Abstract
5-Aminolevulinic acid (5-ALA)-mediated fluorescence does not effectively depict low grade gliomas (LGG) or the infiltrative tumor portion of high-grade gliomas (HGG). While spectroscopy improves sensitivity and precision, this is currently limited by autofluorescence and a second protoporphyrin IX (PpIX) fluorescence state at 620 nm. We investigated the autofluorescence to better characterize the present spectra and thus increase PpIX quantification precision and sensitivity. This study included 128 patients undergoing surgery for malignant glioma. 5-ALA (Gliolan) was administered before anesthesia, and fluorescence was measured using a hyperspectral device. It was found that all 2692 measured spectra consisted of contributions from 620 to 634 nm PpIX, NADH, lipofuscin, and flavins. The basis spectra were characterized and their use in spectral unmixing led to 82.4% lower fitting error for weakly fluorescing areas (p < 0.001), and 92.3% fewer false positive tumor identifications in control measurements (p = 0.0065) compared to previous works. They also decreased the PpIX620 contribution, thus halving the mean Ratio620/634 (p < 0.001). The ratio was approximately 0 for HGGs and increasing for LGGs, as demonstrated previously. Additionally, the Ratio620/634, the MIB-1/Ki-67 proliferation index, and the PpIX peak blue-shift were found to be significantly related to WHO grade, fluorescence visibility, and PpIX contribution (p < 0.001), and the value of these three as quantitative biomarkers is discussed.
Similar content being viewed by others
Introduction
Fluorescence guided-surgery mediated by 5-Aminolevulinic Acid (5-ALA)-induced protoporphyrin IX (PpIX) fluorescence is a useful surgical adjunct in the treatment of malignant glioma. Fluorescence imaging aids in depicting malignant glioma tissue in real-time during surgery. 5-ALA is an naturally occurring metabolite and a non-fluorescent prodrug which takes part in the biosynthesis pathway of hemoglobin in humans. Malignant glioma selectively take up 5-ALA, converting this prodrug into PpIX for reasons to date not fully understood1. 5-ALA is administered in a dose of 20 mg/kg b.w. orally 4 h before induction of anesthesia. PpIX fluorescence can be visualized with commercially available microscopes equipped with special filter systems2. However, the sensitivity of this method is still not high enough to confidently discriminate between the solid and the infiltrative (low cellularity) component in malignant gliomas. Prognosis of patients harboring malignant glioma can be correlated with the extent of resection of tumor tissue3,4. Still, infiltrative tumor cells remain very difficult to detect during surgery, a reason why recurrences often occur within less than 2 cm of the original tumor margin5. Thus, surgical adjuncts such as fluorescence-guidance6 are used in malignant glioma surgery to maximize the extent of resection. In addition to its utility as a photosensitizer agent in photodynamic therapy (PDT)7, 5-ALA, (Gliolan, medac, Germany), a natural heme precursor, is the most established fluorophore in high-grade glioma (HGG) surgery. 5-ALA elicits the expression of fluorescent protoporphyrin IX in HGG cells, which naturally exists in humans and can be intraoperatively visualized with the assistance of commercially available filter systems8,9,10. PpIX exhibits maximum absorption of 405 nm blue light, the Soret Band, which elevates the molecule into an excited state from which it can either react with oxygen to create singlet oxygen that is cytotoxic and thus useful in PDT11, or it can fluoresce. Fluorescence occurs in a characteristic double emission peak at around 635 nm and 705 nm. The difference in wavelength between absorption and emission is called the Stokes shift and allows the excitation light and PpIX fluorescence to be separated using optical filters for visualization of tumors12.
However, only about 20% of LGGs display visible PpIX fluorescence during surgery13, and low density, infiltrative regions of HGGs also often do not fluoresce visibly14. Thus, fluorescence spectroscopy systems are being developed by a number of groups14,15,16 [Kaneko et al. accepted JNS]. In these devices, the emission spectrum is measured at selected points. After normalization for system parameters as well as inhomogeneous scattering and absorption across the tissue17, the relative PpIX fluorescence intensity is extracted using a spectral unmixing routine with known basis spectra18. In spectral unmixing, the optimal linear combination of the basis spectra minimizes the error between the measured and fitted spectrum, thus deriving a relative intensity, or relative concentration, for each present fluorophore including PpIX. PpIX fluorescence intensity is proportional to tumor cell density19,20 so it can be used to locate the tumor and help identify its density and thus malignancy. Such measurement systems greatly increase sensitivity21 as they often allow PpIX overexpression to be measured even when a tumor is not visibly fluorescing.
