Abstract
Primary cells isolated from the human respiratory tract are the state-of-the-art for in vitro airway epithelial cell research. Airway cell isolates require media that support expansion of cells in a basal state to maintain the capacity for differentiation as well as proper cellular function. By contrast, airway cell differentiation at an air–liquid interface (ALI) requires a distinct medium formulation that typically contains high levels of glucose. Here, we expanded and differentiated human basal cells isolated from the nasal and conducting airway to a mature mucociliary epithelial cell layer at ALI using a medium formulation containing normal resting glucose levels. Of note, bronchial epithelial cells expanded and differentiated in normal resting glucose medium showed insulin-stimulated glucose uptake which was inhibited by high glucose concentrations. Normal glucose containing ALI also enabled differentiation of nasal and tracheal cells that showed comparable electrophysiological profiles when assessed for cystic fibrosis transmembrane conductance regulator (CFTR) function and that remained responsive for up to 7 weeks in culture. These data demonstrate that normal glucose containing medium supports differentiation of primary nasal and lung epithelial cells at ALI, is well suited for metabolic studies, and avoids pitfalls associated with exposure to high glucose.
Similar content being viewed by others
Introduction
When isolated, human airway epithelial cells are a mixture reflecting their site of origin, including the nasal, conducting and terminal airway1,2. Nasal and conducting airways show many similarities in morphology and cell type including non-ciliated, ciliated, secretory and multipotent basal progenitor cells. A full complement of mature, location-appropriate and niche appropriate cell types are required to accurately study the airway within a native physiological context. Each cell type contributes to the overall physiology particular to their location in the airway3. Basal cells are largely responsible for the maintenance of a differentiated epithelium, acting as the common airway progenitor cell4. Ciliated cells move mucus unidirectionally out of the airways5. Goblet cells and cells originating from the epithelial lined ducts of submucosal glands secrete heavily glycosylated, mucus-forming proteins6,7. Solitary chemosensory cells sense xenobiotics and other stimuli that induce calcium-mediated intercellular signaling to nearby cells8. Single cell RNAseq analysis has revealed heterogeneity among different cell types and rare subpopulations, such as ionocytes that express high levels of CFTR9,10,11. Each cell type is influenced by environmental niche factors which contribute to collective cellular homeostasis and disease dynamics in tissues12,13.
Cultured human primary cells have proven to be a valuable model to study airway cell differentiation and disease states including cystic fibrosis14,15,16,17,18,19,20,21, asthma22,23, chronic obstructive pulmonary disease (COPD)24 and COVID-1925,26,27. To mimic the native cell environment, cell culture media contain key factors found in tissue fluids in vivo required to support proper cell differentiation4,28,29. Culture media resembling human plasma has supported the premise that nutrient sources in vitro can negatively impact cellular homeostasis which influences assays widely used in research such as cell-based drug screens30. Early passage primary cells grown in traditional epithelial growth medium often do not fully differentiate after expansion which limits their utility as a model to study native tissue physiology. This is even more of a concern when considering human cell samples obtained from non-invasive epithelial sampling techniques, such as nasal or conducting airway brushings, which are valuable for rare disease research but limited by the absolute number of cells that are collected17,31,32. Basal cells derived from human induced pluripotent stem cells (iPSCs) represent another potential source of differentiable cells33. Methods to expand small samples into a large bank of cells with the capacity to be well-differentiated allows better access to primary respiratory epithelial cells for human disease research.
Expanding primary cells while maintaining their capacity to differentiate requires methods that rely on chemical inhibition or stalling of differentiation while still enabling basal progenitor cells to propagate. In addition to retaining multipotency in the expanding pool of basal progenitor cells, expansion methods prolong the differentiation potential of the resulting expanded cells. The cells are then able to undergo more population doublings than using traditional expansion methods, while retaining their ability to differentiate into a properly differentiated cell layer in vitro.
Chief among these is the conditional reprogramming culture (CRC) method where primary epithelial cells are co-cultured with irradiated (non-proliferating) fibroblasts in medium containing the rho-associated protein kinase (ROCK) inhibitor Y-2763228,34,35. These conditions preserve the basal cell phenotype, prevent differentiation and are fully reversible36,37. Epithelial cells expanded using CRC conditions retain their ability to differentiate for at least twice as long as compared to cells grown in traditional epithelial growth medium36.
Another approach, the Dual SMAD inhibitor method, has also proven effective and involves culturing basal cells in the presence of inhibitors that target TGFβ (usually SB431542) and BMP4 signaling (dorsomorphin, LDN193189 or recombinant Noggin)29,38,39. Note that the CRC and Dual SMAD basal cell expansion methods are not completely equivalent. For instance, recent evidence suggests that the CRC method better preserves some aspects of airway cell differentiation potential including ciliation and CFTR channel function40.
Traditional culture techniques rely on expansion media with formulations that are rich in sugars, serum proteins, and supplements that exceed levels found in healthy human serum. This results in medium that fosters cell viability but causes cell overgrowth due to an overreliance in anerobic glycolytic energy metabolism over aerobic oxidative phosphorylation41,42. Given accumulating evidence that the metabolic microenvironment can have a significant impact on airway epithelial cell function14,18,20,43,44,45, it is important to consider medium composition as a variable that can influence behavior of cells in vitro46. Of note, several media commonly used to support airway epithelial cell differentiation have high glucose concentrations, including LHC Basal:DMEM-H 50:5047 and Pneumocult-ALI48 both of which contain ~ 300 mg/dL glucose.
Here we describe a method that can be used to propagate and differentiate basal cells from the upper and conducting airways in medium containing normal resting glucose concentrations. The ALI medium formulation described here supports the ability of airway cells to respond to insulin by stimulating glucose uptake, suggesting that these cultures are well suited for use in studying the impact of energy metabolism on airway cell function.
Results and discussion
Isolation and differentiation of airway epithelial cells
Figure 1 illustrates the workflow of tissue harvest, primary cell isolation, basal cell selection and propagation and differentiation to produce airway cell models to study in vitro. The overall health of the donor tissue directly affects the quality of the resulting isolated cell culture. In our experience, the number of viable epithelial cells able to be isolated is mainly affected by the condition of the tissue when received and the time since resection. Culture of airway epithelial cells consists of two phases: 1) propagation of basal epithelial cells followed by 2) preparation of differentiated cultures using an air liquid interface (ALI).
Workflow for isolation of cells from distinct anatomic regions of the airway tree. (A) Examples of healthy tracheal and bronchial tissue isolates are shown based on donor tissue color, shape, and rigidity as markers for tissue health. (B) Freshly isolated primary (passage 0; P0) cells isolated from nasal or lung tissue contain a mixture of cells, including ciliated (inset) and non-ciliated cells. Bars, 20 μm (left) and 10 μm (right). (C) Basal nasal or airway epithelial cells are selected and expanded using CRC conditions, as imaged by phase contrast microscopy. Bar, 10 μm. (D) Basal airway or nasal cells cultured in E-ALI containing normal glucose properly differentiate as determined by immunofluorescence confocal microscopy using markers for mucus secretion (Muc5AC, red), basal cells (KRT5, green), nuclei (blue, DAPI) and by scanning electron microscopy. Bars, 20 μm (top and middle) and 10 μm (bottom).
