Abstract
Echovirus 9 (E9) belongs to the species Enterovirus B. So far, 12 whole genome sequences of E9 are available in GenBank. In this study, we determined the whole genomic sequences of five E9 strains isolated from the stools of patients with hand-foot-and-mouth disease in Kunming, Yunnan Province, China, in 2019. Their nucleotide and amino acid sequences shared 80.8–80.9% and 96.4–96.8% identity with the prototype Hill strain, respectively, and shared 99.3–99.9% and 99.1–99.8% mutual identity, respectively. Recombination analyses revealed that intertype recombination had occurred in the 2C and 3D regions of the five Yunnan E9 strains with coxsackieviruses B5 and B4, respectively. This study augmented the whole genome sequences of E9 in the GenBank database and extended the molecular characterization of this virus in China.
Similar content being viewed by others
Introduction
The genus Enterovirus, in the family Picornaviridae, is further divided into 15 species: Enterovirus A–K and Rhinovirus A–C, comprising 327 serotypes (https://www.picornaviridae.com). Enterovirus B (EV-B) currently consists of 63 serotypes: coxsackieviruses B (CVB1–6), CVA9,echoviruses (E1–7, 9, 11–21, 24–27, 29–33), and enteroviruses B (EV-B69, 73–75, 77–88, 93, 97–101, 106–107, 110–114) (https://www.picornaviridae.com). Like other enteroviruses (EVs), echoviruses are small nonenveloped, single-stranded positive-sense RNA viruses. The echovirus genomes are about 7400 nucleotides long and contain a long open reading frame (ORF), which encodes a polyprotein that is processed into three polyprotein precursors (P1, P2, and P3). These are further cleaved into four structural proteins (VP4, VP2, VP3, and VP1) and seven nonstructural proteins (2A, 2B, 2C, 3A, 3B, 3C, and 3D). The ORF is flanked by a 5′-untranslated region (UTR) and a 3′-UTR1,2.
EVs are commonly associated with asymptomatic or mild infections, such as fever, irritation, and hand-foot-and-mouth disease (HFMD), although some patients may develop severe diseases, including aseptic meningitis and acute flaccid paralysis. EVs are the main causative pathogen of aseptic meningitis3,4,5. Among the EVs, E9 is often associated with aseptic meningitis6,7,8,9,10,11,12,13 and HFMD14,15,16,17,18, and has been detected in a case of herpangina19,20. Previous studies have reported that E9 was main causative pathogen of aseptic meningitis and HFMD in Yunnan, China, in 2009–201013,14. Twelve whole genome sequences of E9 are currently available in GenBank, which were isolated from children with aseptic meningitis or HFMD, healthy children, and children with other diseases. However, only one is from a Chinese strain. In this study, the whole genomic sequences of five E9 strains recovered from children aged 0.9–4.5 years with HFMD in Kunming, Yunnan Province, China, in 2019 were determined to clarify the molecular characteristics of E9 in China and to increase the number of genome sequences of E9 in the GenBank database.
Results
Primary characterization and whole-VP1 sequence analysis
Four E9 strains (115K3/YN/CHN/2019 [115K3], 123K3/YN/CHN/2019 [123K3], 133K3/YN/CHN/2013 [133K3], and 121K3/YN/CHN/2019 [121K3]) were recovered from human embryonic lung diploid fibroblasts (KMB17) cells, and one strain (115V3/YN/CHN/2019 [115V3]) was recovered from Vero cells, whereas none were from human rhabdomyosarcoma (RD) cells. Of these isolates, 115K3 and 115V3 were isolated from the same sample but from KMB17 and Vero cells rather than human rhabdomyosarcoma (RD) cells. They were isolated from three boys and one girl, ranging in age from 0.9 to 4.5 years.
The whole VP1 sequences (918 nucleotides) of the five Yunnan strains showed the greatest identity (94.4%–94.8%) with E9 strain Echo9/FJPT176/CHN/2016 (MG922545), isolated from a patient with HFMD in China. The five isolates shared 78.3%–78.5% nucleotide identity and 85.7%–86.4% amino acid identity with the whole VP1 sequence of the E9 prototype Hill strain, which was isolated from a healthy child in Cincinnati in 195321, and 80.8%–94.8% nucleotide and 88.1%–99.6% amino acid identity with other E9 strains. The whole-VP1 nucleotide and amino acid identities among the five Yunnan isolates were 99.6%–99.9% and 99.3%–100%, respectively.
