 mRNAs simultaneously in mammary gland of adult virgin mice. Indeed, expression of each transcriptional state marker gene was detected in only a subset of basal cells (Fig. 3A–E; Fig. S3A–H). There did not appear to be any notable spatial patterning of these transcriptional state markers, except that a small number of Tspan8+ cells that are positive for K14 (indicating a basal cell fate2,33) seem to occupy positions between basal and luminal layers (Fig. 3A-D; Fig. S3A–H). Consistent with scRNA-seq data, Acta2 showed the highest coverage in basal cells (~ 39%) (Fig. 3E). This is less than the 99% detection rate in our scRNA-seq data, likely reflecting differential sensitivity of different detection methods. Tspan8 transcripts were detected in ~ 23% of the basal cells (Fig. 3E), and its most abundant expression was actually seen in the luminal layer, a finding confirmed by our scRNA-seq data (Fig. S3I). Egr2 mRNAs were detected in ~ 27% of the basal cells (Fig. 3E) and expression was barely detectable in luminal cells, and this basal-restricted pattern was also seen in our scRNA-seq data (Fig. S3J). Hierarchically clustering basal cells by their expression (number of RNAScope signal foci/per nucleus) of the marker genes revealed groups of single basal cells that expressed a single marker gene (Fig. 3E). Overall, only a small fraction of basal cells (~ 3%) expressed all three markers, and a sizable fraction (~ 35%) of cells did not exhibit detectable expression of any of the makers examined (Fig. 3A′–D′,A″–D″,E). Together, these data uncover the scRNA-seq-predicted transcriptional heterogeneity in the basal compartment of the intact mammary tissue.
RNAScope validation of differential expression of several basal transcriptional state markers. (A–D) Representative 5-channel (A) and 3-channel (B–D) images showing the expression of Acta2 (red), Tspan8 (green), Egr2 (white) in the mammary gland of 10-week-old virgin females. K14 protein immunostaining (blue in all images) marks the basal cells. DAPI stains the nuclei (artificially colored yellow in all images). Red, green, and white arrows indicate a basal cell with only Acta2, Tspan8, and Egr2 expression, respectively. Green arrowhead indicates a Tspan8-expressing K14-positive cell that resides in basal-luminal border. Boxed areas in (A) are enlarged in (Aʹ–Dʹ) and (Aʺ–Dʺ) to highlight basal cells with more than one marker detected (indicated by yellow arrow). Scale bar = 50 μm in (A–D); 10 μm in (Aʹ–Dʹ) and (Aʺ–Dʺ). (E) Quantification results for experiments as in (A-D). Table summarizing the numbers of single, double, and triple-positive cells is shown at the top, and heatmap with hierarchical clustering using the quantified expression of probes shown at the bottom.
Expression of early-response genes such as Egr2 has been reported as an artifact of stresses induced by the tissue dissociation/sequencing protocol21, but our detection of Egr2-expressing basal cells in the mammary tissue suggests the physiological existence of an Egr2High basal transcriptional state. To probe this further, we performed RNAScope on mammary gland of different reproductive stages using the Egr2 probe alone. In adult virgin gland, a significant enrichment of Egr2 foci was again observed in the basal layer relative to the luminal layer, with Egr2 mRNA being detected in ~ 28% of basal cells but in only ~ 2% of luminal cells (Fig. 4A–C). Basal-enriched Egr2 expression was also abundantly detected in the mammary ducts of pregnant mice at day 3 of pregnancy (Fig. 4D). By day 15 of pregnancy, Egr2-expressing cells became less frequent in the ductal basal compartment, and were barely detectable in the alveolar basal compartment (Fig. 4E,F). We also probed the expression of Egr2 using previously published microarray data on whole mammary gland across different stages of the reproductive cycle34, which revealed significantly upregulated Egr2 expression during puberty (5–6 weeks of age) and early pregnancy (day 3) but very low expression during late pregnancy (day 17–19), lactation, and involution (Fig. 4G,H). Taken together, these data demonstrate physiologic and dynamic expression of Egr2 in the mammary gland that coincides with periods of active epithelial morphogenesis and ductal expansion.