Initially, P0 cells were cultured using the CRC method which is submersion culture in medium based on a mixture of DMEM and F12, resulting in normal resting glucose levels (150 mg/dL; 8.3 mM), and including biologic co-factors, the ROCK inhibitor Y-27638 and cholera toxin (FYRM; Table 1). The cells are seeded on collagen coated dishes and co-cultured with 3T3 fibroblasts that were irradiated to inhibit their propagation (Fig. 2A). When ~ 40% or more P0 cells attached to collagen coated dishes seeded with irradiated 3T3 cells, this was an indication that tissue processing was successful. Generally, cells exhibited a 5-day lag period before beginning to proliferate (Fig. 2B). Following the lag period, growth rates of cells isolated from anatomically different areas of the airway were similar (Fig. 2B) with a population doubling rate of roughly one per day (Fig. 2C).
Airway epithelial cells from different tissue sources propagate with similar doubling times. (A) Timeline for processing of cells isolated from lung tissue samples showing frozen cell banking (F), plating for differentiation on Transwell permeable supports (T) or plating for expansion under CRC conditions. Cells from P0 through P3 are banked. Cells beyond P4 are not typically used to generate differentiated cultures for experimental analysis. Detail related to culture on Transwells (T) is shown below, indicating the shift from submerged to air–liquid interface (ALI). Cells are cultured at least 14 days at ALI prior to use in experiments. (B) Representative NhNE, NhTE, and NhBE displayed a lag phase of growth between Day 0 and Day 5 in CRC conditions before replicating at a linear rate. (C) Regardless of anatomical origin or the initial lag phase, airway epithelial cells showed comparable doubling rates under CRC conditions. n = 2 – 3 wells from n = 2 (NhTE, NhBE) or 4 (NhNE) biological replicates plotted as mean ± SD. (D, E) Immunofluorescence analysis of differentiated tracheal epithelial cells (NhTE) at Day 14 in E-ALI. Ciliated cells were identified by immunostaining for acetylated tubulin (Ac-Tubulin, red) and basal cells by cytokeratin 5 (KRT5, green). Nuclei were labeled with DAPI (blue). (D). Mucus producing cells were identified by Muc5AC expression (red) (E). Bar, 20 μm.
Once the cells were isolated and propagated, frozen cell stocks were made at each passage creating a bank of cells with consistent properties that can be used for experimentation (Fig. 2A). Generally, we avoid cryobanking after P3 to ensure that the basal cells maintain their capacity to differentiate. For freshly isolated P0 cells, freeze densities of two million cells per vial allow for at least 500,000 cells per thaw to attach. For P1-P3, freeze densities of one million cells per vial are recommended to facilitate rapid growth from banked vials.
Culture of airway basal cells at ALI is a well-established method to promote their differentiation, however most media used for this purpose contain high glucose concentrations (~ 300 mg/dL; 16.7 mM)47,48, which reflects a hyperglycemic state14,44. Given this, we developed a modified ALI medium, E-ALI, based on a widely used medium formulation47. As shown in Table 2, E-ALI contains normal resting glucose levels (150 mg/dL; 8.3 mM). Otherwise, E-ALI is comparable to other ALI medium formulations47,48, except that it has less insulin (5 µg/ml) and is enriched for the following components: CaCl2 (1 mM), heparin (2 µg/ml), L-glutamine (2.5 mM), hydrocortisone 960 mg/ml, O-phosphorylethanolamine (0.5 µg/ml), bovine pituitary extract (20 µg/ml) and Mg2+ (0.5 µM).
We validated the ability of E-ALI to support the growth and differentiation of freshly isolated P0 normal human tracheal epithelial cells (NhTE cells) plated on collagen-coated Transwell permeable supports as assessed by immunofluorescence microscopy using KRT5 as a marker for basal cells (Fig. 2D-E), acetylated-tubulin as a marker for ciliated epithelia (Fig. 2D), and Muc5AC as a marker for mucus producing cells (Fig. 2E). Cultures routinely contained all three different cell types, indicating that they were well differentiated.
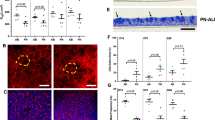
Comparable results were obtained using normal human bronchial epithelial cells (NhBE cells), which are delineated by apical junctions as marked by the tight junction protein ZO-1 (Fig. 3A-F) and also show ciliated cells (Fig. 3A,C), mucus producing cells (Fig. 3B,D), ionocytes (Fig. 3E) and club cells (Fig. 3F). After 14 days of culture in E-ALI, NhBE monolayers had significantly more ciliated cells than NhTE monolayers (32.7 ± 8.9%, n = 3 replicates, 14 fields; 23.2 ± 9.0%, n = 3 replicates, 18 fields) (Fig. 3G). The number of basal cells were comparable for NhBE and NhTE monolayers (38.8 ± 7.2%, n = 2 replicates, 7 fields; 41.9 ± 5.4%, n = 2 replicates, 8 fields). Muc5AC positive cells were also comparable for NhBE and NhTE monolayers (3.9 ± 1.9%, n = 3 replicates, 18 fields; 5.3 ± 2.9%, n = 3 replicates, 14 fields). NhBE monolayers also contained low levels of club cells (2.0 ± 1.1%, n = 2 replicates, 12 fields) and ionocytes (0.1 ± 0.1%, n = 2 replicates, 13 fields). Altogether, we accounted for 77.5 ± 11.7% of the total cells in NhBE monolayers grown using E-ALI. Other cell types likely to be present include suprabasal cells which express KRT4, KRT8 and KRT1349,50, however, suprabasal and other cell populations defined by multiple markers are difficult to detect strictly by immunofluorescence profiling.
Tracheal and bronchial epithelial cells differentiate into mucociliary cultures in E-ALI medium. (A–E) The E-ALI formulation enabled formation of well-differentiated cultured bronchial epithelial cell (NhBE) monolayers in vitro as observed by the presence of tight junctions (green, zonula occludins-1, ZO-1). Cell differentiation was demonstrated by immunofluorescence microscopy measuring acetylated tubulin (red, Ac-Tubulin; A, C), mucin (red, Muc5AC; B, D), an ionocyte marker (red, FoxI1; E) and a club cell marker (red, Scgb1a1; F). Nuclei were labeled with DAPI (blue). Bar, 20 μm. (G) Quantitation of NhTE and NhBE phenotype in cells cultured for 14 days in E-ALI. Data are from n = 2 – 9 fields from n = 2 (KRT5, FoxI1, Scgb1a) or 3 (Muc5AC, AcTub) biological replicates. ***P < 0.0001 by one-way ANOVA. (H) Transepithelial electrical resistance (TER) of NhTE cells was slightly higher than NhBE cells n = 3—21 wells from 4 biological replicates; *P = 0.042, by t test.
Consistent with formation of tight junctions, NhBE and NhTE cells cultured in E-ALI showed high transepithelial resistance (TER) (Fig. 3H), where the barrier formed by NhTE cells after 14 days in E-ALI was slightly, but significantly tighter than NhBE cells (~ 1340 vs ~ 1160 Ohm x cm2). Taken together, these data indicate that the E-ALI formulation containing normal resting glucose supported airway epithelial cell function as determined by apical junction assembly and differentiation of tracheal and bronchial epithelial cells.