In LGGs, however, sensitivity is still limited to only 45%22. The same is likely true for low density regions of HGG. Noise in low-fluorescence measurements, and a high rate of false positive identification of PpIX signals leads to relatively poor delineation of tumor margins. These problems in detection are due in part to the oversimplified nature of the spectral unmixing algorithm. The PpIX fluorescence spectrum is dependent on the chemical microenvironment and the local pH23,24,25 which is due to the formation of different PPIX aggregation states. At acidic to neutral pH it was detected in assemblies of twice the size of those seen at alkaline pH25. Moreover, at high pH values aggregates are deprotonated and fluorescent. Protonated forms are nonfluorescent, but aggregates can encompass fluorescent PPIX molecules. As a result, in brain tissue, two fluorescing states of PpIX are observed: one that peaks at 634 nm, PpIX634, and another that peaks at 620 nm, PpIX620. Their relative intensities vary in different tumor types, densities, and malignancy levels16,26,27,28. The second peak has also been described in other biological context29 and in vitro30. It is presently assumed based on the available data that in low cellularity, alkaline and/or infiltrative zone tumor tissue predominantly PpIX620 dominates, whereas in high cellularity, acidic, HGG tissue PpIX634 is more intense16,26. This is the reason why presumably in low cellularity or low-graded portions of gliomas the PpIX620 state dominates.
To date, most spectroscopic measurements do not reflect the spectral contributions of PpIX620, since they assume that only PpIX634 is relevant in tumor tissues. Fortunately, it is relatively easy to rectify this problem by including separate basis spectra for PpIX620 and PpIX634. However, it becomes difficult to discuss PpIX concentration per se as it is unclear what combination of the two main molecular PpIX forms constitutes the overall PpIX “concentration”. Montcel et al. proposed a ratio between the two states’ intensities, Ratio620/634, which is related to the World Health Organization (WHO) classification31 and density of the measured region and can potentially be used as a biomarker in addition to PpIX intensity16,26.
In addition, the basis spectra used in the unmixing have a profound effect on the resulting PpIX quantification. There are a number of endogenous fluorophores in the brain which fluoresce in the same spectral region as PpIX, making it more difficult to isolate the PpIX signal. The emission spectra of these autofluorescence sources must be considered in spectral unmixing for best results. In their analysis, Montcel et al.16 used NADH, which is modeled as a decaying exponential that is fit to each individual measurement. In later work26, they added an approximate lipofuscin spectrum, modeled as a simple Gaussian curve15. However, by fitting an exponential curve for NADH directly to each measurement the actual shape of the NADH spectrum, not just its magnitude, is inconsistent. In reality, the emission spectrum should be fixed and should only vary in magnitude. Similarly, the Gaussian lipofuscin curve is highly simplified. Mehidine et al. also used simplified spectra, including NADH, tryptophan, tyrosine, and collagen32. Finally, there are other fluorophores present in the brain, including elastin, pyridoxine, flavins, and both free and protein-bound NADH33, which, as far as we know have not been considered in spectral unmixing so far.
While including too many basis spectra can lead to overfitting, especially if they are not needed, the spectral unmixing routine theoretically simply assigns a magnitude of 0 to any spectrum not present in a measurement, ignoring noise and measurement artefacts. On the other hand, if a spectral component is present but not provided as a basis spectrum for the unmixing, the unmixing algorithm is forced to compensate by artificially inflating the contribution of other components. This is observable through large fitting errors in the unmixing and can for example artificially increase one or both PpIX components, thus affecting the Ratio620/634 and leading to false positive PpIX signal interpretation.
In this study, we characterized the spectra of the fluorophores present in the measured fluorescence emission spectra more precisely in order to improve the sensitivity and precision of PpIX quantification.
Methods
Patients undergoing surgery for malignant glioma at the University Hospital of Münster were included in this study. Indication for surgery was discussed in an interdisciplinary tumor board. Biopsies (n = 275) of tumor bulk from 128 patients were obtained intraoperatively and were measured immediately using an ex vivo hyperspectral device, after minimal light exposure to reduce photobleaching. Of these, 12 were control samples: GBM where no 5-ALA was administered, e.g. in an emergency situation, or tissue from epilepsy procedures, where temporal lobe resections were indicated. Approximately 10 fluorescence spectra were extracted from different regions of each tissue, depending on the size of the sample. In total, this study used 2572 measured fluorescence spectra and 120 control measurements.
Informed consent was obtained from each individual in this patient collective. All procedures performed in studies were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Data collection and scientific use of biopsies had previously been approved by the ethics committee of the University of Münster.
Autofluorescence spectra were derived from the measurements by analyzing spectral features consistently present in the data that were independent from other features such as glioma-derived PpIX fluorescence, and comparing them to known fluorophore data available in the literature. The contributions of the PpIX620 state, [PpIX620], PpIX634 state, [PpIX634], and their ratio, Ratio620/634 were analyzed and correlated with pathological parameters including WHO grade, MIB-1/Ki-67 proliferation index, and fluorescence visibility. The latter was assessed by the surgeons as ‘none’, ‘weak’, or ‘strong’ for every biopsy. The breakdown of fluorescence visibility vs. WHO grade is shown in Fig. 1.