The 53 whole VP1 sequences available in GenBank were included in an analysis of the five isolates collected in this study (Fig. 1). According to the approximate mean 15% cutoff divergence value used to genotype enterovirus A71 (EV-A71)22, the E9 strains were divided into eight clusters (A–H). The main epidemic strains belonged to the D, F, and H clusters. Of these, cluster D contained most E9 strains (40 strains), including all the Chinese strains, together with isolates from Poland, Belgium, Australia, France, the USA, and Thailand, collected in 1997–2019. Cluster F included seven strains from France and Russian, collected in 2002–2012. Cluster H included four strains from the USA, Australia, and France, collected in 2013–2017.
Selection pressure analysis of the E9 VP1 gene
For the VP1 gene, no positive selection sites were found in the E9 Yunnan strains. However, only one negative selection site was found at amino acid position 286 of VP1 (position 286, H → R) (Table 1).
Whole-genome sequence analysis
The whole genome sequences of the five strains (115K3, 115V3, 123K3, 133K3, and 121K3) isolated in Yunnan Province in 2019 were determined. The genome sequences were 7445–7450 nucleotides in length, containing an ORF of 6612 nucleotides, which encoded a polyprotein of 2203 amino acids. The ORF sequence was flanked by a noncoding 5′-UTR of 733–738 nucleotides and a noncoding 3′-UTR of 103–106 nucleotides. The whole-genome nucleotide and amino acid identities of the five isolates were 99.3–99.9% and 99.1–99.8%, respectively. The total base compositions were 28.2–28.4% A, 24.6–24.7% G, 23.1–23.2% C, and 23.9–24.1% U. Because the mutual identities of the whole-genome nucleotide and deduced amino acid sequences of the five strains were > 99.1%, strain 115V3 was selected as the representative strain for further analysis.
Pairwise comparisons of the nucleotide and amino acid sequences of strain 115V3 and the E9 prototype Hill strain and other E9 strains are shown in Table 2. Strain 115V3 shares 79.0% and 80.5–88.7% nucleotide identities with the whole genomes of the E9 prototype Hill strain and the other E9 strains, respectively, and derived amino acid sequence identities of 94.4% and 93.5–97.6%, respectively.
Phylogenetic analysis of P1, P2, and P3 regions
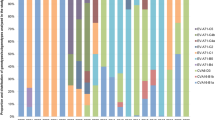
Phylogenetic trees were constructed for P1, P2, and P3 regions of all E9 strains and EV-B prototype strains available in GenBank, the five Yunnan isolates, and three EV-B strains (CV-B5/P727/2013/China, CVB4-B4M063015, and E11-1000/ISR/1999) (Fig. 2). In the P1 region, the Yunnan isolates grouped together with all E9 strains, including the E9 prototype Hill strain, thus further confirming the initial typing results. However, in the P2 and P3 coding regions, the E9 strains clustered with different EV-B strains and formed different clusters (Fig. 2). In the P2 region, the Yunnan isolates clustered with two E9 isolates (MSH/KM812/2010 and E-9/PMKA1322/THA/2011) and one CV-B5 strain, CV-B5/P727/2013/China (KP289438). In the P3 region, the Yunnan isolates clustered with one CVB4 strain, B4M063015 (MG845888). The prototype Hill strain formed a lineage with the E18 prototype strain Metcalf (AF317694), which is a recombinant with the Hill strain23. The results indicated that several potential recombination events have occurred between the E9 strains, including the Yunnan isolates, and other EV-B strains in the non-capsid coding regions, and that these E9 isolates may be the different recombinant strains.
Phylogenetic relationships based on the P1, P2, and P3 coding sequences of all E9 strains, all EV-B prototype strains available in the GenBank database, the five Yunnan isolates, and three EV-B strains (CV-B5/P727/2013/China, CVB4-B4M063015 and E11-1000/ISR/1999), as analyzed by nucleotide sequence alignment with the neighbor joining algorithms implemented in the MEGA 6.06 program. Numbers at nodes indicate bootstrap support for that node (percentage of 1000 bootstrap replicates). The scale bars represent the genetic distance. Only high bootstrap values (> 75%) are shown. Filled triangle: strains isolated in this investigation. Filled circle: E9 strains.