Dynamic Egr2 mRNA expression in mammary gland. (A–D) RNAScope detection of Egr2 expression (green) in mammary gland from 10-week-old virgin mice. K14 antibody (red) stains the base cells and DAPI (blue) stains the nuclei. Representative image of a single mammary duct is shown in (A), and result of quantitative analysis (using multiple ducts as indicated) of the frequency of Egr2-expressing basal or luminal cells is shown in (B). (C) Boxplots displaying the number of Egr2 mRNA (green) foci per cell in basal or luminal cells. (D–F) RNAScope detection of Egr2 mRNA (white) in mammary gland from 3-day (D) or 15-day (E, F) pregnant mice. Scale bar = 50 μm in (A), (D–F). White arrows indicate positive Egr2 signal. (G–H) Bar plots indicating the expression of Egr2 in whole mammary gland across the reproductive cycle using previously published data (GEO accession # for G: GSE6453; accession # for H: GSE8191). p values in (C) and (G–H) were generated using a Student’s t-test.
Genetic evidence for basal-biased expansion of Egr2-expressing cells during pubertal mammary gland development
Egr2 is of interest because its expression marks a population of actively expanding hair follicle progenitor cells15 and it encodes a transcription factor that regulates the expression of Notch1, a critical gene involved in mammary basal-luminal binary differentiation35. To track Egr2-expressing cells in vivo, we crossed Egr2-Cre (Krox20-Cre) mice15 with ROSA26mTmG reporter mice to generate Egr2-Cre; ROSA26mTmG females, where Egr2-expressing cells and their progenies are fluorescently marked by GFP expression (Fig. 5A). To visualize the spatial location of Egr2-expressing cells and progenies, we performed whole-mount imaging analysis of GFP and tdTomato fluorescence in mammary gland of 3-week-old Egr2-Cre; ROSA26mTmG mice. While no GFP+ cells were detected in the ROSA26mTmG control mice as expected, such cells were found throughout the rudimentary ducts as well as in terminal end buds of Egr2-Cre; ROSA26mTmG mice (Fig. 5B).
Genetic labeling of Egr2-expressing cells and progenies in mammary gland. (A) Experimental design. (B) Whole-mount imaging of GFP and tdTomato fluorescence in mammary gland from 3-week-old Egr2-Cre;ROSA26mTmG or ROSA26mTmG mice. Scale bar = 500 μm. (C, D) Representative flow profiles (C) and summary bar plots (D) displaying the number of GFP-positive cells in basal and luminal cells during pubertal mammary development. p value was generated using a Student’s t-test. n = 3 Egr2-Cre;ROSA26mTmG mice per age analyzed.
Flow cytometry analysis using GFP in conjunction with cell lineage surface markers (e.g., Cd49f, Epcam) revealed that 15–27% of the basal cells in mammary gland from 3-week-old (pre-puberty) Egr2-Cre; ROSA26mTmG females were GFP+ (Fig. 5C,D). By mid-puberty (6.5 weeks of age) and in adulthood (10 weeks of age), the number of GFP+ basal cells dramatically increased to near 100% of all basal cells (Fig. 5C,D). The surface expression levels of Cd49f and Epcam in GFP+ and GFP- basal cells were similar (Fig. S4A). At all ages examined, less than 10% of the luminal cells showed GFP expression (Fig. 5C,D). Taken together with the scRNA-seq and RNAScope findings above that fewer than a third of the basal cells in adult virgin gland showed detectable Egr2 mRNA expression, these flow data suggest that virtually all basal cells in the adult gland had transited through an Egr2-expressing state at some point during pubertal mammary gland development and/or are derived from Egr2-expressing basal cells. Further illustrating the dynamic nature of Egr2 expression in basal cells, FACS-sorted GFP- basal cells from mammary glands of 3-week-old Egr2-Cre; ROSA26mTmG females were able to activate GFP expression de novo under organoid culture conditions, whereas GFP+ basal cells remained positive over multiple passages (Fig. S4B).
