Abstract
Treatment of bacterial infections is one of the major challenges of our time due to the evolved resistance mechanisms of pathogens against antibiotics. To circumvent this problem, it is necessary to understand the mode of action of the drug and the mechanism of resistance of the pathogen. One of the most potent antibiotic targets is peptidoglycan (PGN) biosynthesis, as this is an exclusively occurring and critical feature of bacteria. Lipid II is an essential PGN precursor synthesized in the cytosol and flipped into the outer leaflet of the membrane prior to its incorporation into nascent PGN. Antimicrobial peptides (AMPs), such as nisin and colistin, targeting PGN synthesis are considered promising weapons against multidrug-resistant bacteria. However, human pathogenic bacteria that were also resistant to these compounds evolved by the expression of an ATP-binding cassette transporter of the bacitracin efflux (BceAB) type localized in the membrane. In the human pathogen Streptococcus agalactiae, the BceAB transporter SaNsrFP is known to confer resistance to the antimicrobial peptide nisin. The exact mechanism of action for SaNsrFP is poorly understood. For a detailed characterization of the resistance mechanism, we heterologously expressed SaNsrFP in Lactococcus lactis. We demonstrated that SaNsrFP conferred resistance not only to nisin but also to a structurally diverse group of antimicrobial PGN-targeting compounds such as ramoplanin, lysobactin, or bacitracin/(Zn)-bacitracin. Growth experiments revealed that SaNsrFP-producing cells exhibited normal behavior when treated with nisin and/or bacitracin, in contrast to the nonproducing cells, for which growth was significantly reduced. We further detected the accumulation of PGN precursors in the cytoplasm after treating the cells with bacitracin. This did not appear when SaNsrFP was produced. Whole-cell proteomic protein experiments verified that the presence of SaNsrFP in L. lactis resulted in higher production of several proteins associated with cell wall modification. These included, for example, the N-acetylmuramic acid-6-phosphate etherase MurQ and UDP-glucose 4-epimerase. Analysis of components of the cell wall of SaNsrFP-producing cells implied that the transporter is involved in cell wall modification. Since we used an ATP-deficient mutant of the transporter as a comparison, we can show that SaNsrFP and its inactive mutant do not show the same phenotype, albeit expressed at similar levels, which demonstrates the ATP dependency of the mediated resistance processes. Taken together, our data agree to a target protection mechanism and imply a direct involvement of SaNsrFP in resistance by shielding the membrane-localized target of these antimicrobial peptides, resulting in modification of the cell wall.
Similar content being viewed by others
Introduction
Bacterial infections cause over 150,000 deaths every year and are a major threat for humans1,2. The treatment of many infectious diseases is possible due to the development of antibiotics, which have been discovered over the last 100 years, starting with penicillin in 1929. In recent years, however, antibiotic resistance has become a major challenge, as pathogenic bacteria have evolved several resistance mechanisms against antibiotics in use3.
An Achilles heel of bacteria is the synthesis pathway of peptidoglycan (PGN), the main component of the cell wall4. PGN is a heteropolymeric layer that completely encloses the bacterial cell and provides the bacterial shape and integrity. The biosynthesis of PGN requires several steps, which are evolutionarily conserved in all bacterial species but are missing in eukaryotic cells5. Therefore, it is an optimal target for antibacterial agents.
PGN synthesis occurs in three distinctive compartments of the bacterial cell, namely, the cytoplasm, the cytoplasmic membrane, and the cell surface6: (1) In the cytoplasm, lipid II synthesis takes place; lipid II is a PGN precursor composed of an undecaprenyl pyrophosphate (UPP) anchor, the two amino sugars N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) and a covalently attached pentapeptide7. (2) Lipid II is afterwards flipped to the extracellular space (or periplasm for gram-negative bacteria) and is still anchored to the membrane via UPP8. (3) Following this, the GlcNAc-MurNAc-pentapeptide subunit is incorporated into the nascent PGN, leaving UPP attached to the membrane. UPP is subsequently dephosphorylated to undecaprenyl phosphate (UP), which is flipped back into the cytoplasm and implemented into a new PGN synthesis cycle9.
This biosynthetic pathway has been shown to be an ideal target for antimicrobial compounds at any stage of (1)–(3)4,10. In many cases, the incorporation of lipid II into the nascent PGN layer is prevented; antibiotics either bind directly to lipid II or to enzymes that catalyze its incorporation into PGN. Both types of binding lead to nonrecycling of UP and subsequent inhibition of lipid II synthesis. As a consequence, bacterial cell growth is hindered.
Binding of antibiotics occurs to various moieties of lipid II, e.g., to the pyrophosphate moiety (lantibiotics such as nisin and gallidermin)11,12,13 or to the pentapeptide moiety (glycopeptides such as vancomycin)10,14,15 (Fig. 1). A particular class are small antimicrobial peptides (AMPs) that bind specifically to the pyrophosphate-sugar moiety of lipid II11, such as the lipoglycodepsipeptide ramoplanin and the acylcyclodepsipeptide lysobactin10,16,17,18.
Schematic view of peptidoglycan synthesis. Synthesis of lipid II in the cytoplasm and its incorporation into the peptidoglycan. Phosphates are marked with a P, undecaprenyl as a black curved line, uridine phosphate (UDP) in light blue, GlcNAc in blue, MurNAc in red and amino acids of the pentapeptide in orange. Enlarged step of peptidoglycan synthesis showing targets of bacitracin, which is undecaprenyl pyrophosphate (green circle), and targets of many antimicrobial peptides, such as nisin, gallidermin, lysobactin, and ramoplanin: lipid II (violet, dotted circle). GlcNAc N-acetylglucosamine; MurNAc N-acetylmuramic acid. The figure was created using Microsoft Powerpoint Version 16.54.
In addition to lipid II binders, antibiotics are known to inhibit PGN biosynthesis at another stage, e.g., the cyclic peptide bacitracin. The binding of bacitracin to UPP inhibits the dephosphorylation of UP and blocks its regeneration, resulting in the accumulation of intracellular PGN precursors19,20,21 (Fig. 1). The net effect is the destabilization of the cell wall, leading to cell growth inhibition and subsequently to the death of the bacteria.
Bacitracin has been suggested to form a compact ternary 1:1:1 antibiotic-metal-lipid complex that, with its highly amphipathic structure, enhances membrane-binding affinity20. Due to a stabilizing effect and increased antimicrobial activity, a zinc-bacitracin (Zn-bacitracin) complex has been commonly used in human and veterinary medicine in antibiotic formulations22,23.
Bacitracin has also been used as a growth-promoting additive in animal feed24. Additionally, it has been shown to control necrotic enteritis effectively25 and is therefore used as a drug in many countries. Long-term usage of bacitracin in animals leads to an increase in resistance genes in microorganisms. Some molecular bacitracin resistance mechanisms have been reported in bacteria26,27,28,29.
One of the identified resistance mechanisms against AMP in human pathogens is based on the expression of bacitracin efflux (Bce) transporter, a member of the ABC transporter family. Bce confers high-level resistance to bacitracin and/or lantibiotics such as nisin and gallidermin in Bacillus subtilis, Staphylococcus aureus and Streptococcus agalactiae27,30,31,32,33,34. Genomic analysis revealed the presence of homologous transporters (BceAB-type transporters) mostly in bacteria predominantly found in soil and in human pathogenic bacteria35.
The first BceAB-type transporter was identified in B. subtilis. Adjacent to the bceAB genes, the bceRS genes are located, encoding a two-component system (TCS). The TCS regulates the expression of transporters27 and it is hypothesized that the detoxification against peptide antibiotics is functionally linked to it36. Status quo is that upon substrate binding the BceAB type transporter transfers a signal to the histidine kinase that then phosphorylates its cognate response regulator which induces the expression of the ABC transporter genes. This was described i.e. for the GraRS-VraFG system37 in S. aureus and also for several TCS-ABC transporters in B. subtilis (BceRS-AB, YxdJK-LM and YvcPQ-RS)35,38. The direct interaction between the BceAB transporter and the BceS histidine kinase was demonstrated in B. subtilis36. In that study, it was shown that BceAB, which was purified from the membrane, needs to form a complex with BceRS in order to initiate antibiotic resistance signalling36. A characteristic feature of these BceAB-type transporters is an extracellular domain (ECD) of roughly 210–230 amino acid located between transmembrane helices 7 and 8. These domains are supposed to be involved in binding the substrate (e.g., bacitracin). This is hypothesized due to the reason that the cognate histidine kinase, consists only of a short loop which is buried almost entirely in the cytoplasmic membrane and thus cannot detect extracellular stimuli39. So the binding to the ECD is supposed to trigger simultaneously the histidine kinase (HK)36. It has been shown with medically and biotechnologically relevant Gram-positive species that BceS-like HKs require BceAB-type transporters for antibiotic signaling31,36,40,41. Moreover, ATP hydrolysis by the ATPase BceA was shown to play an essential role for lantibiotic signaling23. Additionally, it was found that the associated sensor kinase BceS is unable to detect bacitracin in the absence of the transporter BceAB, which led to proposition that the transporter contains the involved sensory domain of the system40,42. More recently, the binding of AMP LL-37 to the ECD of VraG (a BceAB homologous transporter) was described in B. subtilis37.