Data preprocessing
A hyperspectral measurement device built on a Zeiss OPMI Pico microscope was used to extract spectral information from the fluorescing samples, ex-vivo as described previously18. Spectra were extracted from selected pixels of interest and were corrected for a number of parameters specific to the measurement system such as the camera’s wavelength-dependent sensitivity. They were then normalized according to the two-peak routine proposed by Valdes et al.17 using white-light data. This normalization corrects for inhomogeneous scattering and absorption coefficients across the tissue surface.
The measurements were then decomposed into their spectral constituents, using the known emission spectra of the major sources of fluorescence in the tissue in order to evaluate how much of each component was present. These sources included PpIX as well as autofluorescence, and their spectra form a basis which spans the space of possible fluorescence measurements. To perform the ‘spectral unmixing’, we projected the measured curve onto the basis spectra to find the contribution of each using a least squares optimization routine.
Based on experiments with reference samples (so-called phantoms) of known PpIX concentration (µg/ml), the contributions of the PpIX spectra can be mapped to absolute PpIX concentration in µg/ml. We created phantoms as described by Valdes et al.17 to characterize the measurement system. However, as a result of their composition, the phantoms generally contained only a single PpIX fluorescence state. They lacked most of the autofluorescence sources present in the brain, and a linear correlation of PpIX intensity and concentration was not possible24,34. Moreover, there is large variation between measurement devices. Hence, the calculated concentrations in this study are not intended for comparison with studies from other groups, or known values, but are self-consistent and provide a useful way to compare the relative intensities of the various spectra present in the measured light. They will be referred to as ‘contributions’ not ‘concentrations’ to avoid misinterpretation. Spectral contributions are written as [PpIX6xx] and are given in arbitrary units (a.u.), where the units stem from the normalization procedure described above.
It is important to note that all measurements presented in this paper were carried out on the same device, so the scaling and the measurement system characteristics, artefacts, and imperfections were consistent throughout the study. In addition, any scaling difference between methods and devices should cancel out when finding the [PpIX620]/[PpIX634] ratio, so that the Ratio620/634 results can indeed be compared between groups.
Fluorescence model
In order to discuss the PpIX contributions and the Ratio620/634 further, it is essential to have precisely characterized all basis spectra. The PpIX620 and PpIX634 spectra are well known and available in the literature15,16,17,23,24,26,35. Their shapes were obtained by phantom measurement. However, there is less clarity about the molecular contributions to autofluorescence. Possible candidates include elastin, collagen, pyridoxine, tryptophan, flavins, lipo-pigments, and both free and protein-bound NADH33. The first four on this list do not fluoresce in the same spectral range as PpIX and are of little interest when studying PpIX. However, NADH is apparent in the data and its exponentially decaying tail overlaps with PpIX16,26.
The NADH spectrum was characterized as follows:
-
1.
Spectra (n = 248) with a large NADH component, where the NADH peak magnitude was greater than the PPIX peak, were selected.
-
2.
The spectra were normalized at the NADH peak so that they all had an NADH peak of the same magnitude.
-
3.
The 248 spectra were averaged to find a mean spectrum.
-
4.
An exponential decay curve15 was fit to the section from 450 to 525 nm and a linear curve from 430 to 450 nm as illustrated in Fig. 2 using the ‘fit’ function in MATLAB (Mathworks Inc.).
-
5.
The two curves were concatenated and smoothed using Savitzky-Golay filtering to create the smooth approximate NADH spectrum without losing details (Fig. 2).
(Top) Mean spectrum containing the NADH spectral contribution with the expected NADH curves fitted (left) and the resultant NADH basis spectrum (right). (Middle) Lipofuscin and flavin spectra fitted to the mean measured spectrum (left) and the resulting basis spectra and their sum (right) which closely matches the mean measured spectrum. (Bottom) The resulting 5 basis spectra from this analysis.
The next spectrum, lipofuscin, is of primary interest because it overlaps completely with the PpIX fluorescence and thus biases the calculations for the PpIX contribution. Previous studies of PpIX and the Ratio620/634 for neurosurgery16,26 have mostly neglected this spectrum, but we found it plays a substantial role in brain fluorescence measurements. To characterize the spectrum, the same procedure was followed as for NADH, but using only the 12 control tissues without PpIX. These tissues constituted normal brain from epilepsy patients (n = 6, histological reactive brain changes without tumor cells) and glioblastoma patients where no 5-ALA was given (n = 6, histological WHO grade IV glioblastoma). Additionally, the NADH spectrum was fitted to the measured control spectra in the NADH peak region in order to remove the NADH portion from the spectra and to isolate the remaining fluorophores. When the resulting curves were averaged, it became apparent that there were two fluorophores present (Fig. 2). The lipofuscin spectrum was augmented by an extra peak at ~ 512 nm, which likely originated from flavins36,37,38. Quadratic curves were fitted to both the lipofuscin and the flavin peaks to match the expected shapes of the spectra (Fig. 2, middle left). These were combined with the separated averaged spectrum and smoothed to create a lipofuscin and a flavin spectrum, respectively (Fig. 2, middle right).
