Abstract
India ranks the second in the world in terms of its sheep population with approximately 74.26 million represented by 44 well-described breeds in addition to several non-descript populations. Genetic diversity and phylogeography of Indian sheep breeds remain poorly understood, particularly for south Indian breeds. To have a comprehensive view of the domestication history of Indian sheep, we sequenced the mitochondrial DNA (mtDNA) control region (D-loop) and cytochrome b gene (CYTB) of 16 Indian domestic sheep breeds, most of them (13) from the south India. We analysed these sequences along with published data of domestic and wild sheep from different countries, including India. The haplotype diversity was relatively high in Indian sheep, which were classified into the three known mtDNA lineages, namely A, B and C. Lineage A was predominant among Indian sheep whereas lineages B and C were observed at low frequencies but C was restricted to the breeds of north and east India. The median joining network showed five major expanding haplogroups of lineage A (A1–A5). Out of which, A2, A4 and A5 were more frequent in Indian sheep in contrast to breeds from other parts of the world. Among the 27 Indian sheep breeds analysed, Mandya and Sonadi breeds were significantly different from other Indian breeds in the MDS analyses. This was explained by a very high contribution of lineage B into these two breeds. The Approximate Bayesian Computation (ABC) provided evidence for the domestication of lineage A sheep in the Indian subcontinent. Contrary to the current knowledge, we also found strong support for the introduction of lineage B into Indian subcontinent through sea route rather than from the Mongolian Plateau. The neighbour-joining tree of domestic and wild sheep revealed the close genetic relationship of Indian domestic sheep with Pakistani wild sheep O. vignei blanfordi. Based on our analyses and archaeological evidences, we suggest the Indian subcontinent as one of the domestication centres of the lineage A sheep, while lineage B sheep might have arrived into India from elsewhere via Arabian sea route. To the best of our knowledge, this is the first comprehensive study on Indian sheep where we have analysed more than 740 animals belonging to 27 sheep breeds raised in various regions of India. Our study provides insight into the understanding of the origin and migratory history of Indian sheep.
Similar content being viewed by others
Introduction
Domestic sheep is one of the important livestock species in India and contributes significantly to the livelihood of marginalized small farmers of India. India is the second country with a large population of approximately 74.26 million sheep belonging to 44 described breeds and several non-descript sheep populations1. Mitochondrial DNA (mtDNA) studies of sheep breeds across the world have reported two major lineages of A and B, and three minor lineages of C, D and E. The presence of multiple lineages suggests the possibility of their multiple independent domestication events2,3,4,5,6,7,8. Lineage A, B and C have been identified in Indian sheep with lineage A to be most abundant1,8,9,10. A recent study on the mtDNA control region sequences of Indian sheep proposed that lineage A might have been domesticated in the Indian subcontinent while lineage B might have arrived in the Indian subcontinent through sea route8. In contrast, based on the high levels of genetic diversity in sheep breeds of north China and Mongolian Plateau, Lv et al.11 suggested that lineage A was brought into the Indian subcontinent from the Middle East via Arabia whereas the lineage B and C entered into the Indian subcontinent from Middle East via Mongolian Plateau. However, these studies8,11 have not included south Indian breeds.
Indian sheep breeds have been classified into four major groups based on the agro-ecological regions: (a) north temperate, (b) northwestern arid and semi-arid (c) eastern, and (d) southern peninsular breeds12. The south Indian breeds are mostly hairy and primarily have been developed for meat purposes to be easily distinguished from the woolly type of north Indian breeds. The phylogeographic analysis of Indian sheep would be incomplete if the south Indian sheep is not adequately sampled as they contribute nearly 40% to the sheep population of India12. Therefore, in the present study, we sequenced the mtDNA D-loop and CYTB gene of 16 Indian sheep breeds. Out of these, 13 were from south India while the remaining three from east India. To understand the origin, domestication and migration history of Indian sheep, these new mtDNA sequences were analysed along with published data of sheep breeds of other parts of India and other countries.
Materials and methods
Sample collection
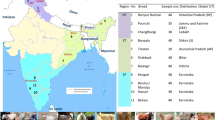
In the present study, 416 animals of 16 Indian sheep breeds, including 13 breeds from south India, were used (Table 1, Fig. 1). The fresh fecal samples were collected from the field/farms within a few minutes of defecation for the purpose of DNA isolation. Sincere efforts were taken to collect only unrelated sheep individuals through interaction with the local herdsmen. The collected feces were stored in absolute ethanol and kept at − 40 °C until DNA extraction. This study used only faecal samples, hence animal ethics committee approval was not required.