Cells cultured in E-ALI medium show insulin-stimulated glucose uptake
To measure the effect of extracellular glucose and medium composition on insulin and glucose clearance, we examined NhBE cells cultured in either E-ALI or 2% Ultroser G containing normal (150 mg/dL) or high (300 mg/dL) glucose (Fig. 4). It is worth noting that E-ALI containing 300 mg/dL glucose is comparable to the glucose content of LHC Basal:DMEM-H 50:5047 and Pneumocult-ALI48. Insulin clearance was not sensitive to medium formulation, where all of the cell culture models tested cleared the majority of insulin within the first 24 h after feeding, causing it to plateau at a low level (~ 0.4 µg/mL; Fig. 4A).
Glucose and insulin sensitivity depends on differentiation medium. (A) Insulin consumption was comparable in all media tested. n = 4 samples, one biological replicate per condition. (B) Glucose consumption of differentiated, expanded P0 airway epithelial cells of mixed tracheal/bronchial origin. Red triangles and squares indicate cells cultured in E-ALI medium and blue triangles and squares indicate cells cultured in 2% Ultroser G (2% UG) medium. High glucose = 300 mg/dL glucose in the base media (triangles); Normal resting glucose = 150 mg/dL glucose in the base media (squares). Glucose consumption was significantly higher for cells in high glucose E-ALI as compared with cells in high glucose 2% UG (n = 4 samples, one biological replicate; ****P < 0.0001; **P = 0.0085; *P = 0.014, by two way ANOVA). (C) Cells differentiated using E-ALI medium containing normal resting glucose elicited a significant increase in insulin stimulated [3H]-2-D-glucose uptake. n = 3–6 samples from n = 1 (E-ALI high glucose, 2% UG normal glucose), 3 (2% UG High glucose) or 4 (E-ALI normal glucose) biological replicates; ***P = 0.0001; ****P < 0.0001, by one-way ANOVA. (D) Cells cultured in E-ALI showed significant increases in relative transepithelial resistance (TER) in response to insulin regardless of glucose content, cells cultured in 2% UG did not. n = 4—11 samples from n = 3 (E-ALI normal glucose) or 2 (all others) biological replicates; *P = 0.015, ****P < 0.0001 by one-way ANOVA. All data is plotted as mean ± SD.
Medium glucose content showed a fast decline within the first 24 h where each culture grown used approximately 50% of the available glucose (Fig. 4B). Glucose clearance was significantly more rapid for cells in high glucose E-ALI as compared with cells in high glucose 2% Ultroser G medium. After 72 h, nearly 125 mg/dL glucose remained in cultures fed with high glucose media, consistent with saturation of uptake. By contrast, cells in normal resting glucose cleared nearly all glucose from the medium after 72 h.
We then determined the impact of medium formulation and glucose content on insulin stimulated glucose uptake, as measured using [3H]-2-deoxy-glucose. Since E-ALI used to culture airway cells contains insulin (5 µg/ml; 0.87 µM), cells were first pre-incubated for 90 min with insulin-free KRH prior to challenge with [3H]-2-deoxy-glucose in the presence or absence of 500 nM (2.9 µg/ml) recombinant human insulin. Of all the conditions tested, only E-ALI medium containing normal resting glucose showed a significant, two-fold increase in glucose uptake in response to added insulin (Fig. 4C). By contrast, cells cultured E-ALI containing high glucose showed elevated glucose uptake that was insulin insensitive and significantly higher than the levels of glucose uptake by cells in E-ALI at normal resting glucose in the absence of insulin. This was not due to an effect of high glucose on insulin signaling, since cells cultured in E-ALI showed an increase in transepithelial resistance (TER) in response to insulin, regardless of glucose concentration and consistent with our previous results20.
Airway cells express multiple glucose transporters, including the insulin regulated Glut4 transporter20. Our data suggest that cells chronically cultured in the presence of high glucose are likely to upregulate constitutive glucose transporters, which would overshadow the impact of insulin stimulated activation of Glut4 mediated by trafficking from secretory vesicles to the plasma membrane20,44. Regardless of mechanism, it is important to note that the glucose concentrations used here represent two extremes as opposed to the physiological glucose levels cells will be exposed to in vivo which significantly vary in response to meals and systemic insulin levels51. Our data suggest that a culture system based on exposing cells to E-ALI containing varying levels of glucose could provide the basis for an in vitro model that mimics in vivo exposure.
Moreover, cells cultured in 2% Ultroser G did not show insulin stimulated glucose uptake or changes in TER (Fig. 4C,D). Considering that constitutive insulin uptake was comparable for cells cultured in E-ALI and 2% Ultroser G, the differences in insulin stimulated glucose uptake and barrier function were not likely to be due to a difference in insulin binding capacity. Instead, the results in Fig. 4C,D and more likely reflect a difference in the capacity for glucose uptake (Fig. 4B) and/or signaling downstream from insulin receptors. For instance, we have observed that the ability of insulin to promote barrier function requires akt signaling, a pathway that is active in primary human airway cells 20. Taken together, these results underscore the importance of medium formulation, especially in studies of airway cell metabolism, and are consistent with the deleterious effects of hyperglycemia on the airway epithelium14,44,52 as well as cell homeostasis in general53.
Expansion and maturation of CF nasal epithelial cells using E-ALI
Nasal cells have proven to be a useful model system that reflects several characteristics of the conducting airway16,54. Unlike primary tracheal and bronchial cells, primary human nasal epithelial (hNE) cells often originate from small samples that require expansion on an appreciable scale for subsequent analysis. We thus evaluated the ability of E-ALI to support differentiation of hNE cells including CFhNE cells harboring both rare and common disease-causing CFTR alleles, as well as cells from non-CF subjects (NhNE cells). Nasal cell isolates (Fig. 5A) initially were expanded using CRC conditions (Fig. 5B) and then differentiated using the protocol illustrated in Fig. 2A. Regardless of genotype, hNE cells showed comparable doubling rates of ~ 0.7–0.9 per day during CRC expansion (Fig. 5C). Ciliated cells were readily detected 21 days after initiating culture in E-ALI by scanning EM (Fig. 5D,E) and by confocal immunofluorescence microscopy (Fig. 5F-H). Taken together these data show that expansion of nasal epithelial cells using the CRC method effectively supported their ability to differentiate in E-ALI.
Expansion and differentiation of non-CF and CF nasal epithelial cells. (A) Ciliated cells (black arrows) are observed in freshly isolated CF nasal cultures by phase contrast microscopy. Bar, 20 μm. (B) Timeline for processing of nasal curettage samples showing frozen cell banking (F), plating for differentiation on Transwell permeable supports (T) or plating for expansion in FYRM submerged culture. Due to the small initial sample size, cells were not banked or plated for differentiation until P4. Cells beyond P7 are not typically used to generate differentiated cultures for experimental analysis. Detail related to culture on Transwells (T) is shown in Fig. 2. (C) Non-CF and CF nasal epithelial cells with 3 different genotypes had a comparable doubling time when cultured in CRC conditions. n = 3 – 6 wells for CF cells; doubling data for NhNE cells is from Fig. 2. (D, E) Cilia and mucus producing CF nasal epithelial cells as observed by scanning electron microscopy at 500x (D) and 2000x (E) magnification. (F–H) Immunofluorescence confocal microscopy of CF nasal airway cells showed tight junctions (green, ZO-1) and cilia (red, Ac-Tubulin). Nuclei were labeled with DAPI (blue). Bar, 20 μm (F, H).