Thus, we arrived at five basis spectra: PpIX620, PpIX634 and the three autofluorescence spectra for lipofuscin, NADH, and flavin. Different combinations of these five spectra (Fig. 2 bottom) constituted every measured spectrum. This was validated as described in the Results section. We could therefore spectrally unmix all the measured, normalized spectra to obtain the desired PpIX contributions [PpIX620] and [PpIX634].
Statistical analysis
Calculations were performed using the commercially available software MATLAB (The Math Works Inc., Natick, MA). p-values were obtained with the non-parametric Wilcoxon Rank Sum Test as the distributions were either discrete or highly skewed. A p-value < 0.05 was considered statistically significant. All reported p-values are two-tailed.
Results
Fluorescence model
A total of 2572 fluorescent and 120 control measurements were obtained intraoperatively. First, all 2692 spectra were unmixed and checked visually, both using all determined basis spectra (Fig. 2 bottom), and using only the PpIX and NADH spectra, as performed previously16, for subsequent comparison. Qualitatively, using the five basis spectra, the fit was significantly improved and more realistic with not a single exception among all samples. Three representative examples are shown in Fig. 3. For quantitative estimation of the improvement, the error between the fit and the measured spectum was calculated using Eq. (1) for every spectrum, where \(\Phi_{meas}\) is the measurement and \(\Phi_{fit}\) the fit generated by the unmixing.
Unmixing of three spectra (A, B, C), each using the five basis spectra (right) and just NADH (left) (orange: PpIX620, blue: PpIX634, purple: lipofuscin, yellow: NADH, green: flavin). The measured spectrum is shown in blue dots, and the fitted curve is bright green. As was the case with all spectra, the fit was better using the five basis spectra than with just NADH. Spectrum C is the control without PpIX showing how the five basis spectra eliminate false positives.
Of course, in strongly fluorescing HGG spectra, which consisted predominantly of PpIX634, the use of the additional basis spectra made little difference. However, in weaker spectra, with [PpIX] < 1, the use of the five basis spectra decreased the error approximately six-fold. Even when considering all spectra, the error was 50% greater when using only NADH. One could argue that only the fit in the PpIX fluorescence region (600 nm to 730 nm) matters since the PpIX contribution is used to recognize tumors. Considering only this region, the error was still approximately halved using the five basis spectra. These results are summarized in Table 1, and show clear improvement in the effectiveness of the spectral unmixing. Some examples are shown in Fig. 3, and the results are further investigated in the Discussion section.
For the neurosurgery application, another important parameter is the rate of false positive PpIX signals, where the system identifies nonzero PpIX contributions in measurements where no PpIX is present. Using the five basis spectra during spectral unmixing, the rate of false positives in the control measurements fell from 97.5% (using only NADH and PpIX) to 7.5% (using five spectra) with a cutoff of 0.001 a.u. below which contributions were set to zero as the signal to noise ratio approached 1. This constitutes a 90% increase in sensitivity (true positive rate) (p = 0.0065). The mean of the calculated [PpIX620] and [PpIX634] of the control spectra was 0.0078 a.u. using just NADH, and 0.0007 a.u. using the five basis spectra (p = 0.015). This is an order of magnitude improvement in specificity (true negative rate).
PpIX results
Having found more realistic basis spectra, it was then possible to explore the PpIX-related results of the measurements including the contributions of each PpIX state, their ratio, and the relation of these values to fluorescence visibility, WHO grade, and proliferation index. In particular, the Ratio620/634 results was compared to the results of Alston, Montcel, et al.16,26. The primary results are summarized in Table 2.
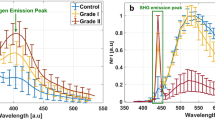
The relations between Ratio620/634 and both WHO grade and fluorescence visibility were significant with > 99.9% confidence. As tumor malignancy increased and thus, for the most part, fluorescence visibility increased (Fig. 1), the 634 nm PpIX state became more dominant compared to PpIX620. Both [PpIX620] and [PpIX634] had a positive correlation to WHO grade and fluorescence visibility, but [PpIX634] grew more quickly, leading to the decreasing ratio.