Similar operon structures have been reported for other BceAB-type transporters, in some cases with an additional gene encoding a membrane-embedding protein such as SaNSR, a BceAB-type transporter in the human pathogen S. agalactiae, conferring resistance against lantibiotics such as nisin A, nisin H and gallidermin32,43. Using a peptide release assay, it was postulated that SaNsrFP transports these peptides via an efflux mechanism back into the medium34.
Several putative mechanisms for BceAB-type transporters have been proposed, ranging from AMP removal from the membrane44, functioning as an exporter34, to flipping the UPP21. Recently, a study proposed a target-AMP dissociative, ATP-hydrolysis-driven mechanism for BceAB-type transporters, in which the target-AMP complex is recognized and UPP physically released from the grip of bacitracin45.
In our study, in order to elucidate the mechanism of SaNsrFP, we expressed the BceAB-type transporter SaNsrFP in L. lactis NZ9000 without its cognate TCS34. We hypothesized that the transporter alone is able to sense an AMP in the surrounding since the Bce type transporters are known to play a crucial role in the signalling process37,42,43,46. It was shown for the related BceAB transporter that signalling is triggered by the activity of the transporter itself and the transporter can autoregulate its own production46. In previous work, it was shown that SaNsrFP is able to confer resistance against nisin without its TCS34. As a control for our study, we also analyzed the ATPase-deficient mutant of the ABC transporter that showed no ATPase function in-vitro47.
Since the BceAB system of B. subtilis is known to confer resistance against bacitracin and also other antibiotics i.e. mersacidin, plectasine and actagardine27,38, we wanted to test whether the SaNsrFP is also able to confer resistance against bacitracin and structurally different antibiotics. By investigating the effect the expression of sansrfp has on the proteome level, we gained further insights into the mode of action. Since here it was observed that genes involved in cell wall biosynthesis were downregulated, we also analyzed the composition of the peptidoglycan layer. Cell wall modification is one of several mechanisms of being involved in antibiotic resistance and has been shown to play an important role in the resistance mechanism in S. aureus, C. difficile, S. pneumoniae and S. agalactiae48,49,50. Also, in recent studies it is suggested that transporters can have a direct or indirect influence on peptidoglycan biosynthesis or peptidoglycan remodeling as was shown recently for the ABC transporter YtrBCDEF in B. subtilis51.
In this study, we aimed to characterize SaNsrFP in larger detail and analyzed the ability of this transporter to confer resistance against different structurally unrelated compounds, such as lysobactin, ramoplanin, vancomycin and bacitracin, as well as its zinc complex Zn-bacitracin. Whole-cell proteome and cytosolic PGN precursor analysis supported our hypothesis that the different antibiotics bind to SaNsrFP inducing an altering of the cell wall. This is relying on the ATP hydrolysis of SaNsrFP since the ATP hydrolysis deficient mutant does not show this phenotype. The transporter is able to protect the target via a first-line and second-line defense, and the energy set free by ATP hydrolysis could be the key to resetting the system. Our study provides new insights into the resistance mechanism of the BceAB-type transporter SaNsrFP. The data presented are in agreement with a mechanism of protection by shielding the target of the antimicrobial peptide.
Results
SaNsrFP enables normal growth in the presence of bacitracin
The BceAB-type transporter NsrFP from the human pathogen S. agalactiae COH1, SaNsrFP, has been shown to confer resistance against the lantibiotic nisin and structurally related compounds such as nisin H and gallidermin by recognizing and binding to the N-terminus of these lantibiotics34.
To investigate whether SaNsrFP confers resistance against bacitracin and Zn-bacitracin, binding to the lipid carrier UPP20,52, we analyzed the influence of the expression of the sansrfp gene. The sensitive L. lactis strain NZ9000, served as an indicator strain for these studies which was transformed with a plasmid encoding the nsrfp gene. We included two controls where the strains were transformed with (I) an empty plasmid (L. lactis NZ9000Cm) (II) a plasmid containing a variant of the nsrfp gene (L. lactis NZ9000SaNsrFH202AP)34. This NZ9000SaNsrFH202AP strain is used since the transporter carries a mutation in the H-loop, a highly conserved region of ABC transporters, and as a result is not able to hydrolyze ATP47. This mutation causes loss of ATP hydrolysis and stabilization of the closed conformation53. Although the substrate still binds to it as the transporter, it cannot be translocated because the required energy cannot be provided34,47. The growth of L. lactis NZ9000Cm, L. lactis NZ9000SaNsrFP, L. lactis NZ9000SaNsrFH202AP and L. lactis NZ9000NisT was monitored online over a time period of 500 min (Fig. 2a,b). After adding the different antibiotics to the culture, the growth curve was determined.
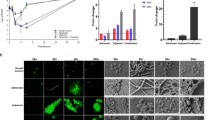
(a) Growth curve of the BceAB type ABC transporter expressing strain L. lactis NZ9000NsrFP (light blue), the ATP-hydrolysis deficient mutant strain L. lactis NZ9000NsrFH202AP (orange), the empty plasmid expressing strain L. lactis NZ9000Cm (black) and the nisin transporter expressing strain L. lactis NZ9000NisT (dark red) were induced with 0.3 nM nisin and treated with 1 µM bacitracin and 1 mM ZnCl2. As a control, L. lactis NZ9000NsrFP (dark blue) was induced with 0.3 nM nisin, and 1 mM ZnCl2 was added. (b) Growth curve of L. lactis NZ9000NsrFP (light blue), L. lactis NZ9000NsrFH202AP (orange), L. lactis NZ9000Cm (black), L. lactis NZ9000NisT (dark red) induced with 0.3 nM nisin and treated with an additional 4 µM bacitracin without ZnCl2. As a control, L. lactis NZ9000NsrFP (dark blue) was induced with 0.3 nM nisin. (c) Growth curve of the control, L. lactis NZ9000NsrFP (dark blue), L. lactis NZ9000Cm (black) and L. lactis NZ9000NisT (dark red) induced with 0.3 nM nisin, and 1 mM ZnCl2 was added. The normalized OD600 was plotted against the time using GraphPad Prism version 9.2.0 for Mac, GraphPad Software, San Diego, California USA, www.graphpad.com.
The expression of the sansrFP and sansrFPH202A genes was induced by adding a sublethal concentration of 0.3 nM nisin to the cells. This subinhibitory concentration of nisin is able to induce the nisA promotor in the pIL-SV plasmids which enables the gene expression of the respective protein. Important to note that this low concentration of nisin is not harming the cells as observed by different growth studies54,55. Simultaneously, either 1 µM bacitracin in combination with 1 mM ZnCl2 (Fig. 2a) or 4 µM bacitracin without zinc (Fig. 2b) was added. As a control, all strains were only induced with 0.3 nM nisin without receiving any additional supplements (Fig. 2c).
Severe growth inhibition was shown for the L. lactis NZ9000Cm and L. lactis NZ9000NisT strains. When using the L. lactis NZ9000SaNsrFP strain, however, the growth behavior was comparable to that of L. lactis NZ9000Cm without the addition of bacitracin (Fig. 2a,b light blue curve and c). Interestingly, L. lactis NZ9000SaNsrFH202AP cells were unable to grow when treated with (Zn)-bacitracin (Fig. 2a orange), whereas reduced growth was observed when bacitracin was added (Fig. 2b orange). Growth retardation, as observed in L. lactis NZ9000Cm upon the addition of bacitracin, has been shown for many bacterial cells, such as methicillin-resistant S. aureus and group B streptococci56,57. It is caused by the binding of bacitracin to UPP, preventing the dephosphorylation reaction and leading to the interruption of PGN biosynthesis20. Intriguingly, L. lactis NZ9000SaNsrFP did not show reduced growth (Fig. 2a,b light blue) in the presence of bacitracin in comparison to the control strain L. lactis NZ9000SaNsrFP (Fig. 2a,b blue) and the sensitive strain L. lactis NZ9000Cm.
These results demonstrated that SaNsrFP is involved in bacitracin resistance with a requirement for ATP hydrolysis. For our study, we expressed sansrfp without its TCS, leading us to the conclusion that the transporter alone is directly involved in bacitracin resistance.
SaNsrFP confers resistance against bacitracin, ramoplanin, vancomycin and lysobactin
Since SaNsrFP conferred resistance in addition to lanthipeptides against bacitracin and Zn-bacitracin, we extended the resistance study to a structurally diverse, rather unrelated group of antibiotics, including ramoplanin, vancomycin and lysobactin, all of which bind to different parts of lipid II10,18.
After adding the different antibiotics to the cultures of the strains L. lactis wild type (WT), L. lactis NZ9000Cm, L. lactis NZ9000SaNsrFP and L. lactis SaNsrFH202AP, the IC50 was determined. By dividing the IC50 obtained for the strains producing SaNsrFP or the inactive variant SaNsrFH202AP by the IC50 value obtained for the sensitive strain NZ9000Cm, the fold change of resistance was calculated, which was independent of small variations in bacterial cell growth behavior.