We further characterized the cell electrophysiology of P0 NhTE cells maintained in E-ALI medium. After 16 weeks at ALI (Fig. 6A) NhTE cells demonstrated measurable ENaC and CFTR currents based on amiloride inhibition and forskolin stimulation, respectively. CFTR currents were also modestly enhanced by Vx-770 (ivacaftor) and curcumin and were inhibited by Inh-172. NhTE cells from the same culture preparation extended to 21 weeks ALI had a comparable electrophysiological profile (Fig. 6B). Primary NhNE cells expanded with the CRC method, then differentiated with E-ALI for either 2 weeks (Fig. 6C) or 7 weeks (Fig. 6D) also showed ENaC and CFTR currents with electrophysiological characteristics comparable to those of NhTE cells. These data demonstrate the utility of E-ALI in supporting long-term cultures that maintained ion channel function.
Representative electrophysiological analysis of primary tracheal and nasal airway epithelia expanded using CRC conditions and differentiated in E-ALI. (A, B) P0 NhTEC cultured in E-ALI for 16 weeks (A) or 21 weeks (B) show comparable response profiles to the ENaC inhibitor amiloride and agents that stimulate or inhibit CFTR currents. (C, D) NhNE cells that were expanded using CRC conditions to P4 and then differentiated with E-ALI for 2 weeks (C) or 7 weeks (D), also exhibited comparable ENaC and CFTR current profiles. FSK, Forskolin; Vx-770, Ivacaftor; CFTRinh172, CFTR channel inhibitor.
We then examined primary CFhNE cells with a G551D/F508del genotype that were isolated, expanded, and differentiated in E-ALI. Differentiated G551D/F508del CFhNE cells produced mature cells capable of eliciting small, but detectable, CFTR currents when treated with forskolin that were modestly enhanced with Vx-770 and Vx-809 (lumacaftor) and inhibited with Inh-172 (Figs. 7A-B) 55,56. Similarly, CFhNE cells with a W1282X/F508del genotype exhibited a small CFTR current in response to forskolin and Vx-770 and were also responsive to curcumin which has shown efficacy in CFTR mutants encoding a premature stop codon (Figs. 7C-D)57. Therefore, E-ALI medium is compatible with CRC expansion and subsequent studying of primary nasal epithelial cells.
Representative electrophysiological analysis of primary nasal CF airway epithelia expanded using CRC conditions and differentiated in E-ALI. (A, B) CFhNE cells with the G551D/F508del genotype were expanded using CRC conditions to P4 and differentiated in E-ALI for 14 days. Shown are two representative traces demonstrating low levels of CFTR currents and modest responses to Vx-770 and Vx-809. (C, D) CFhNE cells with the W1282X/F508del genotype were expanded and differentiated as described above. Shown are two representative traces demonstrating low levels of CFTR currents and responses to Vx-770 and curcumin. FSK, Forskolin; inh172, CFTR channel inhibitor; Vx-770, Ivacaftor; Vx-809, Lumacaftor.
Taken together, these data validate E-ALI as a method to differentiate human airway epithelial cells in medium containing normal resting glucose levels. We also confirmed that expanded nasal epithelial cell isolates have preserved their ability to differentiate and express functional CFTR. E-ALI medium provides a new method amenable to investigation of nasal, tracheal, and bronchial airway epithelia for a variety of applications including ciliation and developmental studies, host pathogen interactions, and drug screening. The ability to differentiate cells in normal resting glucose is expected to facilitate the analysis of airway cell functions that are particularly sensitive to cell metabolism.
Methods
Donor consent
Research involving human research participants was performed in accordance with the Declaration of Helsinki guidelines and samples were de-identified to meet HIPAA requirements. Nasal curettage, tracheal, bronchial, and whole lung tissues were acquired through informed consent via an Emory University IRB approved protocol (protocol #00005792) administered by the Cystic Fibrosis Biospecimen Repository (CFBR). Additional lung tissues were obtained through standardized UNOS consenting procedures for tissue donation for research in conjunction with an IRB-approved waiver from Emory University.
Tracheal epithelial cell isolation
To prepare tracheal epithelial cells, intact human donor tracheal tissue was cut into segments consisting of two to three cartilage rings starting at the carina of the main tracheal bifurcation proceeding distally. The trachealis muscle was removed to simplify the isolation procedure as the muscle tends to disintegrate during enzymatic digestion and increases co-purifying tissue debris. All tracheal segments were placed into a 250 mL sterile plastic bottle and washed in Hanks Balanced Salt Solution (HBSS; Sigma-Aldrich #55021C-1000ML) at least 5 times. Then, the convex outer side of the tracheal segments were cleaned using tweezers and a scalpel to remove excess connective tissue and prevent accumulation of tissue debris.
Epithelial cells were removed from the underlying extracellular matrix by first incubating at 4 °C for least 12–16 h under gentle agitation in 50–150 mL of Conducting Airway Protease Solution (CAPS) consisting of Ham’s F-12 medium (Hyclone #SH30026.FS) supplemented with 1.0% w/v Protease XIV (Sigma-Aldrich #P5147), 0.1% w/v DNAse I (Sigma-Aldrich #DN25), 0.2% Primocin (Invivotech #amt-pm-1), and 0.1% Plasmocin treatment agent (Invivotech #ant-mpt). To loosen and remove intact epithelial cell sheets from the concave inner side of the trachea, the digestion solution containing the tracheal segments was lightly vortexed.
The cell solution was separated from the tissue segments by first decanting into a new conical tube, then scraping the concave side with a scalpel to remove any remaining epithelia. The tracheal segments were washed with HBSS and the solution collected into 50 mL centrifuge tubes. All solutions containing cells were centrifuged at 350 × g for 10 min at RT, resuspended in a total of 20 mL normal glucose DMEM (Sigma-Aldrich # D6046 or Hyclone #SH30021.FS), centrifuged again at 350 × g for 10 min at RT, then resuspended in calcium/magnesium-free PBS supplemented with 1 mM EDTA (PBS/EDTA). Cells in solution were triturated to break apart cell clumps, passed through a 100-micron filter (Corning #352360), followed by a 70-micron filter (Corning #431751). The resulting P0 cells were then either cryopreserved at 1 million cells/mL, cultured for differentiation, or cultured for expansion.
Bronchial epithelial cell isolation
Intact human donor bronchial tissue caudal from the tracheal bifurcation to airways 10 mm in diameter were used to isolate pure bronchial epithelial cells. These tissue segments were generally lined with soft cartilage rings. Bronchial tissue segments were isolated from whole lungs or lobes of intact lungs by carefully removing the surrounding terminal airway tissue working from the tracheal bifurcation towards the caudal end of the bronchi. Isolated bronchial tissue segments were washed at least 5 times in HBSS to remove any accumulated mucus and to loosen any remaining connective tissue. Airways beyond bronchi generally include bronchioles that are not heavily collagenous and are smaller than 3 mm in diameter. These airways take time and effort when isolating to avoid cross contamination with pulmonary arteries and smaller vessels. For bronchi, tissue segments were cleaned of connective tissue, cut longitudinally to expose the epithelium, and then processed to isolate P0 cells as described for tracheal cell isolation. As needed, terminal airway lobe tissue was set aside in normal glucose DMEM for primary fibroblast isolation by established methods58.