Map showing the sampling locations of sheep breeds and distribution of lineage A, B and C of Indian domestic sheep. The map (source: http://www.d-maps.com/carte.php?num_car=24853&lang=en) was edited in Inkscape 1.0 (https://inkscape.org).
DNA extraction, PCR amplification and sequencing
Total genomic DNA was isolated from the feces using QIAamp DNA Stool Mini Kit (Qiagen, Germany) following the procedure of Kamalakkannan et al.13. MtDNA D-loop and CYTB gene were amplified using the following primer sets 5′-CCAGAGAAGGAGAACAACCAA-3′ and 5′-GCATTTTCAGTGCCTTGCTT-3′; 5′-TGTCATCATCATTCTCACATGG-3′ and 5′-GGGAGGTTGGTTGTTCTCCT-3′ respectively8. Amplification was carried out using a thermal cycler with a final volume of 25 µl reaction mixture containing 50 ng of genomic DNA, 12.5 µl of master mix (Promega, Fitchburg, WI) and 2 µl (10 pmol) of each primer under the following PCR conditions: initial denaturation at 94 °C for 5 min, 30 cycles of denaturation at 94 °C for 1 min, annealing at 50 °C for D-loop or 53 °C for CYTB gene for 1 min, extension at 72 °C for 1 min and the final extension at 72 °C for 5 min. Eventually, the PCR products were purified and both strands were sequenced using Big Dye Terminator Cycle sequencing kit (Applied Biosystems, USA) on ABI 3500 Genetic Analyzer (Applied Biosystems, USA) as per the manufacturer’s instructions.
Genetic diversity and phylogenetic analysis
The generated D-loop (GenBank accession numbers: MZ090959–MZ091374) and CYTB gene (GenBank accession numbers: MZ216042–MZ216452) sequences were edited and aligned using the MEGA (version 7) software14. Various population genetics parameters like number of haplotypes, haplotype diversity, AMOVA, pairwise FST values and mismatch distribution were computed using ARLEQUIN 3.115. The calculated pairwise FST values were displayed on the multidimensional scaling (MDS) plot using R program (www.r-project.org). Neighbour-joining (NJ) tree was constructed using MEGA (version 7)14 with 2000 bootstrap and maximum composite likelihood as substitution model to categorize the new sequences based on the reference lineage sequences (HM236174, HM236177, HM236178, HM236180 and HM236182) retrieved from GenBank16. The median-joining (MJ) network was prepared using the program Network 10.2.0.017.
Bayesian inference of sheep domestication history
To better understand the colonization history of domestic sheep lineages, we employed the approximate Bayesian computation (ABC) statistical approach to test different hypotheses of dispersal routes using the D-loop sequences. Simulations were conducted in DIYABC v2.1 software18. The scenarios were built based on the obtained results and previous study11. For lineages of A and B, we considered three metapopulations according to their geographical distributions: The Middle East (ME), the Mongolian Plateau (MP) and the Indian subcontinent (IS). For lineage A, our question was about its origin and colonization routes. To test our hypotheses, we adopted a stepwise procedure to decrease the computational time. In the first round, we built two independent models each with four scenarios, considering the source population identity as the main difference between them (ME and IS, Supplementary Fig. S1). The lineage A scenarios considering ME (named here AME scenarios) as the source were: (1ME) lineage A spread first from ME to MP at time t1, and posteriorly from MP to IS at time t2; (2ME) lineage A expand from ME to IS at time t1, then from IS to MP at time t2; the scenarios 3ME and 4ME occur through two independent colonization events from ME, first MP at time t1 and second IS at time t2 (scenario 3ME), while in the scenario 4ME we considered the first colonization to IS and then to MP. The lineage A scenarios considering IS (hereafter AIS scenarios) as the source were: (1IS) lineage A spread first from IS to MP at time t1, and posteriorly from MP to ME at time t2; (2IS) lineage A expand from IS to ME at time t1, then from ME to MP at t2; in the scenarios 3IS and 4IS, IS was the source population for both MP and ME. In the scenario 3IS the first colonization was to MP and then to ME, while in the 4IS scenario the lineage first spread to ME, and then to MP. The scenarios with high posterior probabilities in each model were tested in the second round (Fig. 2A). For lineage B, we were interested to know the colonization route for the Indian subcontinent (Arabian sea route or Inland route). Two scenarios were hypothesized: (1) lineage B spread from ME to MP at time t1, and posteriorly from MP to IS at time t2 (Inland route) as previously suggested by Lv et al.11; (2) the IS lineage B was originated from ME in an independent colonization event (Arabian sea route) (Fig. 2B). Prior distributions of demographic, historical and mutations parameters are summarized in Supplementary Table S2. For each lineage, we set the mutation model according to its substitution model estimated in JModelTest19. The mean mutation rate was assigned as 10–8 to 10–7 per site per generation20. The statistical summaries (SS) were chosen after we performed a principal components analysis to pre-evaluate the similarity between simulation datasets and the empirical data (data not shown). The SS retained for the generation of the simulated dataset in both lineages were mean of pairwise differences, mean of number of rarest nucleotides at segregating sites, and variance of numbers of the rarest nucleotides at segregating sites, all are considering one-sample SS. Two-sample SS included mean of pairwise differences (B) and FST.