A comparison of these results with those obtained when using only NADH as the autofluorescence basis spectrum is presented in Fig. 4. The lack of the lipofuscin spectrum inflated the [PpIX620] values to 2–5 times their expected value while having little effect on [PpIX634]. As a result, the Ratio620/634 values were approximately twice as high as they should be. The data are compared previous work in the Discussion section, where the anomalously low ratio at WHO grade I is also discussed.
Proliferation index
The proliferation index, Ki-67 (also known as MIB-1), correlated significantly with WHO grade and fluorescence visibility (p < 0.001). As expected, it has a significant (p < 0.001) correlation with Ratio620/634. However, this relationship did not appear to be linear. A line was fitted to the log of the data, with Pearson product-moment correlation r = − 0.514, indicating a medium-strong linear correlation in log-space. The slope of the line of best fit was − 0.35 indicating the approximate functional relationship shown in Eq. (2).
PpIX blue shift
As the Ratio620/634 increased, the location of the main PpIX peak of the measured spectrum shifted from 634 nm towards 620 nm as illustrated in Fig. 5. There was a strong correlation between Ratio620/634 and the PpIX peak wavelength with a Pearson correlation coefficient of -0.76. The PpIX peak position was also significantly related to [PpIX620], [PpIX634], Ki-67, fluorescence visibility, and WHO grade, all with p < 0.001. The latter two are plotted in Fig. 5 (bottom). The value of this potential biomarker is discussed in the Discussion section.
Discussion
In this manuscript we improve the common analysis routine of fluorescence spectra by further characterization of autofluorescence spectra, taking into account not only the contribution of NADH, but also that of lipofuscin and flavins. The newly generated basis spectra matched the data better and led to more accurate spectral unmixing and far lower false positive rates in a diverse population of 128 patients. Using these basis spectra, 2692 measured fluorescence spectra from the 128 patients were spectrally unmixed and the [PpIX620], [PpIX634], and resulting Ratio620/634 were calculated. The results are presented above, and differ from values calculated previously in literature16,26.
Though this is an improved methodology, some caveats merit mention. Firstly, detecting very low PpIX concentrations is limited by the signal to noise ratio of the measurement system, as seen in Fig. 3A. Secondly, this method assumes that the fluorophores have a constant emission spectrum irrespective of possible interactions between them and with their microenvironment. For example, NADH exists in both free and protein-bound form33, and of course PPIX has the two distinct states which we model as two different fluorophores. There could be other such interactions that are not modeled here. We assume, however, that their effect is small. Further, it has been shown that PPIX fluorescence is present in healthy tissue as well32,39. Thus, the presented method of deriving the lipofuscin basis spectrum embeds this slight PPIX fluorescence in the lipofuscin spectrum. However, as we are interested in identifying tumor tissue, not precisely measuring PPIX concentration, this is not a problem. In fact, by leveraging the large amount of data used in this work, it is likely that the difference between healthy and tumor tissue is more effectively captured in this way than it would be by more precisely determining PpIX concentration and trying to impose a healthy tissue cut-off. While this lipofuscin spectrum would tend to underestimate PpIX concentrations, it does so equally for the two different PpIX states, and the magnitude of this underestimation is negligible. This can be seen in Fig. 3, looking at the small details of the lipofuscin basis spectrum which are possibly artefacts of the embedded PPIX signal, and comparing their magnitude to those of the actual PPIX spectra. Thus, the absolute PpIX concentrations are negligibly affected by this fact, and the presented ratios are unaffected.
Spectral unmixing fit quality
The improved fit quality of the spectral unmixing seen in Table 1 is not the result of overfitting, because the chosen fluorophores are known to be present in brain tissue and their fluoresence was clearly present in the data. Although the data were obtained from a large and diverse patient cohort, the use of the five basis spectra consistently provided a very precise fit in every single measurement. In addition, as shown in the Results section, the rate of false positive PpIX signals was greatly reduced, which allowed more precise detection of tumor margins.
Finally, even if a spectrum contained only NADH and PpIX and the use of the two additional basis spectra for lipofuscin and flavin was therefore superfluous, the result of using them was never worse than using only NADH and PpIX. The spectral unmixing routine finds the mathematically optimal fit given the basis spectra, so it simply sets the contributions of the unneeded basis spectra to zero, so they have no effect. This is demonstrated in spectrum A of Fig. 3, where no detectable flavin autofluorescence was present.