3T3 Fibroblast feeder cell preparation
Fibroblast feeder layers required for expansion of primary human basal airway epithelial cells were prepared using the 3T3-J2 fibroblast cell line (ATCC #SCRC-1010)4,36,59,60. 3T3 cells were expanded in DMEM containing high glucose (450 mg/dL) (Sigma-Aldrich #D6429 or Hyclone #SH30243.01) supplemented with 10% FBS (ThermoFisher #26170043), 0.2% Primocin (Invivotech #amt-pm-1) and 0.1% Plasmocin prophylactic agent (Invivotech #ant-mpp) to obtain ten 150 mm culture dishes at 80% confluence. For irradiation, ten 150 mm culture dishes of 3T3 cells were trypsinized, centrifuged at 500 × g, resuspended in 30 mL 3T3 expansion culture medium and x-ray irradiated with a dose of 3000 cGy. The irradiated cells were collected by centrifugation and resuspended in 30 mL 3T3 cell freezing medium consisting of 90% FBS and 10% DMSO (Sigma-Aldrich #D2438) (3 ml freezing medium/150 mm dish of 3T3 cells) and stored in liquid nitrogen. When human feeder cells were needed, MRC-5 cells (ATCC #CCL-171) were expanded, irradiated, stored and used in a comparable manner.
Airway epithelial cell expansion
One day prior to seeding plates with primary epithelial cells, irradiated 3T3 fibroblast feeder layers were plated on plasticware coated with Type IV Collagen (Sigma-Aldrich #C7521) in F + Y Reprogramming Medium (FYRM). FYRM consists of a mixture of DMEM 1.0 g/L glucose w/L-glutamine and w/ sodium pyruvate (Corning 10–014-CV) + Ham’s F-12 medium (Cytiva #SH30026.01; 1.8 g/L glucose) supplemented with 5% FBS, Insulin (5 µg/mL), Epidermal Growth Factor (10 ng/mL), Hydrocortisone (480 ng/mL), Adenine (24 µg/mL), Y-27632 (10 µM), Cholera Toxin (8.33 ng/mL), and antibiotics (Table 1), which was stored in foil-wrapped glass bottles in the dark at 4 °C for up to four weeks. One vial of 3T3 feeder cells was used for one T75 flask, three T25 flasks, or divided evenly in a 6-well tissue culture dish. Epithelial cell plating density is a key parameter in cell expansion; epithelial cells should be seeded at 1.3 × 105 cells/well of a 6 well plate, 3.3 × 105 cells/T25 flask, or 106 cells/T75 flask. FYRM is changed every other day until the cells reach ~ 70% confluence. To remove epithelial cells cultured on 3T3 feeder layers, the 3T3 cells were first detached by washing with PBS/EDTA followed by a 5 min incubation in EDTA/PBS at RT, then light tapping. Rosettes of epithelial cells were detached using Accutase (Sigma-Aldrich #A6964) at room temperature for a maximum of 10 min and then reseeded for further expansion (one well to a T25 flask, one T25 to one T75 flask, or one T75 to three T75 flasks), plated for differentiation, or cryopreserved.
Airway epithelial cell differentiation
E-ALI is based on a 50:50 mixture of DMEM containing 100 mg/dL glucose, w/o L-glutamine and w/sodium pyruvate (Sigma-Aldrich #D5546) and LHC Basal Medium (ThermoFisher #12,677–019) containing the additives summarized in Table 2. This recipe results in medium containing a final glucose concentration of 150 mg/dL. In each case, stock solutions are added to the medium prior to filtering (typically 1 ml of 1000 × stock/L medium). Most biologics were dissolved and aliquoted according to manufacturer’s instructions as 1000 × stock solutions stored frozen at -80 °C. Individual 1000 × stock solutions included 1000 × CaCl2·2H2O (1 M in H2O), 1000 × ZnSO4·7H2O (2 mM in H2O), 1000 × Fe/Mg (1.51 mM FeSO4·7H2O, 300 mM MgCl2·6H2O, 195 mM MgSO4·7H2O, 60 mM HCl in H2O). To make the 1000 × trace element stocks, first 1,000,000 × stocks were made in H2O for each component individually: Na2SeO3 (30 mM); MnCl2·4H2O (1 mM); Na2SiO3·9H2O (500 mM); (NH4)6Mo7O24·4H2O 1 mM; NH4VO3 (5 mM); NiSO4·6H2O (1 mM) and SnCl2·2H2O (0.5 mM). These individual stocks were diluted 1:1000 in H2O and HCl was added to 12 µM to produce the 1000 × trace elements stock. For anti-infectives, 1000 × stocks were made as follows, A: 154 mg Ceftazidime was added to 2 mL Gentamycin sulfate solution. B: 50 mg Cilistatin/Imipenem was added to 2 mL H2O. C: 88 mg Piperacillin and 12 mg Tazobactam were added to 2 mL DMSO. D: 100 mg Azithromycin and 10 mg Voriconazole were added to 2 mL DMSO. E-ALI is stored in foil-wrapped glass bottles in the dark at 4 °C for up to four weeks. Given the light sensitivity of E-ALI, medium changes are done in a biological safety cabinet with the fluorescent light turned off.
To produce differentiated cultures, P0 or expanded epithelial cells were resuspended in 20 mL E-ALI medium and counted. Cells were plated into 0.50 mL of E-ALI onto Type IV collagen coated Transwells at a density of 105 cells/6.5 mm well (Costar #3470) or 3.5 × 105/12 mm well (Costar #3460), with the bottom chamber containing 0.75 ml E-ALI. After 48 h, the basolateral medium was replaced with fresh E-ALI and the apical medium was removed to bring the cells to ALI. Once at ALI, medium was changed every 2–3 days, where the apical surface was washed once with E-ALI that was immediately removed and the basolateral medium was replaced. Benchmarks for differentiation included formation of a high resistance monolayer (> 500 Ohm x cm2) and initiation of cilia growth (day 7). Monolayers were usually fully differentiated 14–21 days after transition to ALI.
For some experiments, 2% Ultroser G medium containing normal resting glucose was made using 50:50 mixture of DMEM containing 100 mg/dL glucose, w/o L-glutamine and w/ sodium pyruvate (Sigma-Aldrich #D5546) and Ham’s F-12 medium (Hyclone #SH30026.FS; 180 mg/dL glucose).
Cell stock cryopreservation and use
Epithelial cells were cryopreserved by diluting them into epithelial cell freezing medium (60% FYRM, 30% FBS, and 10% DMSO) at 1 million cells/mL using a Corning CoolCell according to the manufacturer’s directions. Frozen cell stocks were then transferred to liquid nitrogen for long-term storage. For cell thawing, each frozen vial was separately removed and immediately placed in a 37 °C water bath then left undisturbed until a small sliver of ice remained in the vial. The vials were then removed from the water bath, cleaned with 70% ethanol solution, and transferred to a biosafety cabinet for handling. Each vial was plated into either a single well of a 6-well dish or a T25, either of which was coated with Type IV collagen and containing 3T3 feeder cells, fully supplemented with room-temperature FYRM, avoiding centrifugation.