Recombination analysis
The sequences sharing the greatest identity with strain 115V3 were screened online with BLAST (Table 3). The P3 region of strain 115V3 shared greatest identity with that of CVB4 strain B4 M063015, whereas in the 2C and 3A regions, it shared the greatest identity (90.47%–91.43%) with the CVB5 strain P727/2013/China, and in the 3B, 3C, and 3D regions, it shared the greatest identity (89.39%–94.16%) with CVB4 strain B4M063015. In the 3′-UTR, strain 115V3 shared the greatest identity (92.93%) with E11 strain 1000/ISR/1999.
With a similarity plot and a bootscanning analysis, several recombination events were confirmed in the genomic sequence of strain 115V3 (Fig. 3). In the P1 region, strain 115V3 showed greatest identity (> 90%) with the E9 strain MSH/KM812/2010. However, in the 2C–3A regions and the 3B–3D region, strain 115V3 shared greatest homologies (> 92%) with CVB5 strain P727/2013 and CVB4 strain B4M063015, respectively. Furthermore, RDP4 analysis revealed that the 115V3 isolate underwent a recombination event involving the strain CVB4/B4M063015/MG845888. The recombination event started at approximately 5300 nt and ended at approximately 7350 nt, in the P3 region of the isolates’ genomes (Fig. 4).
Discussion
Since 2008, HFMD caused by EVs has become a serious infectious disease in mainland China, mainly occurring in children under 5 years of age. Many EVs, including echoviruses, cocirculated in both sporadic and epidemic cases of HFMD14,15,16,17,18. Since the introduction of an inactivated EV-A71 vaccine, the prevalent etiological agents of HFMD have changed, and CV-A6 has become the main pathogen in mainland China24,25. EVs, and especially echoviruses, are the major agents of aseptic meningitis3,4,5, and EV-B, including echoviruses, can also cause HFMD18,19,20. The serotypes of the EV pathogens causing HFMD are the same as those causing aseptic meningitis. Therefore, the persistent surveillance of the pathogens responsible for HFMD and aseptic meningitis is very important, and may allow the main serotypes of EVs associated with outbreaks to be predicted.
Because of their error-prone replication, EVs form highly polymorphic populations within their hosts. Since the prototype Hill strain was isolated in 195321 and the Barty strain was isolated from a child with aseptic meningitis in 195726, E9 isolates have evolved into eight clusters, thus indicating that E9 is genetically diverse. On the basis of analysis of the entire VP1 gene, the Chinese E9 strains were divided into D1 (2008–2010) and D3 (2010–2019) clusters, and the five Yunnan isolates in the study formed a single cluster. Furthermore, the average VP1 nucleotide and amino acid sequence divergence was 8.3% (5.3–11.2%) and 2.75% (0.3–5.2%) between the five Yunnan isolates and all whole VP1 gene sequences of other Chinese strains available in GenBank, respectively. In particular, the only whole genome of the Chinese E9 strain available in GenBank, MSH/KM812/2010, which was isolated from a patient with HFMD in 2010 in Yunnan Province, had the nucleotide and amino acid sequence divergence of 12.65% (12.5–12.8%) and 3.1% (2.4–3.8%), respectively. This finding indicated that the Chinese strains have evolved. Therefore, we speculate that the five Yunnan strains have adapted to a changing environment, in response to selection pressures.