Discussion
To date, several scRNA-seq studies have been conducted on human and mouse mammary glands, each presenting a unique perspective on epithelial cellular composition, differentiation dynamics, and stem cell prediction. Previous studies have also delved into basal cell heterogeneity and the presence of rare stem cells in the basal layer6,7,9,10,36,37,38, but the gene expression program that underlies bulk basal cell dynamics and differentiation has not been clearly elucidated. Our study adds to the list of existing datasets and provides a deeper and comprehensive analysis of the transcriptional heterogeneity within mammary basal cells. Our analysis shows that bulk basal cells exist in at least four distinct transcriptional states that can be identified by their unique enrichment for the expression of specific gene signatures associated with potential stemness (e.g., Tspan8high state), differentiation status (e.g., mature myoepithelial state), and/or rapid cellular dynamics and responses (e.g., Egr2High and proliferating states). Intriguingly, our data does not point to a unidirectional, hierarchical differentiation trajectory originating from one state and ending in another. Instead, they suggest that mammary basal cells adopt dynamic non-hierarchical transcriptional states, with the exception that the proliferating state may not readily revert back to any of the “non-proliferating” state. This dynamics implies the inherent plasticity of any given basal cell, a notion that is consistent with the demonstrated functional plasticity of adult basal cells especially upon transplantation, and that mature myoepithelial cells possess regenerative stem cell activity that can manifest under appropriate conditions4,5,10,20. While it may be technically challenging to sort and purify the basal cell subsets in different transcriptional states due to overlapping expression of cell surface markers, future experiments to generate and analyze Cre-expressing alleles driven by temporally controlled promoters of genes encoding select basal transcriptional state markers will enable lineage tracing of the fates and activities of basal cells in each state during mammary development, regeneration, and tumorigenesis.
We were able to confirm the expression of several basal transcriptional state markers in the intact mammary gland using in situ mRNA detection and found them to be largely non-overlapping albeit not mutually exclusive. It has been reported that nearly all basal cells in the mammary gland, including stem cells, express Acta2 at the mRNA and protein levels20. Our scRNA-seq data support this notion by revealing that > 99% of basal cells show detectable expression of Acta2. This said, we found that a fraction of them show heightened expression of Acta2 and other myoepithelial-related genes; these are likely the same cells in which Acta2 expression was over the detection threshold of RNAScope analysis and they may represent mature myoepithelial cells. The Egr2High transcriptional state is of particular interest because of its faster cellular dynamics relative to the other states. Our RNAScope experiments detected the mRNA expression of Egr2 in a subset of basal cells in the intact tissue. Moreover, in scRNA-seq, single-probe, and multi-probe RNAScope experiments, we observed remarkable consistency in the precise frequency (27–30%) of Egr2-positive cells, indicating robustness of the observation.
In the hair follicle, another leading model of adult stem cell biology, Egr2 expression marks the matrix cells, which are highly proliferative but transit-amplifying progenitor cells that terminally differentiate to produce the hair shaft15. In the mammary gland, such transient amplifying progenitor cells remain elusive. Our proof-of principle data based on Egr2-Cre-mediated GFP fluorescence is consistent with the possibility that Egr2-expressing mammary basal cells are such progenitor cells and serve as the workhorse during pubertal mammary gland development to generate nearly the entire basal cell compartment of mature gland. However, an alternative, and perhaps more likely based on our RNA Velocity and organoid culture data, possibility is that Egr2High is simply a transcriptional state that all basal cells transit through at some point during pubertal mammary development. It will be worthwhile to generate Egr2-CreER mice in the future to seek definitive evidence for the function and fate of the Egr2+ basal cell subset during mammary gland development, regeneration, and tumorigenesis.
Overall, our study provides a systematic analysis of mammary basal cell heterogeneity and a useful reference for future investigation into how basal cell gene expression changes during breast cancer initiation and progression. A thorough understanding of the transcriptional heterogeneity of normal mammary basal cells and their malignant counterparts might be particularly relevant in the development of differentiation therapies for basal-like and triple negative breast cancers.
Conclusions
Our results have identified four major transcriptional states within the mammary basal cells that exhibit gene expression signatures suggestive of different functional activity and metabolic preference. Our in vivo data suggest that one of these transcriptional states, marked by Egr2 mRNA expression, represents an actively expanding and/or obligatory transitional state during pubertal mammary morphogenesis. These findings regarding basal cell heterogeneity in the normal tissue lay the foundation for future work to probe the heterogeneity of the malignant counterparts in basal-like and triple negative breast cancer subtypes.