Compared to L. lactis WT, L. lactis NZ9000SaNsrFP exhibited small resistance to the lipid II binders, vancomycin and lysobactin (two to sixfold) (Fig. 3a, SI Fig. S1, Table 1). No significant differences were detected between NZ9000NsrFP and NZ9000NsrFH202AP with ramoplanin. The fold increases of resistance (two, fivefold increases) obtained were significantly lower than the fold changes described for nisin (16-fold) and gallidermin (12-fold)34, suggesting that nisin is a preferred substrate of the transporter. IC50 values decreased to similar levels in L. lactis NZ9000SaNsrFH202AP (one, two and threefold), as observed for the L. lactis NZ9000Cm strain (73.0 nM, 213.5 nM, and 30.7 nM, respectively, Table 1), indicating that resistance was not achieved solely by the expression of the SaNsrFH202AP transporter. To ensure that this loss of resistance was not due to different production levels, we performed Western blot analysis on the purified membrane fractions of the transporter expressing cells using a polyclonal antibody against the ECD of SaNsrP (Fig. 3b). We confirmed that comparable levels of transporters were produced in both strains. Since ATP hydrolysis activity was deleted in the SaNsrFH202AP mutant, the obtained results suggested that SaNsrFP requires ATP hydrolysis to confer resistance.
(a) Fold of resistance of L. lactis NZ9000NsrFP and NZ9000NsrFH202AP (hatched bars) against L. lactis NZ9000Cm calculated with the determined IC50 of ramoplanin A2 (yellow), vancomycin (orange), lysobactin (green), bacitracin (blue) and bacitracin with ZnCl2 (dark red). Values for nisin and gallidermin were taken from Reiners et al.34 and marked with an asterisk. Values were calculated from at least 4 independent measurements and are also listed in Table 1. A two-sided Students t-test was performed with the IC50 data obtained for SaNsrFP and SaNsrFH202AP. Significance was marked with an asterisk. p-values were listed in a separate table (SI Table S1) in the supplement. (b) Expression of SaNsrFP (1) and SaNsrFH202AP (2) and the empty vector pIL-SV (3) in L. lactis NZ9000, monitored via western blot with a polyclonal antibody against the extracellular domain of SaNsrP. Loaded are purified membranes from the corresponding strains. A nonlinear regression curve fit and a two-sided, unpaired Students t-test was performed using Graphpad Prism version 9.2.0 for Mac, GraphPad Software, San Diego, California USA, www.graphpad.com.
However, high-level resistance was observed for L. lactis NZ9000SaNsrFP to bacitracin (350-fold) and (Zn)-bacitracin (132-fold) compared to L. lactis WT (Fig. 3a, SI Fig. S1, Table 1). In contrast, L. lactis NZ9000SaNsrFH202AP displayed only moderate resistance (13-fold), which was completely abolished when the cells were treated with (Zn)-bacitracin (0.7-fold). Resistance at a low level against nisin (2.6-fold) was shown in Khosa et al., in which an inactive variant of the protease SaNSR (SaNSRS236A) was produced32. Even though, it was demonstrated that NZ9000SaNsrFH202AP shows no ATPase activity in-vitro47, it is known that it is difficult to compare in-vitro with in-vivo data since it cannot be excluded that other processes in the bacterial cell might lead to such a residual bacitracin resistance for SaNsrFH202AP. On the other hand, the residual resistance is only observed with bacitracin not with zinc-bacitracin. It has been shown that bacitracin shows a higher attraction to the membranes in the presence of zinc and the target most probably due to the observation that it is forced into an amphiphile conformation20. This could explain why only residual resistance is observed with only bacitracin since it cannot access the membrane as easily as with zinc.
To strengthen this hypothesis and to exclude that the resistance to bacitracin in L. lactis NZ9000SaNsrFH202AP is caused by an altered membrane protein composition due to the overexpression, we performed growth inhibition experiments with another large ABC transporter, namely, NisT from L. lactis, which is not present in the genome of the NZ9000 strain used, using the same plasmid backbone. Recently, it was shown that NisT is produced in high amounts in the used strain58,59. However, NisT is not relevant to bacitracin resistance, as evidenced by similar IC50 values of the strain NZ9000NisT, producing no NisT (Table 1). Therefore, it can be concluded that the production of large membrane proteins, as well as ATP hydrolysis and a possible alteration of the membrane protein composition is not the explanation for the resistant phenotype but is due to the production of SaNsrFP or its inactive variant.
SaNsrFP prevents the accumulation of peptidoglycan precursors after the addition of bacitracin
To further understand the mechanism of action of SaNsrFP, we analyzed PGN precursor accumulation in the cytoplasm of L. lactis NZ9000SaNsrFP, L. lactis NZ9000Cm and L. lactis NZ9000SaNsrFH202AP grown in the presence of bacitracin. HPLC/MS analysis of the PGN extracts revealed the presence of the characteristic PGN precursors UDP-MurNAc-l-Ala-d-iGlu-l-Lys-d-Ala-d-Ala (1148.4 m/z−1) and MurNAc-l-Ala-d-iGlu-l-Lys-(d-Asp)-d-Ala-d-Ala (1263.4 m/z−1) in the L. lactis NZ9000Cm strain (Fig. 4a,b light gray).
Relative abundance of obtained mass in cps against retention time in min of precursor accumulation after treatment with bacitracin. (a) Extracted ion chromatography (EIC) spectrum for UDP-MurNAc-l-Ala-d-iGlu-l-Lys-d-Ala-d-Ala (1148.4 m/z−1) and (b) EIC spectrum for UDP-MurNAc-l-Ala-d-iGlu-l-Lys(d-Asp)-d-Ala-d-Ala (1263.4 m/z−1) of the strains NZ9000Cm (light gray), NZ9000SaNsrFH202AP (gray) and NZ9000SaNsrFP (black). Extracted ion chromatograms (EICs) in negative ion mode for UDP-MurNAc-l-Ala-d-iGlu-l-Lys-d-Ala-d-Ala (m/z−1 1148.34 ± 0.1) and UDP-MurNAc-l-Ala-d-iGlu-l-Lys-(d-Asp)-d-Ala-d-Ala (m/z−1 1263.37 ± 0.1) were analyzed with Data Analysis (Bruker), exported and presented with GraphPad Prism 6.0, GraphPad Software, San Diego, California USA, www.graphpad.com. UDP undecaprenyl-phosphate, MurNAc N-acetylmuramic acid, Ala alanine, iGlu isoglutamic acid, Glu glutamine, Asp aspartate/aspartic acid, Lys lysine.
Interestingly, no accumulation of the PGN precursors was observed in the L. lactis NZ9000SaNsrFP strain (Fig. 4a,b black line), whereas L. lactis NZ9000SaNsrFP without bacitracin treatment revealed the accumulation of the PGN precursor UDP-MurNAc-l-Ala-d-iGlu-l-Lys-d-Ala-d-Ala (1148.4 m/z−1) (SI Fig. S6a,b). The fact that bacitracin was not able to block UPP recycling, together with the results obtained in the resistance test, clearly suggests that SaNsrFP prevents the binding of bacitracin to UPP and thus the accumulation of PGN precursors. This hypothesis is further supported by the results obtained for the L. lactis NZ9000SaNsrFH202AP strain. Here, reduced PGN precursor accumulation was observed compared to the bacitracin-sensitive L. lactis NZ9000Cm strain (Fig. 4a,b gray line), implicating that the availability of UPP for bacitracin binding is decreased in the L. lactis NZ9000SaNsrFH202AP strain. Considering that bacitracin did not inhibit PGN synthesis in L. lactis NZ9000SaNsrFP, SaNsrFP might protect the bacitracin target and directly interact with a component of PGN, most likely UPP, to evade the accumulation of PGN precursors.
Binding of bacitracin to the UPP or UP normally results in the accumulation of lipid II precursors in the cytosol. The lack of this accumulation in L. lactis SaNsrFP suggests that bacitracin is unable to bind to its membrane-localized target UPP. This, together with the observation that the cell growth of L. lactis SaNsrFP in the presence of bacitracin is similar to that of cells without bacitracin, supports the shielding mechanism proposed by Kobras et al.45.
SaNsrFP causes downregulation of proteins involved in peptidoglycan synthesis
To get an insight to the mode of action initiated by SaNsrFP expression, we analyzed the whole proteome of L. lactis NZ9000SaNsrFP, L. lactis NZ9000Cm and L. lactis NZ9000SaNsrFH202AP grown under identical growth conditions (at 30 °C in GM17 medium containing 5 µg/ml chloramphenicol and induced with 0.3 nM nisin) by mass spectrometry. The analyses led to the identification of 894 proteins (identified by at least two unique peptides in each strain). The comparison between L. lactis NZ9000Cm and NZ9000SaNsrFP revealed 315 with differential abundances (Fig. 5a) and 339 proteins showing differential abundances between the L. lactis strains NZ9000SaNsrFH202AP and NZ9000SaNsrFP (Fig. 5b). In 231 proteins there was no change of abundance. Here, we took the slightly different OD after 5 h of cell growth into account and adjusted the whole cell protein concentration accordingly. In particular, the latter highlights that these up- or downregulation of the proteins do not arise from the expression of the transporter since they are expressed at similar levels (see Fig. 2b). This high number of differentially produced proteins implied that the L. lactis NZ9000SaNsrFP strain has to respond significantly to counteract the effects mediated by the presence and activity of the SaNsrFP BceAB-type ABC transporter.