The VP1 capsid protein of the EVs is located together with many immunodominant-serotype-specific epitopes in the exposed B-C loop. Although VP1 is the most variable of the capsid proteins, the N-terminus of VP1, which contains the B-C loop, is highly conserved within individual enteroviral serotypes27. The B-C loop is located inside the viral particle and is negligibly influenced by the immune pressure exerted by the host. The sequence of the E9 B-C loop, GDPESTDRFDA (amino acids 83–93 of the VP1 protein)28, in the five Yunnan strains is the same as that in other recent China strains, such as HY/F4/17/E9 (2017, aseptic meningitis), HY/F3/17/E9 (2017, aseptic meningitis), Echo9/FJSM296/CHN/2016 (2016, HFMD), Echo9/FJPT176/CHN/2016 (2016, HFMD), 060/LS/CHN/AM/08/E9 (aseptic meningitis), 051/LS/CHN/AM/08/E9, and 61253–70985 (aseptic meningitis). This indicates that the B-C loop is highly conserved in the Chinese strains. Moreover, amino acid A81, which is exposed at the surface near the B-C loop in the VP1 protein, is shared by most E9 strains, including the five Yunnan strains. A previous study reported that an E9 strain carrying the T81A substitution in VP1 had lytic potential toward pancreatic cells28. However, the non-lytic E9 strains Hill and Barty, including the five Yunnan strains in the study (dada not shown), contain alanine at this site. Thus, additional genetic substitutions in the viral genome may be very important in the pathogenicity toward pancreatic islets, and further research is required29.
The RGD motif at the C-terminus of the VP1 protein is very important to the pathogenicity of E930. The interaction between the virus and the cell occurs via the contact between the RGD motif and the host cell receptor29. The Barty strain, which is highly virulent in newborn mice, and other E9 strains, including three of the Yunnan strains, contain this motif, whereas the Hill strain, which is nonpathogenic to newborn mice, does not. This suggests that these three Yunnan strains would display the same pathogenicity as the Barty strain. However, the RGD → GGD substitution is present in strains 115V3 and 133K3, although the five Yunnan strains were all isolated from patients with HFMD, without aseptic meningitis. Therefore, the RGD → GGD substitution requires further study. For EV, VP1 is the most immunogenic protein and is involved in its host cell with receptor-mediated entry 31. However, one negative selection site (VP1 286 H → R) was found in the study. We speculate that although the amino acids are basic, this substitution may affect viral function, although this possibility requires further study.
Recombination is a major mechanism of enteroviral evolution, particularly that of EV-B32,33. The E9 prototype Hill strain is a recombinant between the E9 strain Barty and the E18 prototype strain Metcalf23. Therefore, we used BLAST online to screen the strains sharing the highest identity with the E9 strain 115V3 in different regions of the E9 genome. We consequently identified several putative recombination events in different coding regions between strain 115V3 and the CVB5 strain CV-B5-P727/2013/China, the CVB4 strain B4M063015, and the E11 strain 1000/ISR/1999. On the basis of the very high (approximately 90%) sequence identities, we inferred that these viruses are recombination partners. Among them, CBV5 strain CV-B5-P727/2013/China was isolated from patients with HFMD34, the CVB4 strain B4M063015 was isolated from raw sewage35, and the E11 strain 1000/ISR/1999 was isolated from a chronically infected immunodeficient patient. Es play major roles in natural recombination events among the coxsackieviruses B (CVBs)36. CVB infections are associated with HFMD, aseptic meningitis, acute myocarditis, and fatal neonatal infections37,38,39. CVB5 is one of the five most common EVs in the USA40, and it has the highest prevalence in Germany and Spain3,41. CVB5 was also involved in an outbreak of neurological HFMD in China42. The numbers of cases of HFMD caused by CVB4 and CVB5 have been reported to be increasing in China36. These viruses also frequently cocirculate in patients with HFMD in China43. Therefore, their epidemiological characteristics may offer these viruses sufficient opportunities for mixing and recombination.
In conclusion, the E9 strains were highly genetically diverse, and intertypic recombination events have occurred in the genomic regions encoding nonstructural proteins. This serotype is globally widespread, and frequently cocirculates with other EVBs. Recombination and mutation drive the evolution of E9. Although the five Yunnan E9 strains analyzed in this study were isolated from HFMD patients without aseptic meningitis, other possible pathogenicities, including aseptic meningitis, cannot be ruled out, and the pathogenicity of these strains warrants further study. Systematic epidemiological surveillance is required to assess the links between E9 and its associated diseases.