Nasal epithelial cell isolation and expansion
Twenty-four hours prior to nasal cell collection, irradiated 3T3 fibroblast feeder cells were plated into 6-well cell culture dishes in Collection Medium consisting of DMEM containing 100 mg/dL glucose, w/o L-glutamine and w/ sodium pyruvate (Sigma-Aldrich #D5546) supplemented with 10% FBS (R&D Systems #S11150), 0.2% Primocin, and 0.1% Plasmocin treatment agent (Invivotech #ant-mpt), stored at 4 °C for up to two weeks. Nasal cell curettage was performed by a trained otorhinolaryngologist. A curettage was used to gently scrape each inferior nasal turbinate on both sides of the nose. Two separate scrapings were performed for each nostril to increase the number of isolated epithelial cells. Each curettage was placed into a 15 mL conical tube containing 3 mL Collection Medium. Nasal scrapes were then transported on ice for processing. The cells were dislodged from the curette by brief vortex. Nasal curettage samples from a single donor were then combined, centrifuged at 350 g for 5 min at RT, resuspended in PBS/EDTA, dissociated for 5 min at RT prior to straining through a 100 μM filter mesh and then centrifuged at 350 g for 10 min at RT. The dissociated cells were then resuspended in FYRM and placed onto irradiated 3T3 feeder cells at a density of one combined donor sample per well of a six-well culture plate that was precoated with Type IV collagen. After the initial two days of culture, the medium was changed with fresh FYRM daily until 60% confluence was reached. The cells were isolated as described above and then re-seeded in a T25 tissue culture flask containing irradiated 3T3 feeder cells in FYRM. At each passage, a portion of the cells were frozen in epithelial cell freezing medium (60% FYRM, 30% FBS, 10% DMSO) as illustrated in Fig. 5. Nasal epithelial cell differentiation was done using E-ALI as described above.
Immunofluorescence and imaging
Antibodies used for immunofluorescence included: mouse monoclonal antibody (mAb) anti-acetylated tubulin at 1:100 (Sigma-Aldrich clone 6-11-B1; #T7451); mouse monoclonal antibody anti-Mucin 5AC at 1:500 (Abcam clone 2-11M1; #ab24071); rabbit polyclonal antibody anti-ZO-1 at 1:250 (ThermoFisher; #40-2300); mouse anti ZO-1 at 1:100 (ThermoFisher #33-9100), rabbit monoclonal antibody anti-Cytokeratin 5 at 1:250 (Abcam clone EP1601Y; #ab52635); rat monoclonal anti-Uteroglobin/SCGB1A1 at 1:50 (R&D Systems; MAB4218) and mouse monoclonal anti-FOXI1 clone OTI1D4 at 1:100 (Origene; TA800144). Cells were fixed in 4% paraformaldehyde in Dulbecco’s PBS containing Ca2+ Mg2+ (DPBS) for 10 min at RT, washed three times with DPBS, incubated with 1:1 MeOH:acetone for 2 min at RT, washed three times with DPBS, washed once with DPBS containing 0.5% Triton-X 100, blocked with DPBS supplemented with 2% (wt/vol) BSA and 5% (wt/vol) goat serum for 1 h at RT, and then incubated with primary cell phenotype marker antibodies overnight in DPBS containing 2% BSA and 5% goat serum at 4 °C with mixing. The next day, cells were incubated with primary ZO1 antibody for 1 h at RT. Fluorescent secondary antibodies used were Cy2 Goat anti-mouse AffiniPure IgG (1:500; Jackson Immuno #115–165-166), Cy3 goat anti-rabbit AffiniPure IgG (1:500; Jackson Immuno #111–225-144), AlexaFluor568 goat anti-mouse IgG (1:1500; Invitrogen A-11031), AlexaFluor488 goat anti-rabbit IgG (1:1500; Invitrogen A-11034), or AlexaFluor488 donkey anti-rat IgG (1:500; Jackson Immuno #712–545-150). Secondary antibodies were diluted in DPBS supplemented with 2% BSA and 5% goat serum and incubated with cells for 1 h at RT. Cells were mounted in ProLong Gold Antifade Reagent with DAPI (Invitrogen; #P36931). Images were collected using a Zeiss FV1000 confocal microscope in the Emory University Integrated Cellular Imaging Microscopy Core.
Transepithelial resistance and electrophysiology
To measure confluence and tight junction formation, transepithelial resistance (TER) was measured using an EVOM voltmeter, as previously described61. CFTR currents of cells on Transwells in physiologic Krebs-Ringers HEPES (KRH) buffer (1 g/l D-glucose, 50 mM HEPES, 137 mM NaCl, 4.7 mM KCl, 1.85 mM CaCl2, and 1.3 mM MgSO4 at pH 7.4) were measured as previously described61 with an Ussing chamber system (VCC MC-8 Multichannel Voltage/Current Clamp controller and analyzed using Acquire & Analyze software (Physiological Instruments)). Ion channel inhibitors and activators used for Ussing chamber analysis included amiloride (100 µM), forskolin (5 µM), 3-isobutyl-1-methylxanthine (IBMX; 100 µM), Vx-770 (5 µM), Vx-809 (3 µM), and CFTR inh172 (10 µM) and curcumin (40 µM).
Insulin stimulation and glucose uptake
Medium glucose was measured using a colorimetric glucose quantification kit (Cayman Chemical; #10009582) and medium insulin was measured using by ELISA (Alpco Diagnostics; #80-INSHU-CH01). Uptake of 2-deoxy-D-[3H] glucose was measured as previously described20, with modifications. In brief, cells on Transwell permeable supports were washed with KRH and then incubated for 90 min at 37 °C in KRH. The cells were washed and incubated for 30 min at 37 °C with apically added KRH containing 0.5 μCi 2-deoxy-D-[3H]glucose (NEN Radiochemicals, PerkinElmer #NET328250UC) and 2-deoxy-D-glucose (Sigma-Aldrich #D6134) adjusted to 5.6 mM total final concentration in either the presence or absence of 500 nM (2.9 µg/ml) recombinant human insulin with zinc (Gibco #12585014). The cells were washed with cold KRH, Transwell filters were removed and placed in a scintillation vial containing 200 µL 0.1 M NaOH to lyse the cells. Scintillation fluid was added and the samples were measured for 3H using a Beckman-Coulter LS6500 scintillation counter.
Statistics
All statistics were calculated using GraphPad Prism v6 for Windows with methods indicated in each figure legend. Cell doubling rate was calculated as (log(total cells)-log(number seeded cells)/log(2))/(time in culture).
References
Schlingmann, B., Molina, S. A. & Koval, M. Claudins: Gatekeepers of lung epithelial function. Semin. Cell Dev. Biol. 42, 47–57. https://doi.org/10.1016/j.semcdb.2015.04.009 (2015).
Whitsett, J. A. & Alenghat, T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat. Immunol. 16, 27–35. https://doi.org/10.1038/ni.3045 (2014).
Tam, A., Wadsworth, S., Dorscheid, D., Man, S. F. & Sin, D. D. The airway epithelium: more than just a structural barrier. Ther. Adv. Respir. Dis. 5, 255–273. https://doi.org/10.1177/1753465810396539 (2011).
Suprynowicz, F. A. et al. Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc. Natl. Acad. Sci. U S A 109, 20035–20040. https://doi.org/10.1073/pnas.1213241109 (2012).
Nawroth, J. C., van der Does, A. M., Ryan Firth, A. & Kanso, E. Multiscale mechanics of mucociliary clearance in the lung. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190160. https://doi.org/10.1098/rstb.2019.0160 (2020).
Jaramillo, A. M., Azzegagh, Z., Tuvim, M. J. & Dickey, B. F. Airway Mucin secretion. Ann. Am. Thorac. Soc. 15, S164–S170. https://doi.org/10.1513/AnnalsATS.201806-371AW (2018).
Widdicombe, J. H. & Wine, J. J. Airway gland structure and function. Physiol. Rev. 95, 1241–1319. https://doi.org/10.1152/physrev.00039.2014 (2015).