(a) Volcano plot of the proteome analysis of NZ9000SaNsrFP against NZ9000Cm and (b) NZ9000SaNsrFP against NZ9000SaNsrFH202AP. Proteins involved in cell wall synthesis are highlighted in orange if upregulated in NZ9000Cm (a) and NZ9000SaNsrFH202AP (b) and highlighted in blue if upregulated in NZ9000SaNsrFP. Proteins with a p-value ≤ 0.05 and a fold change ≥ 2 were considered as statistically significant. Proteome data was plotted using GraphPad Prism 6.0 for Mac, GraphPad Software, San Diego, California USA, www.graphpad.com.
In-depth analysis showed that the production of proteins involved in PGN synthesis was reduced in L. lactis NZ9000SaNsrFP (Fig. 4a,b, SI Fig. S3). Among them the UDP-N-acetylglucosamine 1-carboxyvinyltransferase MurA (ADJ59532), was produced 6.2-fold less in the L. lactis NZ9000SaNsrFP compared to L. lactis NZ9000Cm; UDP-N-acetylmuramate-l-alanine ligase MurC (ADJ61283), 4.0-fold, UDP-N-acetylmuramoyl-l-alanyl-d-glutamate synthetase MurD (ADJ59924) the 3.5-fold, UDP-N-acetylmuramoylalanyl-d-glutamate-2,6-diaminopimelate ligase MurE (ADJ60966), 4.0-fold less produced in the control strain as well as UDP-N-acetylmuramoylalanyl-d-glutamyl-2,6-diaminopimelate-d-alanyl-d-alanine ligase MurF (ADJ59966). Furthermore, proteins involved in the synthesis of components of lipid II synthesis, such as uracil phosphoribosyl transferase or glucosamine-fructose-6-phosphate aminotransferase, responsible for UMP and glucosamine-6-phosphate synthesis, respectively, were downregulated in L. lactis NZ9000SaNsrFP (Table 2, SI Fig. S3).
We included in the analysis proteins from other metabolic pathways, such as amino sugar metabolism and translation, represented by α-d-glucosamine-1,6-phosphomutase or phenylalanyl-tRNA synthetase beta subunit in order to show that the expression of the ABC transporter SaNsrFP is not toxic to the cells and has an influence on other important cell processes. These proteins did not reveal any differences in production among all three tested strains (Table 2), confirming that the observed differences for the other proteins were due to the expression of the active SaNsrFP transporter.
In summary, upregulated proteins when SaNsrFP was produced, such as UDP-glucose-4-epimerase, N-acetylmuramic acid-6-phosphate etherase MurQ and RodA, were found to be associated with AMP resistance and cell wall modification (Fig. 6).
Schematic view of the proposed mechanism of SaNsrFP. Phosphates are marked with a P, undecaprenyl as a black curved line, GlcNAc in blue, MurNAc in red and amino acids of the pentapeptide in orange. The transporter SaNsrFP is shown in blue, showing its functions of sensing antibacterial attack, shielding the target most likely by releasing the target from the grip of bacitracin and initiating a secondary defense leading to possible cell wall thickening, modifying the electrostatic charge of the cell wall by integrating lipoteichoic acids and increasing d-alanylation in the cell wall. Subsequentially, the released target can enter a new cell wall synthesis cycle and be incorporated into the peptidoglycan (not shown fully here but in Fig. 1). GlcNAc N-acetylglucosamine, MurNAc N-acetylmuramic acid. The figure was created using Microsoft Powerpoint Version 16.54.
Additionally, the components of the nascent cell wall of L. lactis NZ9000Cm-, SaNsrFP- and SaNsrFH202AP-expressing L. lactis cells were analyzed via LC–MS. The comparison of their chromatograms revealed that some peaks, which are occurring only in the sensitive strain and the inactive mutant (Fig. 7, SI Fig. S5a–f). These peaks correspond to components consisting of GlcNAc-MurNAc-l-Ala-d-iGln-l-Lys-(d-Asn) with m/z 938.37 [M + H]+ (RT 27.0–28.3 min) (Fig. 7, SI Fig. S5a, peak 1), to GlcNAc-MurNAc-l-Ala-d-iGln-l-Lys-(d-Asp) with m/z 939.37 [M + H]+ (RT 29.8–30.6 and 32.4–33.1 min) (Fig. 7, SI Fig. S5b, peak 2), to GlcNAc-MurNAc-l-Ala-d-iGln-l-Lys-(d-Asn)-d-Ala with m/z 1009.45 [M + H]+ (RT 35.8–36.9 min) (Fig. 7, SI Fig. S5c, peak 3), to GlcNAc-MurNAc-l-Ala-d-iGln-l-Lys-(d-Asn)-d-Ala-d-Ala with m/z 1080.50 [M + H]+ (RT 38.0–39.3 min) (Fig. 7, SI Fig. S5d, peak 4). The obtained masses are in agreement with the data described in the literature60,61.
LC–MS chromatogram of isolated muropeptides from L. lactis NZ9000Cm (purple), L. lactis NZ9000NsrFP (red) and L. lactis NZ9000NsrFH202AP (blue). The peaks that only occur in L. lactis NZ9000Cm and L. lactis NZ9000NsrFH202AP are framed in green. Black framed is the peak, which can especially be observed in L. lactis NZ9000NsrFP. Peak 1: GlcNAc-MurNAc-l-Ala-d-iGln-l-Lys-(d-Asn); peak 2: GlcNAc-MurNAc-l-Ala-d-iGln-l-Lys-(d-Asp); peak 3: GlcNAc-MurNAc-l-Ala-d-iGln-l-Lys-(d-Asn)-d-Ala; peak 4: GlcNAc-MurNAc-l-Ala-d-iGln-l-Lys-(d-Asn)-d-Ala-d-Ala; peak 5: GlcNAc-MurNAc-l-Ala-d-iGln-l-Lys-(Ala)-d-Ala-d-Ala, and peaks 6–8: GlcNAc-MurNAc-l-Ala-d-iGln-l-Lys-(Ala-Ala)-d-Ala-d-Ala. UDP undecaprenyl-phosphate, GlcNAc N-acetylglucosamine, MurNAc N-acetylmuramic acid, MurN N-deacetylated muramic acid, Ala alanine, iGlu isoglutamic acid, iGln isoglutamine, Glu glutamine, Asp aspartate/aspartic acid, Asn asparagine, Lys lysine.
In contrast, one peak at RT 40.4–41.4 min with mass m/z 1037.49 [M + H]+ (Fig. 7, SI Fig. S5e, peak 5) was detected only in L. lactis SaNsrFP-expressing cells. The detected mass may indicate the presence of GlcNAc-MurNAc-l-Ala-d-iGln-l-Lys-(Ala)-d-Ala-d-Ala muropeptide fragment, that could contain an Ala residue attached to Lys, forming the crosslinking bridge. In addition, we detected three peaks (Fig. 7, SI Fig. S5f, peaks 6–8) whose masses presumably corresponded to GlcNAc-MurNAc-l-Ala-d-iGln-l-Lys-(Ala-Ala)-d-Ala-d-Ala (RT 44.4–45.5 min, 47.0–51.8 min and 54.4–55.7 min) with m/z 1108.53 [M + H]+.These masses are in accordance with the masses described in the literature62,63,64. Intriguingly, the SaNsrFH202AP-expressing strain also showed some double alanine muropeptide species, albeit with a lower percentage than in the active transporter. These findings suggest that the transporter not only confers resistance by defending the target but also induces modulation of the cell wall.