Materials and methods
Ethics statement
The research content of this study was approved by the Institutional Review Boards of the Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College and complied with the ethical regulations of that institution. The experimenters and staff involved in the project gave their informed consent, and informed consent was obtained from the patients’ families. The samples used in this experiment were obtained from stools collected from four children with HFMD, aged 0.9–4.5 years.
Viral isolation
The viruses were isolated from the samples with three different cell lines (KMB17, Vero, and RD)44. These cells were provided by the Chinese Academy of Medical Sciences. Positive cultures (those with cytopathic effects after three passages) were stored at −80 °C.
Viral identification and sequencing
The culture supernatants were collected and freeze-thawed three times to allow the complete release of the virus from the cells. The viral RNAs were extracted from the culture supernatants with a QIAamp Viral RNA Mini Kit (QIAGEN, CA, USA) according to the instruction manual. Reverse transcription-polymerase chain reaction (RT-PCR)44 was performed with a PrimeScript™ One Step RT-PCR Kit Ver.2 (TaKaRa, Dalian, China) with primer pairs AN88 (TACTGGACCACCTGGNGGNA) and AN89 (CCAGCACTGACAGCAGYNG). The PCR-positive products were sequenced with an ABI 3130 Genetic Analyzer (Applied Biosystems, USA) at Kunming Tsingke Biotechnology Co., Ltd. (Kunming, Yunnan Province, China). The sequences were compared with BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
The serotype was identified by comparison of the nucleotide sequence with known sequences using BLAST45. Two long-distance PCR amplifications were performed with a PrimeScript™ One Step RT-PCR Kit Ver.2 (TaKaRa, Dalian, China) and the following primer pairs: E201F (TTAAAACAGCCTGTGGGTTG) and E93R (TCCACATCAAAGCGCAAGTA) for 5´ end amplification, and E93F (AGGCATGTGAAAAATTACCA) and E98R (ACCGAATGCGGAGAATTTAC) for 3’ end amplification. The primers used for sequencing of the whole-length genome were designed by a primer-walking” strategy46. All primers used in the study are listed in Table 2. The PCR products were purified with a QIAquick PCR purification kit (Qiagen, Germany), and sequenced in both directions at least twice with an ABI 3130 Genetic Analyzer (Applied Biosystems, USA).
Selection pressure analysis of the E9 VP1 gene
The selection pressure of E9 VP1 was predicted with the Datamonkey online Application46 (http://www.datamonkey.org) and calculated with the following four methods: mixed-effects model of evolution (MEME); fixed-effects likelihood (FEL); fast unbiased Bayesian approximation (FUBAR); and single likelihood ancestor count (SLAC). The significance level was set at 0.05 for MEME, FEL, and SLAC and at 0.9 for FUBAR. Validated results obtained from both methods were considered credible (Table 4).
Sequence analysis and recombination analysis
Nucleotide and amino acid sequence analyses was performed with the Geneious Basic 5.4.1 Beta software. MEGA 7.0 was used for the phylogenetic analysis47, with a Kimura two-parameter model and the neighbor-joining method, with 1000 bootstrap replicates. The Recombination Detection Program (RDP) package Beta 4.101 was used to identify recombinant sequences in the default mode. Recombination analysis was performed with the seven algorithms in the following programs: RDP, GENECONV, BootScan, Maxchi, Chimaera, SiScan, and 3Seq. Recombination events with significance p < 0.01 obtained by three or more algorithms were considered reliable. The Jukes-Cantor method was used48 in Simplot 3.5.1, with each 200-nucleotide (nt) window shifted in 20 nt steps, and a similarity plot was constructed and a bootscanning analysis performed. Strain 115V3/YN/CHN/2019 was compared with other GenBank-listed sequences to identify the sequences that shared the highest homology with 115V3/YN/CHN/2019 in each genomic segment, and the most similar strains for each segment are listed in Table 3.
Nucleotide sequence accession numbers
The whole genomes of the five E9 strains isolated in this study have been submitted to GenBank under accession numbers MZ488277–MZ488281.
References
Song, Y. et al. Phylogenetic characterizations of highly mutated EV-B106 recombinants showing extensive genetic exchanges with other EV-B in Xinjiang, China. Sci. Rep. 7, 43080. https://doi.org/10.1038/srep43080 (2017).