Sell, E. A., Ortiz-Carpena, J. F., Herbert, D. R. & Cohen, N. A. Tuft cells in the pathogenesis of chronic rhinosinusitis with nasal polyps and asthma. Ann. Allergy Asthma Immunol. 126, 143–151. https://doi.org/10.1016/j.anai.2020.10.011 (2021).
Plasschaert, L. W. et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381. https://doi.org/10.1038/s41586-018-0394-6 (2018).
Montoro, D. T. et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324. https://doi.org/10.1038/s41586-018-0393-7 (2018).
Schuler, B. A. et al. Age-determined expression of priming protease TMPRSS2 and localization of SARS-CoV-2 in lung epithelium. J. Clin. Invest. https://doi.org/10.1172/JCI140766 (2021).
Bergeron, C. & Cantin, A. M. Cystic fibrosis: pathophysiology of lung disease. Semin. Respir. Crit. Care Med. 40, 715–726. https://doi.org/10.1055/s-0039-1694021 (2019).
Wallmeier, J. et al. Motile ciliopathies. Nat. Rev. Dis. Primers 6, 77. https://doi.org/10.1038/s41572-020-0209-6 (2020).
Garnett, J. P. et al. Hyperglycaemia and Pseudomonas aeruginosa acidify cystic fibrosis airway surface liquid by elevating epithelial monocarboxylate transporter 2 dependent lactate-H(+) secretion. Sci. Rep. 6, 37955. https://doi.org/10.1038/srep37955 (2016).
Suzuki, S. et al. Highly efficient gene editing of cystic fibrosis patient-derived airway basal cells results in functional CFTR correction. Mol. Ther. 28, 1684–1695. https://doi.org/10.1016/j.ymthe.2020.04.021 (2020).
Brewington, J. J. et al. Brushed nasal epithelial cells are a surrogate for bronchial epithelial CFTR studies. JCI Insight https://doi.org/10.1172/jci.insight.99385 (2018).
Harris, C. M. et al. Assessment of CFTR localisation in native airway epithelial cells obtained by nasal brushing. J. Cyst. Fibros 3(Suppl 2), 43–48. https://doi.org/10.1016/j.jcf.2004.05.009 (2004).
Bengtson, C. D. et al. Hyperglycaemia in cystic fibrosis adversely affects BK channel function critical for mucus clearance. Eur. Respir. J. https://doi.org/10.1183/13993003.00509-2020 (2021).
Bedi, B. et al. UPR modulation of host immunity by Pseudomonas aeruginosa in cystic fibrosis. Clin. Sci. (Lond) 134, 1911–1934. https://doi.org/10.1042/CS20200066 (2020).
Molina, S. A. et al. Insulin signaling via the PI3K/Akt pathway regulates airway glucose uptake and barrier function in a CFTR-dependent manner. Am. J. Physiol. Lung Cell Mol. Physiol. 312, L688–L702. https://doi.org/10.1152/ajplung.00364.2016 (2017).
Wu, Y. S. et al. ORKAMBI-mediated rescue of mucociliary clearance in cystic fibrosis primary respiratory cultures is enhanced by arginine uptake, arginase inhibition, and promotion of nitric oxide signaling to the cystic fibrosis transmembrane conductance regulator channel. Mol. Pharmacol. 96, 515–525. https://doi.org/10.1124/mol.119.117143 (2019).
Lan, B. et al. Airway epithelial compression promotes airway smooth muscle proliferation and contraction. Am. J. Physiol. Lung Cell Mol. Physiol. 315, L645–L652. https://doi.org/10.1152/ajplung.00261.2018 (2018).
Mitchel, J. A. et al. In primary airway epithelial cells, the unjamming transition is distinct from the epithelial-to-mesenchymal transition. Nat. Commun. 11, 5053. https://doi.org/10.1038/s41467-020-18841-7 (2020).
Kim, M. D. et al. Losartan reduces cigarette smoke-induced airway inflammation and mucus hypersecretion. ERJ Open Res. 7, 149. https://doi.org/10.1183/23120541.00394-2020 (2021).
Hou, Y. J. et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370, 1464–1468. https://doi.org/10.1126/science.abe8499 (2020).
Vanderheiden, A. et al. Type I and Type III interferons restrict SARS-CoV-2 infection of human airway epithelial cultures. J. Virol. https://doi.org/10.1128/JVI.00985-20 (2020).
Schweitzer, K. S. et al. Influenza virus infection increases ACE2 expression and shedding in human small airway epithelial cells. Eur. Respir. J. https://doi.org/10.1183/13993003.03988-2020 (2021).
Liu, X. et al. Conditional reprogramming and long-term expansion of normal and tumor cells from human biospecimens. Nat. Protoc. 12, 439–451. https://doi.org/10.1038/nprot.2016.174 (2017).
Mou, H. et al. Dual SMAD signaling inhibition enables long-term expansion of diverse epithelial basal cells. Cell Stem Cell 19, 217–231. https://doi.org/10.1016/j.stem.2016.05.012 (2016).
Cantor, J. R. et al. Physiologic medium rewires cellular metabolism and reveals uric acid as an endogenous inhibitor of UMP synthase. Cell 169, 258-272 e217. https://doi.org/10.1016/j.cell.2017.03.023 (2017).
Clancy, J. P. et al. CFTR modulator theratyping: current status, gaps and future directions. J. Cyst. Fibros 18, 22–34. https://doi.org/10.1016/j.jcf.2018.05.004 (2019).
Zuo, W. L. et al. Ontogeny and biology of human small airway epithelial club cells. Am. J. Respir. Crit. Care Med. 198, 1375–1388. https://doi.org/10.1164/rccm.201710-2107OC (2018).
Hawkins, F. J. et al. Derivation of airway basal stem cells from human pluripotent stem cells. Cell Stem Cell 28, 79-95 e78. https://doi.org/10.1016/j.stem.2020.09.017 (2021).
Martinovich, K. M. et al. Conditionally reprogrammed primary airway epithelial cells maintain morphology, lineage and disease specific functional characteristics. Sci. Rep. 7, 17971. https://doi.org/10.1038/s41598-017-17952-4 (2017).
Wolf, S. et al. Conditional reprogramming of pediatric airway epithelial cells: a new human model to investigate early-life respiratory disorders. Pediatr. Allergy Immunol. 28, 810–817. https://doi.org/10.1111/pai.12810 (2017).
Reynolds, S. D. et al. Airway progenitor clone formation is enhanced by Y-27632-dependent changes in the transcriptome. Am. J. Respir. Cell Mol. Biol. 55, 323–336. https://doi.org/10.1165/rcmb.2015-0274MA (2016).
Zhang, Z. et al. Conditionally reprogrammed human normal bronchial epithelial cells express comparable levels of cytochromes p450 and are sensitive to BaP induction. Biochem. Biophys. Res. Commun. 503, 2132–2138. https://doi.org/10.1016/j.bbrc.2018.08.001 (2018).
Tadokoro, T., Gao, X., Hong, C. C., Hotten, D. & Hogan, B. L. BMP signaling and cellular dynamics during regeneration of airway epithelium from basal progenitors. Development 143, 764–773. https://doi.org/10.1242/dev.126656 (2016).
Miller, A. J. et al. In vitro and in vivo development of the human airway at single-cell resolution. Dev. Cell 53, 117-128 e116. https://doi.org/10.1016/j.devcel.2020.01.033 (2020).