PpIX blue shift
The more PpIX620 is present relative to PpIX634 in a measurement, the more the peak shifts towards shorter wavelengths. The benefit of using this blue shift as a biomarker instead of the Ratio620/634 is that it is independent of the choice of autofluorescence basis spectra. Interestingly, the three plots in Fig. 5 also appear more linear than the Ratio620/634 curves in Fig. 4, and the WHO grade I tumors are not outliers as in Fig. 4B. Thus, the PpIX peak position could be a more robust and objective measure than Ratio620/634. However, it does not have the same dynamic range as the Ratio620/634 and therefore cannot resolve small differences in malignancy or tumor cell density. In addition, small variations in the PpIX peak due to noise can lead to changes in the detected peak position. Thus, noise-robust methods must be used to determine the peak. In this work, the signal was smoothed first using a moving mean filter before the peak was determined. This process is imperfect, though. An alternative would be to fit a PpIX like signal to the noisy one and determine its peak. However, this relies on knowing the PpIX basis spectrum precisely, which negates the primary benefit of this non-model-based method. The spectral unmixing method, on the other hand, is relatively immune to noise, except at very low fluorescence levels. Ultimately, further studies are needed before this can be used in practice.
Comparison to the literature
Alston et al.26 measured the Ratio620/634 from solid and infiltrating parts of HGG and LGG tumors. While we did not differentiate between solid and infiltrative parts of tumors and only imaged the solid part, we still evaluated HGGs and LGGs, which have similar ratios to the low density infiltrative regions of HGGs26. Alston et al. found, as did we, that the ratio was close to 0 for HGGs and increased in LGGs. The results are compared in Table 3.
While the HGG results were similar, differences can be explained by the slightly different NADH and PpIX spectra used. In addition, Montcel et al.’s HGGs consisted only of WHO grade IV while the present study included 490 grade III and 1604 grade IV measurements. The average value dropped to 0.048 ± 0.002 when considering only grade IV tumors.
Among the low-grade tumors a greater difference was observed. This is likely due to the large differences in [PpIX620], which was inflated when not using the five basis spectra, as seen in Fig. 4 (bottom left). Without the lipofuscin and flavin contributions, the mean Ratio620/634 more than doubled from 0.123 to 0.285. This accounts for some of the difference between the Montcel and Alston values and those found in the present study. Additionally, the differing measurement system and normalization would play a role. Finally, Alston et al. had only grade II tumors in their LGG group while we had 40 grade I measurements. Perhaps due to the extremely weak PpIX signal strength of these measurements and the small signal to noise ratio, the mean Ratio620/634 was anomalously low as seen in the top middle panel of Fig. 4. Because there were so many more grade II tumors than grade I, this had little effect on the mean LGG ratio; it nonetheless drags the ratio down slightly. Without the grade I tumors, the mean LGG Ratio620/634 was 0.43.
With a decrease in Ratio620/634 from grade II to I (Fig. 4B), the PPIX blue shift should decrease as well. However, the PPIX peak wavelength for grade I tumors in Fig. 5 may be artificially low because the small signal to noise ratio in the grade I tumors affects how accurately the PpIX peak location can be determined. Furthermore, in these extremely weak spectra, the lipofuscin begins to dominate the PPIX, which drives the apparent blue shift further towards 620 nm.
Finally, we did not use a photoproduct spectrum in this work because the tissues were removed and immediately imaged with minimal light exposure and thus minimal photobleaching. No photoproduct was detected in the measurements.
Conclusion
This work aimed to add to the discussion in fluorescence guided neurosurgery, especially with regard to concepts introduced in previous articles16,26, and thus to increase the sensitivity and precision of PpIX quantification in fluorescence spectroscopy. It was found that improved basis spectra decreased the artificial inflation of [PpIX620], thus approximately halving the average Ratio620/634 values. The ratio was nonetheless close to 0 for HGGs and increasing for LGGs. Additionally, the proliferation index, Ki-67, significantly related to WHO grade, fluorescence visibility, PpIX contribution, and Ratio620/634. Finally, the blue shift of the peak wavelength of PpIX, showed potential as biomarker. Both this shift and the Ratio620/634 will have to be studied in more depth before they can serve as biomarkers. However, the use of the derived autofluorescence spectra in combination with the two PpIX fluorescence states in spectral unmixing routines offer promising results towards increased sensitivity and precision in PpIX quantification and delineation of tumor margins. Future work can also investigate if the other present fluorophores such as NADH, lipofuscin, and flavins have any predictive value in recognizing tumor tissue.
References
Colditz, M. J., Leyen, K. & Jeffree, R. L. Aminolevulinic acid (ALA)-protoporphyrin IX fluorescence guided tumour resection. Part 2: Theoretical, biochemical and practical aspects. J. Clin. Neurosci. 19(12), 1611–1616 (2012).
Stummer, W. & Suero Molina, E. Fluorescence imaging/agents in tumor resection. Neurosurg. Clin. N. Am. 28, 569–583 (2017).
Sanai, N., Polley, M. Y., McDermott, M. W., Parsa, A. T. & Berger, M. S. An extent of resection threshold for newly diagnosed glioblastomas. J. Neurosurg. 115, 3–8 (2011).