Nasri, D. et al. Basic rationale, current methods and future directions for molecular typing of human enterovirus. Expert Rev. Mol. Diagn. 7, 419–434. https://doi.org/10.1586/14737159.7.4.419 (2007).
Trallero, G. et al. Enteroviruses in Spain over the decade 1998–2007: Virological and epidemiological studies. J. Clin. Virol. 47, 170–176. https://doi.org/10.1016/j.jcv.2009.11.013 (2010).
Cabrerizo, M. et al. Molecular characterization of enteroviruses associated with neurological infections in Spain, 2008. J. Med. Virol. 85, 1975–1977. https://doi.org/10.1002/jmv.23693 (2013).
Li, W. et al. Molecular epidemiological study of enteroviruses associated with encephalitis in children from Hangzhou, China. Medicine (Baltimore) 95, e4870. https://doi.org/10.1097/MD.0000000000004870 (2016).
Lee, H. Y. et al. Clinical features of echovirus 6 and 9 infections in children. J. Clin. Virol. 49, 175–179. https://doi.org/10.1016/j.jcv.2010.07.010 (2010).
Graf, J. et al. Meningitis gone viral: Description of the echovirus wave 2013 in Germany. BMC Infect. Dis. 19, 1010. https://doi.org/10.1186/s12879-019-4635-6 (2019).
Danthanarayana, N. et al. Acute meningoencephalitis associated with echovirus 9 infection in Sri Lanka, 2009. J. Med. Virol. 87, 2033–2039. https://doi.org/10.1002/jmv.24267 (2015).
Tao, Z. et al. Molecular epidemiology of human enterovirus associated with aseptic meningitis in Shandong Province, China, 2006–2012. PLoS ONE 9, e89766. https://doi.org/10.1371/journal.pone.0089766 (2014).
Krasota, A., Loginovskih, N., Ivanova, O. & Lipskaya, G. Direct identification of enteroviruses in cerebrospinal fluid of patients with suspected meningitis by nested PCR amplification. Viruses https://doi.org/10.3390/v8010010 (2016).
Nkosi, N. et al. Molecular characterisation and epidemiology of enterovirus-associated aseptic meningitis in the Western and Eastern Cape Provinces, South Africa 2018–2019. J. Clin. Virol. 139, 104845. https://doi.org/10.1016/j.jcv.2021.104845 (2021).
Smuts, H. et al. Molecular characterization of an outbreak of enterovirus-associated meningitis in Mossel Bay, South Africa, December 2015-January 2016. BMC Infect. Dis. 18, 709. https://doi.org/10.1186/s12879-018-3641-4 (2018).
Zhu, Y. et al. Molecular identification of human enteroviruses associated with aseptic meningitis in Yunnan province, Southwest China. Springerplus 5, 1515. https://doi.org/10.1186/s40064-016-3194-1 (2016).
Ma, S. et al. Dynamic constitution of the pathogens inducing encephalitis in hand, foot and mouth disease in Kunming, 2009–2011. Jpn. J. Infect. Dis. 68, 504–510. https://doi.org/10.7883/yoken.JJID.2014.428 (2015).
Zhou, Y. et al. Genetic variation of multiple serotypes of enteroviruses associated with hand, foot and mouth disease in Southern China. Virol. Sin. 36, 61–74. https://doi.org/10.1007/s12250-020-00266-7 (2021).
Zhou, Y. et al. Molecular strategy for the direct detection and identification of human enteroviruses in clinical specimens associated with hand, foot and mouth disease. PLoS ONE 15, e0241614. https://doi.org/10.1371/journal.pone.0241614 (2020).
Yao, X. et al. Epidemiological and etiological characteristics of herpangina and hand foot mouth diseases in Jiangsu, China, 2013–2014. Hum. Vaccin. Immunother. 13, 823–830. https://doi.org/10.1080/21645515.2016.1236879 (2017).
Du, Z. et al. Ongoing change of severe hand, foot, and mouth disease pathogens in Yunnan, China, 2012 to 2016. J. Med. Virol. 91, 881–885. https://doi.org/10.1002/jmv.25393 (2019).