Discussion
To elucidate the mechanism of the SaNsrFP resistance mechanism, we showed that this transporter is able to circumvent reduced cell growth when cells are treated with bacitracin and/or Zn-bacitracin. Furthermore, we determined that resistance occurs against lipid II-binding AMPs. However, the highest resistance was observed for bacitracin and its Zn-bacitracin counterpart. This suggesting that this is the main substrate for NsrFP. The previously observed nisin resistance (Reiners et al.34) appears to be a side effect of the resistance mechanism. Here, our data implies that this resistance is ATP hydrolysis dependent and therefore is an active process, something that has been underestimated until now. Bacitracin resistance has been shown for several other BceAB-type transporters and appears to be conserved within this protein family. Examples include the AnrAB transporter from Listeria monocytogenes30, VraDE from S. aureus31,33, and the ABC transporter BceAB from B. subtilis38. However, similar to SaNsrFP, these transporters additionally exhibited a certain degree of resistance to nisin and gallidermin, suggesting a general resistance mechanism rather than specific resistance to one type or even to specific antibiotics. Therefore, considering that SaNsrFP confers resistance to structurally unrelated compounds, we concluded that SaNsrFP is neither able to inactivate nor bind various compounds but rather that the resistance is based on a more general mechanism, such as shielding PGN biosynthesis components, including lipid II, UPP or UP, which are exposed on the outer surface of the bacterial membrane. The high-level resistance observed for the UPP binders bacitracin and (Zn)-bacitracin suggested that SaNsrFP could shield either UPP or lipid II. (Figs. 1, 3a). Current hypotheses explain the resistance mechanism by the inaccessibility of the target UPP to bacitracin in this strain by either (i) target removal21, (ii) target protection45 or (iii) the combination of an active AMP defense mechanism that also mediates a multifactorial AMP defense response. The AMP defense mechanism does not involve only a higher expression of ABC transporter encoding genes but also modifications of the cell wall structure or the membrane lipid composition, PGN thickening, changes in net charge and degrading enzymes4. Given that L. lactis SaNsrFP cells are still growing at high (Zn-) bacitracin concentrations, PGN synthesis was not completely inhibited. Since SaNsrFP cells repel structurally diverse antibiotics, as is known for the related BceAB transporter of B. subtilis27, we hypothesize that SaNsrFP could mediate resistance by shielding UPP and subsequent modification of PGN synthesis. These mechanisms have also been postulated by recent studies45. Based on previous studies and transporter activity studies in the presence of accumulated UPP or C35 isoprenoid heptaprenyl diphosphate (HPP), the authors proposed that the BceAB transporter detects UPP-bacitracin complexes and shields the target (e.g., lipid II or UPP or HPP) by severing the bond between them65. This further excludes other mechanisms, such as UPP flipping for BceAB in B. subtilis21 and import and inactivation of the target23.
Current opinion of researchers investigating antibiotic resistance conferring ABC transporters is that detoxification against peptide antibiotics is functionally linked to a two-component system36. It is hypothesized that upon sensing the antibiotic, the histidine kinase phosphorylates its cognate response regulator which induces the expression of the ABC transporter genes. Such a scenario was described i.e., for the GraRS-VraFG system37 in S. aureus and also for several TCS-ABC transporters in B. subtilis (BceRS-AB, YxdJK-LM and YvcPQ-RS)36,38. Moreover, a direct interaction of the BceRS and BceAB was shown in in vitro and in vivo studies36. In their study, it is claimed that BceAB and the TCS need to form a complex in order to be able to sense the AMP.
In our study, in order to elucidate the mechanism of the BceAB-type transporter SaNsrFP, we expressed it without its cognate TCS. It has been shown that the ABC transporter without its TCS can confer resistance against nisin34. The large extracellular domain is the hallmark of BceAB-type transporter which is hypothesized to be involved in extracellular detection of antibiotics43. Interestingly, the cognate histidine kinase consists only of a short loop which is buried almost entirely in the cytoplasmic membrane and thus cannot detect extracellular stimuli39. The crucial role of a Bce-type transporter for lantibiotic signalling has been shown in various studies already23,42. For the BceAB transporter it was shown that signalling is triggered by the activity of the transporter itself and the transporter can autoregulate its own production45,46. This is the reason we hypothesize that the ABC transporter SaNsrFP should also be able to sense the AMP via its large extracellular domain. Therefore, we strived to investigate the role SaNsrFP plays together with its 221 amino acid large extracellular domain in conferring resistance without its cognate TCS.
Since SaNsrFP is able to confer resistance against bacitracin and other AMP’s, we can show that the transporter is directly involved in sensing the antibiotic and the resistance process.
By the expression of sansrfp, adjustments within the bacterial cells occur. For example, the downregulation of proteins involved in lipid II biosynthesis. The reduced production of the key enzymes of the lipid II cycle was remarkable and suggested that the biosynthesis of new lipid II molecules occurred with less efficiency in the L. lactis NZ9000SaNsrFP strain. This could be the case if lipid II or UPP might be the actual substrate of SaNsrFP, but this hypothesis remains controversial, as it does not correspond to the growth behavior observed in the growth analysis. Here, the SaNsrFP-expressing strain showed similar growth to the control strains L. lactis NZ9000NisT and L. lactis NZ9000Cm (see above Fig. 2c).
This strengthens the idea that BceAB-type transporters interact with precursors of cell wall synthesis or its recycling by binding. Therefore, the lipid II cycle might be inhibited, and the bacteria react by downregulating its lipid II pool. This has been directly shown by the analyses of the cell wall precursors that were clearly reduced in the SaNsrFP strain. This would not only result in growth inhibition but would also lower the number of available targets at the membrane surface. Controversially, we could not see reduced growth in SaNsrFP-expressing cells, and thus far, it was not possible to measure a difference in targets at the membrane surface. Less target on the surface could explain the moderate resistance observed for the lipid II binding AMPs like ramoplanin, vancomycin and others but it is not at all clear if this is a possible scenario since the removal of the target would likely lead to growth inhibition which would be toxic for the cell. Therefore, we cannot entirely exclude that the heterologous expression of the BceAB-type transporter influences the cell wall synthesis of L. lactis or whether the transporter itself is responsible for the alteration of its cell wall. Nonetheless, the NICE expression system (nisin-controlled gene expression system) that we used for the overexpression of SaNsrFP is a tightly regulated system which can be turned on by adding a subinhibitory concentration of nisin to the media. It has been shown for the system that genes of closely related Gram-positive organisms (e.g., Streptococcus, Enterococcus, Staphylococcus, and low-GC Lactobacillus) are expressed effectively usually without any problems66. Interestingly, in the work of Marreddy et al.67, overexpression of a membrane protein led to an upregulation of cell wall synthesis in the membrane protein expressing strain. In our data we detected a downregulation of involved proteins of cell wall synthesis but a slight upregulation of cell wall modification proteins. Moreover, we could not observe a significant change of expression of proteins responsible for a general stress response. Nonetheless, we chose the best possible system for heterologous expression of SaNsrFP to overcome possible bottlenecks.
We show evidence that cells expressing SaNsrFP obtain a modified cell wall: instead of an apartate/asparagine bridge in the pentapeptide found for the sensitive mutant, a species with two alanines was detected. In SaNsrFH202AP, a mixture was found, although the two alanine species were present in only minor amounts (Fig. 7). This suggests that the transporter might already sense and mediate a second line of defense ATP-independently. ABC transporters that confer resistance against cationic antimicrobials are hypothesized to be involved or mediate modification processes of peptidoglycan in Gram-positive bacteria. d-alanylation of teichoic acids is assumed to diminish electrostatic attraction based on the observation that a lack of alanylation leads to increased binding to several positively charged molecules, e.g., gallidermin and vancomycin41. Additionally, the upregulation of the gal operon, especially of UDP-glucose-4-epimerase (GalE), influences the lipoteichoic acid (LTA) structure. GalE is responsible for the synthesis of \(\alpha\)-galactose, which is transported across the membrane to become a part of LTA68.
We can see this in our data with the upregulation of the MurQ which is responsible for the intracellular conversion of MurNAc-6P to N-acetylglucosamine-6-phosphate and d-lactate for the SaNsrFP and SaNsrFH202AP mutants in comparison to the sensitive strain. For transporter-expressing cells, we also observed an upregulation of proteins associated with antimicrobial resistance, such as UDP-glucose-4-epimerase and RodA68.
In the case that SaNsrFP might mediate cell wall modifications upon receiving information on the cell wall targeting AMP, altered expression of genes could be the consequence. This finding might also reduce the number of proteins in the cytosol that are involved in lipid II biosynthesis, as seen by the whole proteome data where the expression of the genes is downregulated but not completely abolished. It needs to be verified whether SaNsrFP is directly responsible for this or whether the L. lactis strain is reacting since its lipid II cycle is severely changed and, as a consequence, alters its cell wall composition.
Based on all results from this study, a joint activity of the transporter as a first-line defender and initiator for a second-line defense is very likely and results in resistance against compounds targeting the lipid II cycle and thus cell wall synthesis. By shielding the target UPP and lipid II from the extracellular space, e.g., by PGN modification that alters electrostatic attraction, less antibiotic, e.g., bacitracin, can be bound, and increased antibiotic concentrations can be detected in the supernatant. Our findings are in agreement with the previous conclusions for an export mechanism and further assumptions on the removal of AMPs from the membrane34,44. The tendency for upregulation of proteins associated with antimicrobial resistance and cell wall modification in SaNsrFP-producing cell proteins indicates the activation of a second-line defense system.
Conclusively, BceAB-type transporters such as SaNsrFP are evolutionarily conserved in human pathogenic and nonpathogenic strains. Although they are less conserved at the sequence level, the topology of the protein and their encoding operons are conserved. The resistance observed in different BceAB-type transporter studies indicates a common mechanism. The findings in this study are in line with a target protection mechanism, as was postulated for the BceAB transporter. Our data implies that AMP resistance is a far more complex process that involves a combination of an active target mechanism, which enables continuous growth, and a second line of defense, which could be initiated after sensing the AMP directly by the SaNsrFP transporter.