Awatade, N. T. et al. Significant functional differences in differentiated conditionally reprogrammed (CRC)- and feeder-free dual SMAD inhibited-expanded human nasal epithelial cells. J. Cyst. Fibros https://doi.org/10.1016/j.jcf.2020.12.019 (2021).
Gewolb, I. H. & Torday, J. S. High glucose inhibits maturation of the fetal lung in vitro. Morphometric analysis of lamellar bodies and fibroblast lipid inclusions. Lab. Invest. 73, 59–63 (1995).
Mboge, M. Y. & Bissell, M. J. The not-so-sweet side of sugar: influence of the microenvironment on the processes that unleash cancer. Biochim. Biophys. Acta Mol. Basis Dis. 1866, 165960. https://doi.org/10.1016/j.bbadis.2020.165960 (2020).
Garnett, J. P. et al. Proinflammatory mediators disrupt glucose homeostasis in airway surface liquid. J. Immunol. 189, 373–380. https://doi.org/10.4049/jimmunol.1200718 (2012).
Bearham, J., Garnett, J. P., Schroeder, V., Biggart, M. G. S. & Baines, D. L. Effective glucose metabolism maintains low intracellular glucose in airway epithelial cells after exposure to hyperglycemia. Am. J. Physiol. Cell Physiol. 317, C983–C992. https://doi.org/10.1152/ajpcell.00193.2019 (2019).
Li, K. et al. Airway epithelial regeneration requires autophagy and glucose metabolism. Cell Death Dis. 10, 875. https://doi.org/10.1038/s41419-019-2111-2 (2019).
Saint-Criq, V. et al. Choice of differentiation media significantly impacts cell lineage and response to CFTR modulators in fully differentiated primary cultures of cystic fibrosis human airway epithelial cells. Cells https://doi.org/10.3390/cells9092137 (2020).
Fulcher, M. L. & Randell, S. H. Human nasal and tracheo-bronchial respiratory epithelial cell culture. Methods Mol. Biol. 945, 109–121. https://doi.org/10.1007/978-1-62703-125-7_8 (2013).
Bovard, D. et al. A lung/liver-on-a-chip platform for acute and chronic toxicity studies. Lab. Chip 18, 3814–3829. https://doi.org/10.1039/c8lc01029c (2018).
Zoso, A., Sofoluwe, A., Bacchetta, M. & Chanson, M. Transcriptomic profile of cystic fibrosis airway epithelial cells undergoing repair. Sci. Data 6, 240. https://doi.org/10.1038/s41597-019-0256-6 (2019).
Deprez, M. et al. A single-cell atlas of the human healthy airways. Am. J. Respir. Crit. Care Med. 202, 1636–1645. https://doi.org/10.1164/rccm.201911-2199OC (2020).
Inman, T. B., Proudfoot, J. A., Lim, M. & Demeterco-Berggren, C. Continuous glucose monitoring in a cystic fibrosis patient to predict pulmonary exacerbation?. J. Cyst. Fibros 16, 628–630. https://doi.org/10.1016/j.jcf.2017.05.009 (2017).
Hunt, W. R. et al. Hyperglycemia impedes lung bacterial clearance in a murine model of cystic fibrosis-related diabetes. Am. J. Physiol. Lung Cell Mol. Physiol. 306, L43-49. https://doi.org/10.1152/ajplung.00224.2013 (2014).
Giri, B. et al. Chronic hyperglycemia mediated physiological alteration and metabolic distortion leads to organ dysfunction, infection, cancer progression and other pathophysiological consequences: an update on glucose toxicity. Biomed. Pharmacother. 107, 306–328. https://doi.org/10.1016/j.biopha.2018.07.157 (2018).
Ghosh, B. et al. Strong correlation between air-liquid interface cultures and in vivo transcriptomics of nasal brush biopsy. Am. J. Physiol. Lung Cell Mol. Physiol. 318, L1056–L1062. https://doi.org/10.1152/ajplung.00050.2020 (2020).
Mall, M. A., Mayer-Hamblett, N. & Rowe, S. M. Cystic fibrosis: emergence of highly effective targeted therapeutics and potential clinical implications. Am. J. Respir. Crit. Care Med. 201, 1193–1208. https://doi.org/10.1164/rccm.201910-1943SO (2020).
Joshi, D., Ehrhardt, A., Hong, J. S. & Sorscher, E. J. Cystic fibrosis precision therapeutics: emerging considerations. Pediatr. Pulmonol. 54(Suppl 3), S13–S17. https://doi.org/10.1002/ppul.24547 (2019).
Wang, W., Hong, J. S., Rab, A., Sorscher, E. J. & Kirk, K. L. Robust stimulation of W1282X-CFTR channel activity by a combination of allosteric modulators. PLoS ONE 11, e0152232. https://doi.org/10.1371/journal.pone.0152232 (2016).
Baglole, C. J. et al. Isolation and phenotypic characterization of lung fibroblasts. Methods Mol. Med. 117, 115–127. https://doi.org/10.1385/1-59259-940-0:115 (2005).
Liu, X. et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 180, 599–607. https://doi.org/10.1016/j.ajpath.2011.10.036 (2012).
Terunuma, A., Limgala, R. P., Park, C. J., Choudhary, I. & Vogel, J. C. Efficient procurement of epithelial stem cells from human tissue specimens using a Rho-associated protein kinase inhibitor Y-27632. Tissue Eng. Part A 16, 1363–1368. https://doi.org/10.1089/ten.TEA.2009.0339 (2010).
Molina, S. A. et al. Junctional abnormalities in human airway epithelial cells expressing F508del CFTR. Am. J. Physiol. Lung Cell Mol. Physiol. 309, L475-487. https://doi.org/10.1152/ajplung.00060.2015 (2015).
Acknowledgements
We thank Kirsten Cottrill and Ryan Chance Reed for critical reading of the manuscript. Cells were provided jointly by the Children’s Healthcare of Atlanta, the Emory University CF Discovery Core and the CF@LANTA RDP Experimental Models Core. This research project was supported by the Emory University Integrated Cellular Imaging Microscopy Core, the Winship Research Pathology Core and SEM was done by the Robert P. Apkarian Integrated Electron Microscopy Core (IEMC) of Emory University School of Medicine. This project was also supported by CF@LANTA RDP Center Grant (MCCART15R0), R01-HL116958 and R01-AA025854 to M.K., T32-HL116271 to S.A.M., R01-HL139876 and R01-HL136414 to E.J.S., F31-AA029000 to K.F.E., and by the Emory+Children’s Center for Cystic Fibrosis and Airways Disease Research.
Author information
Authors and Affiliations
Contributions
S.A.M., E.J.S. and M.K. designed the scope of the study. S.L.G. and W.R.H. performed nasal curettage. W.R.H. obtained tissues from the Emory Transplant Center and CF@LANTA Clinical and Translational CF Biorepository. R.M., K.F.E., L.D.W. and S.A.M. cultured cells, tested medium formulations, performed immunofluorescence microscopy and barrier function measurements. S.A.M. and L.D.W. measured glucose uptake. C.M. and S.A.M. performed electrophysiological measurements. S.A.M., M.K. and K.F.E. analyzed the data, compiled the figures, and wrote the first draft of manuscript. All of the co-authors edited and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Morgan, R., Manfredi, C., Easley, K.F. et al. A medium composition containing normal resting glucose that supports differentiation of primary human airway cells. Sci Rep 12, 1540 (2022). https://doi.org/10.1038/s41598-022-05446-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-05446-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.