Li, W. et al. Molecular epidemiology of enterovirus from children with herpangina or hand, foot, and mouth disease in Hangzhou, 2016. Arch. Virol. 164, 2565–2571. https://doi.org/10.1007/s00705-019-04356-0 (2019).
Peng, Q. et al. Molecular epidemiology of the enteroviruses associated with hand, foot and mouth disease/herpangina in Dongguan, China, 2015. Arch. Virol. 161, 3463–3471. https://doi.org/10.1007/s00705-016-3058-6 (2016).
Zimmermann, H., Eggers, H. J., Zimmermann, A., Kraus, W. & Nelsen-Salz, B. Complete nucleotide sequence and biological properties of an infectious clone of prototype echovirus 9. Virus Res. 39, 311–319. https://doi.org/10.1016/0168-1702(95)00078-x (1995).
Chan, Y. F., Sam, I. C. & AbuBakar, S. Phylogenetic designation of enterovirus 71 genotypes and subgenotypes using complete genome sequences. Infect. Genet. Evol. 10, 404–412. https://doi.org/10.1016/j.meegid.2009.05.010 (2010).
Andersson, P., Edman, K. & Lindberg, A. M. Molecular analysis of the echovirus 18 prototype: Evidence of interserotypic recombination with echovirus 9. Virus Res. 85, 71–83. https://doi.org/10.1016/s0168-1702(02)00019-9 (2002).
Jiang, H. et al. The epidemiological characteristics of enterovirus infection before and after the use of enterovirus 71 inactivated vaccine in Kunming, China. Emerg. Microbes Infect. 10, 619–628. https://doi.org/10.1080/22221751.2021.1899772 (2021).
Meng, X. D. et al. Epidemical and etiological study on hand, foot and mouth disease following EV-A71 vaccination in Xiangyang, China. Sci. Rep. 10, 20909. https://doi.org/10.1038/s41598-020-77768-7 (2020).
Zimmermann, H., Eggers, H. J. & Nelsen-Salz, B. Molecular cloning and sequence determination of the complete genome of the virulent echovirus 9 strain barty. Virus Genes 12, 149–154. https://doi.org/10.1007/BF00572953 (1996).
Norder, H., Bjerregaard, L. & Magnius, L. O. Homotypic echoviruses share amino terminal VP1 sequence homology applicable for typing. J. Med. Virol. 63, 35–44 (2001).
Hara, K. et al. Molecular evolution of human echovirus 9 isolated from patients with aseptic meningitis in northern Kyushu during the summer of 1997. Microbiol. Immunol. 45, 717–720. https://doi.org/10.1111/j.1348-0421.2001.tb01306.x (2001).
Paananen, A. et al. A single amino acid substitution in viral VP1 protein alters the lytic potential of clone-derived variants of echovirus 9 DM strain in human pancreatic islets. J. Med. Virol. 85, 1267–1273. https://doi.org/10.1002/jmv.23574 (2013).
Zimmermann, H., Eggers, H. J. & Nelsen-Salz, B. Cell attachment and mouse virulence of echovirus 9 correlate with an RGD motif in the capsid protein VP1. Virology 233, 149–156. https://doi.org/10.1006/viro.1997.8601 (1997).
Meng, T., Wong, S. M. & Chua, K. B. A novel attenuated enterovirus A71 mutant with VP1-V238A, K244R exhibits reduced efficiency of cell entry/exit and augmented binding affinity to sulfated glycans. J. Virol. 95, e0105521. https://doi.org/10.1128/JVI.01055-21 (2021).
Zhang, J. et al. Molecular characterization of a new human coxsackievirus B2 associated with severe hand-foot-mouth disease in Yunnan Province of China in 2012. Arch. Virol. 162, 307–311. https://doi.org/10.1007/s00705-016-3075-5 (2017).
Zheng, H. et al. Isolation and characterization of a highly mutated Chinese isolate of enterovirus B84 from a patient with acute flaccid paralysis. Sci. Rep. 6, 31059. https://doi.org/10.1038/srep31059 (2016).
Guo, W. P. et al. Fourteen types of co-circulating recombinant enterovirus were associated with hand, foot, and mouth disease in children from Wenzhou, China. J. Clin. Virol. 70, 29–38. https://doi.org/10.1016/j.jcv.2015.06.093 (2015).