Materials and methods
Cloning and expression
The plasmids pIL-SV SaNsrFP and pIL-SV SaNsrFH202AP, the latter harboring a point mutation in the H-loop, known to be crucial for ATP hydrolysis, were generated by cloning nsrfp from S. agalactiae COH1 as described in Alkhatib et al.69 and Reiners et al.34. Each plasmid and the empty vector pIL-SVCm was transformed into electrocompetent L. lactis NZ9000 cells70, and the resulting strains were termed NZ9000SaNsrFP, NZ9000SaNsrFH202AP and NZ9000Cm.
All strains used in this study have been described in previous publications34,69.
The L. lactis strains NZ9000SaNsrFP and NZ9000SaNsrFH202AP were cultured in GM17 medium containing 5–10 µg/ml chloramphenicol. Expression was induced by adding 0.3 nM nisin, and cultures were grown at 30 °C.
To analyze the expression, cultures were grown for 5 h and subsequently harvested using a centrifugation step for 30 min at 5000×g. The pellets were resuspended to an OD600 of 200 in resuspension buffer (50 mM HEPES pH 8.0, 150 mM NaCl, 10% glycerol), then 1/3 (w/v) 0.5 mm glass beads were added. The cells were lysed, and the supernatant was separated from cell debris as well as glass beads by centrifuging at 10,000×g. Subsequently, the membranes were harvested from the supernatant by a 100,000×g centrifugation step. Membrane fractions were mixed with SDS-loading dye (0.2 M Tris–HCl, pH 6.8, 10% (w/v) SDS, 40% (v/v) glycerol, 0.02% (w/v) bromophenol and β-mercaptoethanol) and used for SDS-PAGE and western blot analysis. A polyclonal antibody against the extracellular domain of SaNsrP was used to detect the expressed SaNsrFP protein (Davids Biotechnologie, Regensburg, Germany).
Biological assays
Purification of nisin
Nisin was purified with ion-exchange chromatography as previously described71, and the concentration was determined with RP-HPLC according to Abts et al.72.
Determination of the half-maximal inhibitory concentration (IC50)
The half maximal inhibitory concentration was determined according to Abts et al.71. Briefly, L. lactis NZ9000Cm, L. lactis NZ9000SaNsrFP and L. lactis NZ9000SaNsrFH202AP cells were grown in GM17 medium containing 5 µg/ml chloramphenicol and 0.3 nM nisin at 30 °C overnight. Fresh GM17Cm medium with a sublethal amount of nisin (0.3 nM) was inoculated with overnight cultures to an OD600 of 0.1. A 96-well plate was prepared with a serial dilution of examined antibiotics (concentration ranges ramoplanin 0.014 nM–3.75 µM; lysobactin 0.002 nM–10 µM; vancomycin 0.02 nM–80 µM; nisin 0.0001 nM–0.5 µM) and subsequently the cell culture was added and plates were incubated at 30 °C for 5 h. Afterwards, the optical density was measured, and the IC50 values for each strain and antibiotic were calculated34. To make those values more comparable, the fold of resistance was determined by dividing the IC50 values of L. lactis NZ9000SaNsrFP and L. lactis NZ9000SaNsrFH202AP by the corresponding value for L. lactis NZ9000Cm.
Growth curves
To detect the growth behavior of the different strains, precultures of L. lactis NZ9000Cm, L. lactis NZ9000SaNsrFP and L. lactis NZ9000SaNsrFH202AP cells were grown in GM17 medium with 5 or 10 µg/ml chloramphenicol and 0.3 nM nisin at 30 °C overnight. Freshly prepared GM17Cm medium with 0.3 nM nisin was inoculated with overnight cultures to an OD600 of 0.1 and grown to an OD600 of 0.4–0.5 at 30 °C. Afterwards, the cells were diluted to an OD600 of 0.05 in GM17Cm medium containing 0.3 nM nisin. Cells were treated with either 1 µM bacitracin and 1 mM ZnCl2 or 4 µM bacitracin without ZnCl2. Growth was detected at OD584 every 10 min with a FLUOstar OPTIMA (BMG Lab technology).
Cell wall precursor analysis
Growth condition and sample preparation
Cells were grown in M17 medium supplemented with 0.5% glucose and 0.3 nM nisin overnight at 30 °C without shaking. The next day, 100 ml with 0.5% glucose and 0.3 nM nisin was inoculated with overnight cultures to OD600 = 0.1. When OD600 = 1.2 bacitracin (100 µg/ml) was added to the cultures to enrich cell wall precursors, and the cultures were incubated for an additional 30 min at 30 °C. This step was repeated once. (As a control, a second culture each was harvested before bacitracin was added at an OD600 = 1.2, and cell pellets were stored at − 20 °C) After incubation with bacitracin, the cells were harvested, and the cell pellets were stored at − 20 °C. The next day, the cell pellets were resuspended in 25 ml water and cooked for 60 min in boiling water to break the cells. Cell debris was removed by centrifugation (15 min, 500×g, 4 °C). The supernatant, containing the cell wall precursors, was lyophilized overnight. Cell pellets were resuspended in 150 µl water and used for LC/MS analysis.
LC/MS analysis of cell wall fragments
Five microliters of each sample were injected into an XCT6330 LC/MSD ultratrap system (Agilent Technologies) equipped with a Nucleosil 100 C18 column (3 μm × 100 mm × 2 mm internal diameter, Dr. Maisch GmbH). The column was used at 40 °C. A linear gradient was performed from 0 to 10% eluent B (0.06% formic acid in acetonitrile) over 25 min with a flow rate of 400 µl/min. The column was re-equilibrated for 10 min with 100% buffer A (0,1% formic acid in water). Ionization alternated between positive and negative ion modes with a capillary voltage of 3.5 kV at 350 °C. Extracted ion chromatograms (EICs) in negative ion mode for UDP-MurNAc-l-Ala-d-iGlu-l-Lys-d-Ala-d-Ala (m/z−1 1148.34 ± 0.1) and UDP-MurNAc-l-Ala-d-iGlu-l-Lys-(d-Asp)-d-Ala-d-Ala (m/z−1 1263.37 ± 0.1) were analyzed with Data Analysis (Bruker), exported and presented with GraphPad Prism 6.0.
Peptidoglycan analysis
Isolation of peptidoglycan
600 ml main culture of L. lactis NZ9000Cm, L. lactis SaNsrFP and L. lactis SaNsrFH202AP were inoculated with overnight culture and incubated to an OD600 of 0.1 at 30 °C. After reaching the late exponential growth phase, the cells were harvested. To isolate the peptidoglycan, the cells were thawed on ice and resuspended in 15 ml of 50 mM Tris/HCl buffer pH 7.0. The cell suspension was added dropwise to 60 ml of boiling, stirred 4% SDS solution. After boiling for another 15 min, the suspension was cooled to room temperature and centrifuged at 13,000×g for 10 min. The pellet was washed twice with 1 M NaCl followed by water until no SDS was detectable in the supernatant. Pellet was resuspended in 1 ml water, and 1/3 volume of glass beads (Æ 0.5 mm) were added. After cell lysis the glass beads were harvested at 2000×g for 5 min. The supernatant was centrifuged at 25,000×g for 15 min, and the pellet containing the cell walls were resuspended in 100 mM Tris/HCl pH 8.5 buffer with 20 mM MgSO4. After addition of 10 µg/ml DNase I and 50 µg/ml RNase, the samples were incubated at 37 °C with 180 rpm for 2 h. Following the addition of 10 mM CaCl2 and 100 µg/ml trypsin, an 18 h incubation was performed under the same conditions. Enzymatic activities were stopped by the addition of 1% SDS and incubation at 80 °C for 15 min. The suspension was diluted to 20 ml with water and centrifuged at 25,000×g for 30 min. The pellet was resuspended and incubated at 37 °C for 15 min with 10 ml 8 M LiCl and 10 ml 100 mM EDTA pH7, respectively. The peptidoglycan pellet was washed with water, acetone and water and was lyophilized.
Samples were treated as follows: 150 µl of resuspended peptidoglycan were mixed with 60 µl of mQ water and with 75 µl of TES buffer (200 mM TES, 4 mM MgCl2, pH 7.0 with final concentration in sample: 150 mM TES, 3 mM MgCl2, pH 7.0) and 15 µl of mutanolysine (75U) (Sigma-Aldrich, 5 kU/ml, dissolved in mQ water). Samples were incubated overnight at 37 °C and then centrifuged at RT for 5 min at 14,000 rpm. 90 µl of the supernatant were used for HPLC–MS analysis.
HPLC–MS analysis of muropeptides
90 µl of the sample were injected for HPLC–MS analyses (XCT 6330 LC/MSD Ultra Trap system; Agilent Technologies) and Reprosil-Gold 300 C18 column (5 µm by 250 mm by 4.6 mm internal diameter). The HPLC parameters were as follows: Holding with 5% of solvent B (methanol + 0.06% HCOOH) for 5 min and then start with a linear gradient from 30% solvent B to 70% solvent A (water + 0.1% HCOOH) for 150 min with additional holding with 30% solvent B over 30 min at a flow rate of 500 µl/min. The MS parameters were as follows: Ionization alternating positive and negative, capillary voltage 3.5 kV, and temperature 350 °C.