Colonization scenarios tested using DIYABC v2.1 in (A) lineage A and (B) lineage B of domestic sheep. Colours represent the populations: ME (the Middle East, red), MP (the Mongolian Plateau, blue) and IS (the Indian subcontinent, green). In lineage B, the second scenario has a different time divergence designation because it is unknown which lineage diverged first from the Middle East.
After simulating one million datasets for each competing scenario, we performed a principal components analysis to pre-evaluate the similarity between simulation datasets and the empirical data. The posterior probability for each scenario were assessed with polychotomous weighted logistic regression20 on the 1% of simulated datasets that were closest to the observed data. Winner scenarios had the highest significant posterior probability value with a nonoverlapping 95% confidence interval (CI). To evaluate the confidence of the winner scenario, the posterior predictive error was calculated using 1000 simulated datasets in the logistic approach. Parameter estimation was conducted for the scenario with high posterior probabilities using local linear regression21 on the 1% closest simulated datasets, applying logit transformation to all parameter values. The precision of each parameter estimation was evaluated by calculating the relative median of the absolute error (RMAE)20. Finally, we assessed a model verification step by evaluating the goodness-of-fit of the winner scenario concerning the observed dataset.
Results
Genetic diversity of south Indian sheep breeds
D-loop
A total of 359 mtDNA D-loop sequences were generated from 13 south Indian sheep breeds. The 950 bp D-loop sequences revealed 233 haplotypes which resulted from 101 polymorphic sites. Of the 233 haplotypes, 180 were unique and the most frequent haplotype was present in 22 individuals from 10 breeds. The haplotype diversity ranged from 0.8659 ± 0.05 in Mandya breed to 1.0 in Katchaikatty Black breed with only six samples (Table 1). Besides Katchaikatty Black, Madras Red had the second highest haplotype diversity (0.9935 ± 0.01). The AMOVA showed 26.44% of variation among the south Indian breeds. Further, the calculated pairwise FST values were plotted on MDS where all the south Indian sheep breeds formed a single cluster except Nilgiri and Mandya breeds (Supplementary Fig. S2A). To determine whether the high variation among the breeds was due to the effect of Mandya and Nilgiri breeds, the AMOVA was reperformed in three different combinations (1) Both Mandya and Nilgiri breed were excluded, (2) Only Nilgiri breed was excluded and (3) Only Mandya breed was excluded. The variation among breeds remained to be similar in combination 1 and 2. However, in combination 3, the variation among breeds was reduced down to 7.22%, suggesting Mandya breed to be distinct from other south Indian breeds.
CYTB gene
A total of 352 mtDNA CYTB gene sequences were generated from 13 south Indian sheep breeds. The 856 bp CYTB gene showed 36 haplotypes defined by 25 polymorphic sites. The maximum haplotype diversity was found for the Madras Red breed (0.6488 ± 0.0907) while only one haplotype was present among 30 Ramnad White sheep sampled from different flocks and places. The AMOVA showed 24.11% of variation among the south Indian breeds. Further, the pairwise FST values were plotted on MDS which showed Mandya breed to be distinct from others (Supplementary Fig. S2B). Based on the MDS result, the AMOVA was repeated by excluding Mandya breed. Similar with the D-loop data, the variation among breeds was decreased to 10.09%, confirming the unique maternal lineage of Mandya breed compared with other south Indian breeds.