Meister, S., Verbyla, M. E., Klinger, M. & Kohn, T. Variability in disinfection resistance between currently circulating enterovirus B serotypes and strains. Environ. Sci. Technol. 52, 3696–3705. https://doi.org/10.1021/acs.est.8b00851 (2018).
Pu, X., Qian, Y., Yu, Y. & Shen, H. Echovirus plays a major role in natural recombination in the coxsackievirus B group. Arch. Virol. 164, 853–860. https://doi.org/10.1007/s00705-018-4114-1 (2019).
Tracy, S. & Gauntt, C. Group B coxsackievirus virulence. Curr. Top. Microbiol. Immunol. 323, 49–63. https://doi.org/10.1007/978-3-540-75546-3_3 (2008).
Wikswo, M. E. et al. Increased activity of Coxsackievirus B1 strains associated with severe disease among young infants in the United States, 2007–2008. Clin. Infect. Dis. 49, e44-51. https://doi.org/10.1086/605090 (2009).
Chen, W. et al. Molecular epidemiology of Coxsackievirus B1–5 associated with HFMD in Fujian Province, China, 2011–2016. Biomed. Environ. Sci. 32, 633–638. https://doi.org/10.3967/bes2019.082 (2019).
Khetsuriani, N. et al. Enterovirus surveillance–United States, 1970–2005. MMWR Surveill. Summ. 55, 1–20 (2006).
Roth, B., Enders, M., Arents, A., Pfitzner, A. & Terletskaia-Ladwig, E. Epidemiologic aspects and laboratory features of enterovirus infections in Western Germany, 2000–2005. J. Med. Virol. 79, 956–962. https://doi.org/10.1002/jmv.20917 (2007).
Han, J. F. et al. Recombination of human coxsackievirus B5 in hand, foot, and mouth disease patients, China. Emerg. Infect. Dis. 18, 351–353. https://doi.org/10.3201/eid1802.111524 (2012).
Xiao, J. et al. Coxsackievirus B4: An underestimated pathogen associated with a hand, foot, and mouth disease outbreak. Arch. Virol. 166, 2225–2234. https://doi.org/10.1007/s00705-021-05128-5 (2021).
Prim, N. et al. Combining cell lines to optimize isolation of human enterovirus from clinical specimens: Report of 25 years of experience. J. Med. Virol. 85, 116–120. https://doi.org/10.1002/jmv.23426 (2013).
Kroneman, A. et al. An automated genotyping tool for enteroviruses and noroviruses. J. Clin. Virol. 51, 121–125. https://doi.org/10.1016/j.jcv.2011.03.006 (2011).
Pond, S. L. & Frost, S. D. Datamonkey: Rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21, 2531–2533. https://doi.org/10.1093/bioinformatics/bti320 (2005).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. https://doi.org/10.1093/molbev/msw054 (2016).
Lole, K. S. et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73, 152–160. https://doi.org/10.1128/JVI.73.1.152-160.1999 (1999).
Acknowledgements
This work was supported by the Research Projects of Yunnan Province, China (grant numbers 20200AA100009, 2017FA006) and Innovation Team Project of Yunnan Science and Technology Department (202105AE160020) and CAMS Innovation Fund for Medical Sciences(CIFMS) (2021-I2M-1-043).
Author information
Authors and Affiliations
Contributions
M.Z. and W.G. contributed equally to this work as joint first authors of this study. M.Z., W.G., D.X. and S.M. conceived the study and prepared the manuscript; M.Z., C.F., D.X. and W.G. performed the experiments. G.B., S.M., W.G., Z.Y. and H.S. performed the data analysis. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, M., Guo, W., Xu, D. et al. Molecular characterization of echovirus 9 strains isolated from hand-foot-and-mouth disease in Kunming, Yunnan Province, China. Sci Rep 12, 2293 (2022). https://doi.org/10.1038/s41598-022-06309-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-06309-1
This article is cited by
-
Complete genome analysis of echovirus 30 strains isolated from hand-foot-and-mouth disease in Yunnan province, China
Virology Journal (2023)
-
Genetic Characteristics and Phylogeographic Dynamics of Echovirus
Journal of Microbiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.