Proteome analysis
Sample preparation
The L. lactis strains NZ9000SaNsrFP and NZ9000SaNsrFH202AP were grown at 30 °C in GM17 medium containing 5 µg/ml chloramphenicol and 0.3 nM nisin. Precultures were inoculated to an OD600 of 0.1 and grown to the exponential growth phase before a main culture was inoculated to an OD600 of 0.1. The cells were harvested using 5000×g, and the pellets were resuspended in phosphate buffer pH 7 to an OD600 of 200. Then, 1/3 (w/v) 0.5 mm glass beads were added. The cells were lysed, and the supernatant was separated by centrifugation at 10,000×g.
Protein concentration was determined by means of a Pierce 660 nm Protein Assay (Fischer Scientific, Schwerte, Germany), and 10 µg protein per sample was loaded on an SDS-PAGE gel for in-gel digestion. The isolated gel pieces were reduced, alkylated and underwent tryptic digestion. The peptides were resolved in 0.1% trifluoracetic acid and subjected to liquid chromatography.
LC–MS analysis
For the LC–MS analysis, a QExactive plus (Thermo Scientific, Bremen, Germany) connected with an Ultimate 3000 Rapid Separation liquid chromatography system (Dionex/Thermo Scientific, Idstein, Germany) equipped with an Acclaim PepMap 100 C18 column (75 µm inner diameter, 25 cm length, 2 mm particle size from Thermo Scientific, Bremen, Germany) was applied. The length of the LC gradient was 120 min. The mass spectrometer was operated in positive mode and coupled with a nano electrospray ionization source. The capillary temperature was set to 250 °C, and the source voltage was set to 1.4 kV. In the QExactive plus mass spectrometer for the survey scans, a mass range from 200 to 2000 m/z at a resolution of 70,000 was used. The automatic gain control was set to 3,000,000, and the maximum fill time was 50 ms. The 10 most intensive peptide ions were isolated and fragmented by high-energy collision dissociation (HCD).
Computational mass spectrometric data analysis
Proteome Discoverer (version 2.1.0.81, Thermo Fisher Scientific, Bremen, Germany) was applied for peptide/protein identification by applying Mascot (version 2.4, Matrix Science, London, UK) as a search engine employing the EnsemblBacteria database (Lactococcus lactis subsp. cremoris NZ9000; date 03-11-2019). A false discovery rate of 1% (p ≤ 0.01) at the peptide level was set as the identification threshold. Proteins were quantified with Progenesis QI for Proteomics (Version 2.0, Nonlinear Dynamics, Waters Corporation, Newcastle upon Tyne, UK). Only proteins containing at least two unique peptides were taken into consideration. For the calculation of enriched proteins in the groups, a 5% false discovery rate and a minimum fold change of two were used.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD017318.
The protein lists, which have been uploaded to PRIDE, are also provided as Supplementary Material.
Abbreviations
- UDP:
-
Undecaprenyl-phosphate
- GlcNAc:
-
N-acetylglucosamine
- MurNAc:
-
N-acetylmuramic acid
- MurN:
-
N-deacetylated muramic acid
- Ala:
-
Alanine
- iGlu:
-
Isoglutamic acid
- iGln:
-
Isoglutamine
- Glu:
-
Glutamine
- Asp:
-
Aspartate/aspartic acid
- Asn:
-
Asparagine
- Lys:
-
Lysine
References
Laxminarayan, R. et al. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 13, 1057–1098 (2013).
Li, B. & Webster, T. J. Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopedic infections. J. Orthop. Res. 36, 22–32 (2018).
Dever, L. A. & Dermody, T. S. Mechanisms of bacterial resistance to antibiotics. Arch. Intern. Med. 151, 886–895 (1991).
Muller, A., Klockner, A. & Schneider, T. Targeting a cell wall biosynthesis hot spot. Nat. Prod. Rep. 34, 909–932 (2017).
Bugg, T. D. & Walsh, C. T. Intracellular steps of bacterial cell wall peptidoglycan biosynthesis: Enzymology, antibiotics, and antibiotic resistance. Nat. Prod. Rep. 9, 199–215 (1992).
Typas, A., Banzhaf, M., Gross, C. A. & Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123–136 (2011).
Dik, D. A. et al. A synthetic 5,3-cross-link in the cell wall of rod-shaped gram-positive bacteria. Proc. Natl. Acad. Sci. U.S.A. 118, e2100137118 (2021).
Kuk, A. C. Y., Hao, A., Guan, Z. & Lee, S. Y. Visualizing conformation transitions of the lipid II flippase MurJ. Nat. Commun. 10, 1736 (2019).
Lovering, A. L., Safadi, S. S. & Strynadka, N. C. Structural perspective of peptidoglycan biosynthesis and assembly. Annu. Rev. Biochem. 81, 451–478 (2012).
Breukink, E. & de Kruijff, B. Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 5, 321–332 (2006).
Wiedemann, I. et al. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276, 1772–1779 (2001).
Hsu, S. T. et al. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat. Struct. Mol. Biol. 11, 963–967 (2004).
Bonelli, R. R., Schneider, T., Sahl, H. G. & Wiedemann, I. Insights into in vivo activities of lantibiotics from gallidermin and epidermin mode-of-action studies. Antimicrob. Agents Chemother. 50, 1449–1457 (2006).
Perkins, H. R. Specificity of combination between mucopeptide precursors and vancomycin or ristocetin. Biochem. J. 111, 195–205 (1969).
Malin, J. J. & de Leeuw, E. Therapeutic compounds targeting lipid II for antibacterial purposes. Infect. Drug Resist. 12, 2613–2625 (2019).
Hu, Y., Helm, J. S., Chen, L., Ye, X. Y. & Walker, S. Ramoplanin inhibits bacterial transglycosylases by binding as a dimer to lipid II. J. Am. Chem. Soc. 125, 8736–8737 (2003).
Hamburger, J. B. et al. A crystal structure of a dimer of the antibiotic ramoplanin illustrates membrane positioning and a potential lipid II docking interface. Proc. Natl. Acad. Sci. U.S.A. 106, 13759–13764 (2009).
Lee, W. et al. The mechanism of action of lysobactin. J. Am. Chem. Soc. 138, 100–103 (2016).
Storm, D. R. & Strominger, J. L. Complex formation between bacitracin peptides and isoprenyl pyrophosphates. The specificity of lipid-peptide interactions. J. Biol. Chem. 248, 3940–3945 (1973).
Economou, N. J., Cocklin, S. & Loll, P. J. High-resolution crystal structure reveals molecular details of target recognition by bacitracin. Proc. Natl. Acad. Sci. U.S.A. 110, 14207–14212 (2013).
Kingston, A. W., Zhao, H., Cook, G. M. & Helmann, J. D. Accumulation of heptaprenyl diphosphate sensitizes Bacillus subtilis to bacitracin: Implications for the mechanism of resistance mediated by the BceAB transporter. Mol. Microbiol. 93, 37–49 (2014).
Lightbown, J. W., Kogut, M. & Uemura, K. The Second International Standard for Bacitracin. Bull. World Health Organ. 31, 101–109 (1964).
Rietkotter, E., Hoyer, D. & Mascher, T. Bacitracin sensing in Bacillus subtilis. Mol. Microbiol. 68, 768–785 (2008).
Wei, S., Gutek, A., Lilburn, M. & Yu, Z. Abundance of pathogens in the gut and litter of broiler chickens as affected by bacitracin and litter management. Vet. Microbiol. 166, 595–601 (2013).
Collinder, E. et al. Influence of zinc bacitracin and Bacillus licheniformis on microbial intestinal functions in weaned piglets. Vet. Res. Commun. 27, 513–526 (2003).
Manson, J. M., Keis, S., Smith, J. M. & Cook, G. M. Acquired bacitracin resistance in Enterococcus faecalis is mediated by an ABC transporter and a novel regulatory protein, BcrR. Antimicrob. Agents Chemother. 48, 3743–3748 (2004).
Ohki, R. et al. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49, 1135–1144 (2003).
Radeck, J. et al. Anatomy of the bacitracin resistance network in Bacillus subtilis. Mol. Microbiol. 100, 607–620 (2016).
Ma, J. et al. Bacitracin resistance and enhanced virulence of Streptococcus suis via a novel efflux pump. BMC Vet. Res. 15, 377 (2019).
Collins, B., Curtis, N., Cotter, P. D., Hill, C. & Ross, R. P. The ABC transporter AnrAB contributes to the innate resistance of Listeria monocytogenes to nisin, bacitracin, and various beta-lactam antibiotics. Antimicrob. Agents Chemother. 54, 4416–4423 (2010).
Hiron, A., Falord, M., Valle, J., Debarbouille, M. & Msadek, T. Bacitracin and nisin resistance in Staphylococcus aureus: A novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol. Microbiol. 81, 602–622 (2011).
Khosa, S., Lagedroste, M. & Smits, S. H. Protein defense systems against the lantibiotic nisin: Function of the immunity protein nisi and the resistance protein NSR. Front. Microbiol. 7, 504 (2016).