Data availability
The single-cell datasets used and analyzed during the current study are available under Accession #GSE155636 (https://www.ncbi.nlm.nih.gov/gds). Code will be provided upon request. All other data generated or analyzed during this study are included in this published article and its supplementary information files. All relevant information about materials used in the study is provided in the Methods section of the text.
Abbreviations
- FACS:
-
Fluorescence activated cell sorting
- GO:
-
Gene ontology
- Myo:
-
Myoepithelial
- PPI:
-
Protein–protein interaction
- scRNA-seq:
-
Single-cell RNA-sequencing
- UMAP:
-
Uniform manifold approximation projection
References
Macias, H. & Hinck, L. Mammary gland development. Rev. Dev. Biol. https://doi.org/10.1002/wdev.35 (2012).
Inman, J. L., Robertson, C., Mott, J. D. & Bissell, M. J. Mammary gland development: Cell fate specification, stem cells and the microenvironment. Development (Cambridge) https://doi.org/10.1242/dev.087643 (2015).
Faulkin, L. J. & Deome, K. B. Regulation of growth and spacing of gland elements in the mammary fat pad of the C3H mouse. J. Natl. Cancer Inst. 24, 953–969 (1960).
Shackleton, M. et al. Generation of a functional mammary gland from a single stem cell. Nature https://doi.org/10.1038/nature04372 (2006).
Stingl, J. et al. Purification and unique properties of mammary epithelial stem cells. Nature https://doi.org/10.1038/nature04496 (2006).
Wang, D. et al. Identification of multipotent mammary stemcells by protein C receptor expression. Nature https://doi.org/10.1038/nature13851 (2015).
Cai, S. et al. A quiescent Bcl11b high stem cell population is required for maintenance of the mammary gland. Cell Stem Cell https://doi.org/10.1016/j.stem.2016.11.007 (2017).
Fu, N. Y. et al. Identification of quiescent and spatially restricted mammary stem cells that are hormone responsive. Nat. Cell Biol. https://doi.org/10.1038/ncb3471 (2017).
Sun, H. et al. Single-cell RNA-Seq reveals cell heterogeneity and hierarchy within mouse mammary epithelia. J. Biol. Chem. https://doi.org/10.1074/jbc.RA118.002297 (2018).
Van Amerongen, R., Bowman, A. N. & Nusse, R. Developmental stage and time dictate the fate of Wnt/β-catenin- responsive stem cells in the mammary gland. Cell Stem Cell https://doi.org/10.1016/j.stem.2012.05.023 (2012).
Nguyen, Q. H. et al. Profiling human breast epithelial cells using single cell RNA sequencing identifies cell diversity. Nat. Commun. https://doi.org/10.1038/s41467-018-04334-1 (2018).
Pal, B. et al. Construction of developmental lineage relationships in the mouse mammary gland by single-cell RNA profiling. Nat. Commun. https://doi.org/10.1038/s41467-017-01560-x (2017).
Giraddi, R. R. et al. Single-cell transcriptomes distinguish stem cell state changes and lineage specification programs in early mammary gland development. Cell Rep. 24, 1653-1666.e7 (2018).
Pervolarakis, N. et al. Integrated single-cell transcriptomics and chromatin accessibility analysis reveals regulators of mammary epithelial cell identity. Cell Rep. 33, 10058 (2020).
Liao, C. P., Booker, R. C., Morrison, S. J. & Le, L. Q. Identification of hair shaft progenitors that create a niche for hair pigmentation. Genes Dev. https://doi.org/10.1101/gad.298703.117 (2017).
Gu, B., Watanabe, K., Sun, P., Fallahi, M. & Dai, X. Chromatin effector Pygo2 mediates wnt-notch crosstalk to suppress luminal/alveolar potential of mammary stem and basal cells. Cell Stem Cell 13, 48–61 (2013).
Watanabe, K. et al. Mammary morphogenesis and regeneration require the inhibition of EMT at terminal end buds by ovol2 transcriptional repressor. Dev. Cell 29, 59–74 (2014).
Han, Y. et al. Coordinate control of basal epithelial cell fate and stem cell maintenance by core EMT transcription factor Zeb1. Cell Rep. 38, 110240 (2022).