Hervey-Jumper, S. L. & Berger, M. S. Evidence for improving outcome through extent of resection. Neurosurg. Clin. N. Am. 30, 85–93 (2019).
Petrecca, K., Guiot, M. C., Panet-Raymond, V. & Souhami, L. Failure pattern following complete resection plus radiotherapy and temozolomide is at the resection margin in patients with glioblastoma. J. Neurooncol. 111, 19–23 (2013).
Stummer, W. et al. Fluorescence-guided surgery with 5-aminolevulinic acid for resection of malignant glioma: A randomised controlled multicentre phase III trial. Lancet Oncol. 7, 392–401 (2006).
Schipmann, S. et al. Combination of ALA-induced fluorescence-guided resection and intraoperative open photodynamic therapy for recurrent glioblastoma: Case series on a promising dual strategy for local tumor control. J. Neurosurg. 134, 1–11 (2020).
Stummer, W. et al. Fluorescence-guided resection of glioblastoma multiforme by using 5-aminolevulinic acid-induced porphyrins: A prospective study in 52 consecutive patients. J. Neurosurg. 93, 1003–1013 (2000).
Stummer, W. et al. Technical principles for protoporphyrin-IX-fluorescence guided microsurgical resection of malignant glioma tissue. Acta Neurochir. (Wien) 140, 995–1000 (1998).
Stummer, W. et al. Intraoperative detection of malignant gliomas by 5-aminolevulinic acid-induced porphyrin fluorescence. Neurosurgery 42, 518–526 (1998).
De Rosa, F. S. & Bentley, M. V. L. B. Photodynamic therapy of skin cancers: Sensitizers, clinical studies and future directives. Pharm. Res. 17, 1447–1455 (2000).
Rollakanti, K. R., Kanick, S. C., Davis, S. C., Pogue, B. W. & Maytin, E. V. Techniques for fluorescence detection of protoporphyrin IX in skin cancers associated with photodynamic therapy. Photonics Lasers Med. 2, 287–303 (2013).
Jaber, M. et al. Is visible aminolevulinic acid-induced fluorescence an independent biomarker for prognosis in histologically confirmed (World Health Organization 2016) low-grade gliomas?. Clin. Neurosurg. 84, 1214–1224 (2019).
Bravo, J. J. et al. Hyperspectral data processing improves PpIX contrast during fluorescence guided surgery of human brain tumors. Sci. Rep. 7, 1–13 (2017).
Li, Y., Rey-Dios, R., Roberts, D. W., Valdés, P. A. & Cohen-Gadol, A. A. Intraoperative fluorescence-guided resection of high-grade gliomas: A comparison of the present techniques and evolution of future strategies. World Neurosurg. 82, 175–185 (2014).
Montcel, B., Mahieu-Williame, L., Armoiry, X., Meyronet, D. & Guyotat, J. Two-peaked 5-ALA-induced PpIX fluorescence emission spectrum distinguishes glioblastomas from low grade gliomas and infiltrative component of glioblastomas. Biomed. Opt. Express 4, 548 (2013).
Valdés, P. A. et al. A spectrally constrained dual-band normalization technique for protoporphyrin IX quantification in fluorescence-guided surgery. Opt. Lett. 37, 1817 (2012).
Kaneko, S., Suero Molina, E., Ewelt, C., Warneke, N. & Stummer, W. Fluorescence-based measurement of real-time kinetics of protoporphyrin IX after 5-aminolevulinic acid administration in human in situ malignant gliomas. Neurosurgery 85, 739–746 (2019).
Johansson, A. et al. 5-aminolevulinic acid-induced protoporphyrin IX levels in tissue of human malignant brain tumors. Photochem. Photobiol. 86, 1373–1378 (2010).
Yoneda, T. et al. Spectral radiance of protoporphyrin IX fluorescence and its histopathological implications in 5-aminolevulinic acid-guided surgery for glioblastoma. Photomed. Laser Surg. 36, 266–272 (2018).
Valdés, P. A. et al. Quantitative spectrally resolved intraoperative fluorescence imaging for neurosurgical guidance in brain tumor surgery: Pre-clinical and clinical results. In Optical Techniques in Neurosurgery, Neurophotonics, and Optogenetics Vol. 8928 (eds Hirschberg, H. et al.) 892809 (SPIE, 2014).
Valdés, P. A. et al. Quantitative fluorescence using 5-aminolevulinic acid-induced protoporphyrin IX biomarker as a surgical adjunct in low-grade glioma surgery. J. Neurosurg. 123, 771–780 (2015).
Lozovaya, G. I., Masinovsky, Z. & Sivash, A. A. Protoporphyrin ix as a possible ancient photosensitizer: Spectral and photochemical studies. Orig. Life Evol. Biosph. 20, 321–330 (1990).