Popella, P. et al. VraH is the third component of the Staphylococcus aureus VraDEH system involved in Gallidermin and Daptomycin resistance and pathogenicity. Antimicrob. Agents Chemother. 60, 2391–2401 (2016).
Reiners, J. et al. The N-terminal region of nisin is important for the BceAB-type ABC transporter NsrFP from Streptococcus agalactiae COH1. Front. Microbiol. 8, 1643 (2017).
Dintner, S. et al. Coevolution of ABC transporters and two-component regulatory systems as resistance modules against antimicrobial peptides in Firmicutes Bacteria. J. Bacteriol. 193, 3851–3862 (2011).
Dintner, S., Heermann, R., Fang, C., Jung, K. & Gebhard, S. A sensory complex consisting of an ATP-binding cassette transporter and a two-component regulatory system controls bacitracin resistance in Bacillus subtilis. J. Biol. Chem. 289, 27899–27910 (2014).
Cho, J., Costa, S. K., Wierzbicki, R. M., Rigby, W. F. C. & Cheung, A. L. The extracellular loop of the membrane permease VraG interacts with GraS to sense cationic antimicrobial peptides in Staphylococcus aureus. PLoS Pathog. 17, e1009338 (2021).
Staron, A., Finkeisen, D. E. & Mascher, T. Peptide antibiotic sensing and detoxification modules of Bacillus subtilis. Antimicrob. Agents Chemother. 55, 515–525 (2011).
Mascher, T. Intramembrane-sensing histidine kinases: A new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol. Lett. 264, 133–144 (2006).
Gebhard, S. ABC transporters of antimicrobial peptides in Firmicutes bacteria—Phylogeny, function and regulation. Mol. Microbiol. 86, 1295–1317 (2012).
Revilla-Guarinos, A., Gebhard, S., Mascher, T. & Zuniga, M. Defence against antimicrobial peptides: Different strategies in Firmicutes. Environ. Microbiol. 16, 1225–1237 (2014).
Bernard, R., Guiseppi, A., Chippaux, M., Foglino, M. & Denizot, F. Resistance to bacitracin in Bacillus subtilis: Unexpected requirement of the BceAB ABC transporter in the control of expression of its own structural genes. J. Bacteriol. 189, 8636–8642 (2007).
Clemens, R., Zaschke-Kriesche, J., Khosa, S. & Smits, S. H. J. Insight into two ABC transporter families involved in lantibiotic resistance. Front. Mol. Biosci. 4, 91 (2017).
Gebhard, S. & Mascher, T. Antimicrobial peptide sensing and detoxification modules: Unravelling the regulatory circuitry of Staphylococcus aureus. Mol. Microbiol. 81, 581–587 (2011).
Kobras, C. M. et al. BceAB-type antibiotic resistance transporters appear to act by target protection of cell wall synthesis. Antimicrob. Agents Chemother. https://doi.org/10.1128/AAC.02241-19 (2020).
Fritz, G. et al. A new way of sensing: Need-based activation of antibiotic resistance by a flux-sensing mechanism. MBio 6, e00975 (2015).
Furtmann, F. et al. Characterization of the nucleotide-binding domain NsrF from the BceAB-type ABC-transporter NsrFP from the human pathogen Streptococcus agalactiae. Sci. Rep. 10, 15208 (2020).
Draper, L. A., Cotter, P. D., Hill, C. & Ross, R. P. Lantibiotic resistance. Microbiol. Mol. Biol. Rev. 79, 171–191 (2015).
Poyart, C., Lamy, M. C., Boumaila, C., Fiedler, F. & Trieu-Cuot, P. Regulation of D-alanyl-lipoteichoic acid biosynthesis in Streptococcus agalactiae involves a novel two-component regulatory system. J. Bacteriol. 183, 6324–6334 (2001).
Peschel, A., Vuong, C., Otto, M. & Gotz, F. The D-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob. Agents Chemother. 44, 2845–2847 (2000).
Benda, M., Schulz, L. M., Stulke, J. & Rismondo, J. Influence of the ABC transporter YtrBCDEF of Bacillus subtilis on competence, biofilm formation and cell wall thickness. Front. Microbiol. 12, 587035 (2021).
Storm, D. R. Mechanism of bacitracin action: A specific lipid-peptide interaction. Ann. N. Y. Acad. Sci. 235, 387–398 (1974).
Zaitseva, J., Jenewein, S., Jumpertz, T., Holland, I. B. & Schmitt, L. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 24, 1901–1910 (2005).
Alkhatib, Z., Abts, A., Mavaro, A., Schmitt, L. & Smits, S. H. Lantibiotics: How do producers become self-protected? J. Biotechnol. 159, 145–154 (2012).
Mierau, I. & Kleerebezem, M. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68, 705–717 (2005).
Klinzing, D. C. et al. The two-component response regulator LiaR regulates cell wall stress responses, pili expression and virulence in group B Streptococcus. Microbiology (Reading) 159, 1521–1534 (2013).
Vemula, H., Ayon, N. J., Burton, A. & Gutheil, W. G. Antibiotic effects on methicillin-resistant Staphylococcus aureus cytoplasmic peptidoglycan intermediate levels and evidence for potential metabolite level regulatory loops. Antimicrob. Agents Chemother. 61, e02253 (2017).
Lagedroste, M., Reiners, J., Smits, S. H. J. & Schmitt, L. Impact of the nisin modification machinery on the transport kinetics of NisT. Sci. Rep. 10, 12295 (2020).
Lagedroste, M., Smits, S. H. J. & Schmitt, L. Importance of the leader peptide sequence on the lanthipeptide secretion level. FEBS J. 288, 4348 (2021).
Courtin, P. et al. Peptidoglycan structure analysis of Lactococcus lactis reveals the presence of an L, D-carboxypeptidase involved in peptidoglycan maturation. J. Bacteriol. 188, 5293–5298 (2006).
Bellais, S. et al. Aslfm, the D-aspartate ligase responsible for the addition of D-aspartic acid onto the peptidoglycan precursor of Enterococcus faecium. J. Biol. Chem. 281, 11586–11594 (2006).
Bouhss, A. et al. Synthesis of the L-alanyl-L-alanine cross-bridge of Enterococcus faecalis peptidoglycan. J. Biol. Chem. 277, 45935–45941 (2002).
Vollmer, W., Blanot, D. & de Pedro, M. A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167 (2008).
Chapot-Chartier, M. P. & Kulakauskas, S. Cell wall structure and function in lactic acid bacteria. Microb. Cell Fact 13, S9 (2014).
Kobras, C. M. et al. BceAB-type antibiotic resistance transporters appear to act by target protection of cell wall synthesis. Antimicrob. Agents Chemother. https://doi.org/10.1101/835702 (2019).
Kunji, E. R., Slotboom, D. J. & Poolman, B. Lactococcus lactis as host for overproduction of functional membrane proteins. Biochim. Biophys. Acta 1610, 97–108 (2003).
Marreddy, R. K. et al. The response of Lactococcus lactis to membrane protein production. PLoS ONE 6, e24060 (2011).
Kramer, N. E., van Hijum, S. A., Knol, J., Kok, J. & Kuipers, O. P. Transcriptome analysis reveals mechanisms by which Lactococcus lactis acquires nisin resistance. Antimicrob. Agents Chemother. 50, 1753–1761 (2006).
AlKhatib, Z. et al. Lantibiotic immunity: Inhibition of nisin mediated pore formation by NisI. PLoS ONE 9, e102246 (2014).
Holo, H. & Nes, I. F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55, 3119–3123 (1989).
Abts, A. et al. Easy and rapid purification of highly active nisin. Int. J. Pept. 2011, 175145 (2011).
Abts, A., Montalban-Lopez, M., Kuipers, O. P., Smits, S. H. & Schmitt, L. NisC binds the FxLx motif of the nisin leader peptide. Biochemistry 52, 5387–5395 (2013).
Acknowledgements
The authors are thankful to Jelle Postma and Stefanie Weidtkamp-Peters from the Center for Advanced Imaging, Heinrich-Heine-University Germany, for valuable suggestions and great support during fluorophore studies. This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—270650915/GRK 2158 TP4a to S.S. E.S. gratefully acknowledges financial support from the DFG, TRR261, Project ID 398967434.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
J.G. and J.Z.K. performed biological assays and wrote the manuscript. S.U., I.V. and A.K. performed cell wall precursor analysis. L.V.B. performed biological assays. N.O. performed proteome analysis. K.S. and E.S. contributed to the writing. S.H.J.S. initiated and supervised the study. S.H.J.S. and J.G. wrote the manuscript with input of all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gottstein, J., Zaschke-Kriesche, J., Unsleber, S. et al. New insights into the resistance mechanism for the BceAB-type transporter SaNsrFP. Sci Rep 12, 4232 (2022). https://doi.org/10.1038/s41598-022-08095-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-08095-2
This article is cited by
-
Bacteriocin diversity, function, discovery and application as antimicrobials
Nature Reviews Microbiology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.