Haensel, D., Jin, S., Gratton, E., Nie, Q. & Correspondence, X. Defining epidermal basal cell states during skin homeostasis and wound healing using single-cell transcriptomics. Cell Rep. 30, 3932-3947.e6 (2020).
Prater, M. D. et al. Mammary stem cells have myoepithelial cell properties. Nat. Cell Biol. https://doi.org/10.1038/ncb3025 (2014).
Van Den Brink, S. C. et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14, 935–936 (2017).
Mahendralingam, M. et al. Mammary epithelial cells have lineage-restricted metabolic identities. biorxiv 79, 8173 (2019).
Casey, A. E. et al. Mammary molecular portraits reveal lineage-specific features and progenitor cell vulnerabilities. J. Cell Biol. 217, 2951–2974 (2018).
Heiden, M. G. V., Cantley, L. C. & Thompson, C. B. Understanding the warburg effect: The metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Szklarczyk, D. et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Chen, E. Y. et al. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 14, 1005 (2013).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Svensson, V. & Pachter, L. RNA velocity: Molecular kinetics from single-cell RNA-Seq. Mol. Cell 72, 7–9 (2018).
Zywitza, V., Misios, A., Bunatyan, L., Willnow, T. E. & Rajewsky, N. Single-cell transcriptomics characterizes cell types in the subventricular zone and uncovers molecular defects impairing adult neurogenesis. Cell Rep. 25, 2457-2469.e8 (2018).
Suoqin, J. et al. Inference and analysis of cell-cell communication using Cell Chat. bioRxiv. 2, 1008 (2011).
Hsu, Y. C., Pasolli, H. A. & Fuchs, E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell https://doi.org/10.1016/j.cell.2010.11.049 (2011).
Rios, A. C., Fu, N. Y., Lindeman, G. J. & Visvader, J. E. In situ identification of bipotent stem cells in the mammary gland. Nature https://doi.org/10.1038/nature12948 (2014).
Williams, M. M. et al. ErbB3 drives mammary epithelial survival and differentiation during pregnancy and lactation. Breast Cancer Res. https://doi.org/10.1186/s13058-017-0893-7 (2017).
Bouras, T. et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell https://doi.org/10.1016/j.stem.2008.08.001 (2008).
Liu, W., Wu, T., Dong, X. & Zeng, Y. A. Neuropilin-1 is upregulated by Wnt/β-catenin signaling and is important for mammary stem cells. Sci. Rep. https://doi.org/10.1038/s41598-017-11287-w (2017).
Soady, K. J. et al. Mouse mammary stem cells express prognostic markers for triple-negative breast cancer. Breast Cancer Res. https://doi.org/10.1186/s13058-015-0539-6 (2015).
Regan, J. L. & Smalley, M. J. Integrating single-cell RNA-sequencing and functional assays to decipher mammary cell states and lineage hierarchies. NPJ Breast Cancer. https://doi.org/10.1038/s41523-020-00175-8 (2020).
Acknowledgements
The authors would like to thank Lu Le’s lab for the Egr2-Cre mice, the Genomics High Throughput Facility, the Institute for Immunology FACS Core Facility, and the Optical Biology Core at UC Irvine for expert service.
Funding
This work was supported by NIH Grant R01-GM123731 and R01-AR068074 (X.D.). G.G. was supported by a diversity supplement (R01GM123731-02S2) and was a NSF-Simons Center for Multiscale Cell Fate Research predoctoral fellow through NSF Grants DMS1763272 and Simons Foundation Grant 594598 (Q.N.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
G.G. and X.D. conceived the study and designed the analysis. G.G. carried out the computational/bioinformatics analysis. G.G., P.S., and Y.H. performed the experiments. G.G. and X.D. wrote the manuscript, and P.S. and Y.H. read, edited, and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gutierrez, G., Sun, P., Han, Y. et al. Defining mammary basal cell transcriptional states using single-cell RNA-sequencing. Sci Rep 12, 4893 (2022). https://doi.org/10.1038/s41598-022-08870-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-08870-1
This article is cited by
-
Dependence of human cell survival and proliferation on the CASP3 prodomain
Cell Death Discovery (2024)
-
Gene expression in organoids: an expanding horizon
Biology Direct (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.