Alston, L., Rousseau, D., Hebert, M. & Mahieu-Williame, L. Nonlinear relation between concentration and fluorescence emission of protoporphyrin IX in calibrated phantoms. J. Biomed. Opt. 23, 1 (2018).
Melø, T. B. & Reisæter, G. The physicochemical state of protoporphyrin IX in aqueous solution investigated by fluorescence and light scattering. Biophys. Chem. 25, 99–104 (1986).
Alston, L. et al. Spectral complexity of 5-ALA induced PpIX fluorescence in guided surgery: A clinical study towards the discrimination of healthy tissue and margin boundaries in high and low grade gliomas. Biomed. Opt. Express 10, 2478 (2019).
Ando, T. et al. Precise comparison of protoporphyrin IX fluorescence spectra with pathological results for brain tumor tissue identification. Brain Tumor Pathol. 28, 43–51 (2011).
Zanello, M. et al. Multimodal optical analysis discriminates freshly extracted human sample of gliomas, metastases and meningiomas from their appropriate controls. Sci. Rep. 7, 1–11 (2017).
Dietel, W., Fritsch, C., Pottier, R. H. & Wendenburg, R. 5-Aminolaevulinic-acid-induced formation of different porphyrins and their photomodifications. In Lasers in Medical Science, Vol. 12, 226–236 (Springer, 1997).
Barron, G. A. et al. Porphyrin profile in four human cell lines after supplementation with 5-aminolaevulinic acid and its methyl ester. Photodiagn. Photodyn. Ther. 10, 654–663 (2013).
Louis, D. N. et al. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 131, 803–820 (2016).
Mehidine, H. et al. Optical signatures derived from deep UV to NIR excitation discriminates healthy samples from low and high grades Glioma. Sci. Rep. 9, 8786 (2019).
Schaefer, P. M., Kalinina, S., Rueck, A., von Arnim, C. A. F. & von Einem, B. NADH autofluorescence-a marker on its way to boost bioenergetic research. J. Quant. Cell Sci. 95, 34–46 (2019).
Marois, M., Bravo, J., Davis, S. C. & Kanick, S. C. Characterization and standardization of tissue-simulating protoporphyrin IX optical phantoms. J. Biomed. Opt. 21, 035003 (2016).
Valdés, P. A. et al. δ-aminolevulinic acid-induced protoporphyrin IX concentration correlates with histopathologic markers of malignancy in human gliomas: The need for quantitative fluorescence-guided resection to identify regions of increasing malignancy. Neuro Oncol. 13, 846–856 (2011).
Chen, A. X. et al. Functional imaging of mitochondria in retinal diseases using flavoprotein fluorescence. Nat. Eye https://doi.org/10.1038/s41433-020-1110-y (2020).
Reinert, K. C., Gao, W., Chen, G. & Ebner, T. J. Flavoprotein autofluorescence imaging in the cerebellar cortex in vivo. J. Neurosci. Res. 85, 3221–3232 (2007).
Reinert, K. C., Dunbar, R. L., Gao, W., Chen, G. & Ebner, T. J. Flavoprotein autofluorescence imaging of neuronal activation in the cerebellar cortex in vivo. J. Neurophysiol. 92, 199–211 (2004).
Nazeer, S. S., Saraswathy, A., Gupta, A. K. & Jayasree, R. S. Fluorescence spectroscopy as a highly potential single-entity tool to identify chromophores and fluorophores: Study on neoplastic human brain lesions. J. Biomed. Opt. 18(6), 067002 (2013).
Acknowledgements
We would like to thank Carl Zeiss Meditec (Oberkochen, Germany) for providing us with the required BLUE400 filter and the OPMI PICO system.
Funding
Open Access funding enabled and organized by Projekt DEAL. We acknowledge support from the Open Access Publication Fund of the University of Muenster.
Author information
Authors and Affiliations
Contributions
Conception and design: D.B., E.S.-M. Acquisition of data: S.Ka., E.S.-M. Analysis and Interpretation: D.B., A.W., S.Kö., W.S., E.S.-M. Drafting the article. D.B., E.S.-M. Critically revising the article: S.Ka., A.W., S.Kö., W.S. Statistical analysis: D.B., E.S.-M. Approved the final version of the manuscript on behalf of all authors: E.S.-M. Study supervision: E.S.-M.
Corresponding author
Ethics declarations
Competing interests
Walter Stummer has received speaker and consultant fees from Medac, Zeiss, Leica, Photonamic and NXDC and research support from Zeiss. All other authors have no competing interest to declare (financial/non-financial).
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Black, D., Kaneko, S., Walke, A. et al. Characterization of autofluorescence and quantitative protoporphyrin IX biomarkers for optical spectroscopy-guided glioma surgery. Sci Rep 11, 20009 (2021). https://doi.org/10.1038/s41598-021-99228-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-99228-6
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.