Phylogenetic structure of south Indian sheep breeds
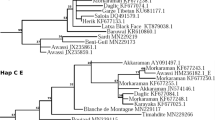
To understand the phylogenetic relationship of south Indian sheep breeds, we constructed NJ trees using D-loop and CYTB gene sequences along with representative lineage sequences. The NJ trees showed two distinct major clades for south Indian sheep which corresponded to the lineages of A and B (Fig. 3). The lineage A was found to be predominant, which consisted of 92.2% of south Indian sheep while the lineage B consisted of only 7.8% of south Indian sheep.
Neighbour-joining phylogenetic tree of south Indian domestic sheep. The NJ tree was based on 359 and 352 sequences for (A) D-loop (950 bp) and (B) CYTB gene (856 bp), respectively. The NJ tree was constructed using the software MEGA (version 7)14 and the final output was edited in Inkscape 1.0 (https://inkscape.org).
Median-joining networks were constructed using D-loop and CYTB gene sequences to understand the distribution pattern of south Indian sheep haplotypes. The MJ network of D-loop was found to be complex but clearly distinguished the lineages of A and B (Supplementary Fig. S3). The lineage A was predominant in all the breeds except Mandya. Notably, seven breeds of Bellary, Coimbatore, Hassan, Katchaikatty Black, Nilgiri, Ramnad White and Vembur were fully encompassed with lineage A while Kenguri, Kilakarsal, Madras Red, Mecheri and Tiruchy Black breeds all had very low occurrences (0.28–0.84%) of lineage B. In contrast, majority of the Mandya sheep carried lineage B (79%). Lineage A displayed five major expanding haplogroups (A1, A2, A3, A4 and A5), indicating extensive expansion of lineage A among south Indian sheep. Also, there was a large number of singletons in the lineage A. On the other hand, lineage B showed no star like expansion among south Indian sheep breeds, indicating the lack of expansion of lineage B. There was no difference between the MJ networks of CYTB gene and D-loop haplotypes, other than the complexity of network (Supplementary Fig. S4).
Phylogeography of Indian sheep breeds
To have a comprehensive view on the genetic diversity and phylogeography of Indian sheep breeds, we also sequenced three breeds (Chottanagpuri, Balangir and Ganjam), from north and east India, retrieved the mtDNA sequences of 12 Indian sheep breeds from GenBank and analysed them along with all new sequences from this study, and thus we included mtDNA sequences of 27 Indian sheep breeds in our analyses (746 D-loop and 738 CYTB gene sequences). The AMOVA showed 17.27% and 14.11% of variations among Indian breeds for D-loop and CYTB gene, respectively. However, when AMOVA was reperformed by excluding Sonadi and Mandya breeds, which were found to be distinct from others in the MDS plots (Fig. 4) the variations among the breeds were dropped down to 10.28% and 6.29% for D-loop and CYTB gene, respectively. Further MDS drawn using only lineage A sequences confirmed Sonadi and Mandya breeds to be distinct from other Indian sheep breeds due to the higher contribution of lineage B in these two breeds (Supplementary Fig. S5). Further, these 27 breeds were divided into four groups based on the agro-ecological regions of India for AMOVA to examine the effect of geographical locations on breed structuring. But this classification was not supported by the variation between these regions (2.42%). MJ networks were constructed separately for lineages of A, B and C using D-loop (Figs. 5, 6) and CYTB gene (Fig. 7) sequences to unveil the distribution patterns of different haplotypes in Indian sheep. There was no significant difference between south Indian and Indian (includes all Indian sequences) networks of lineages A and B while lineage C was only present in five animals belonging to Chottanagpuri (two), Jaisalmeri (two) and Nali (one) breeds (Fig. 6).
Multidimensional scaling plot (MDS) of Indian sheep breeds. The MDS plot was constructed using the pairwise FST values calculated for 746 D-loop (612 bp; (A)) and 738 CYTB gene (737 bp; (B)). BE Bellary, CO Coimbatore, HA Hassan, KE Kenguri, KI Kilakarsal, MN Mandya, MR Madras Red, KB Katchaikatty Black, ME Mecheri, NI Nilgiri, RW Ramnad White, TB Tiruchy Black, VE Vembur, CN Chottanagpuri, BA Balangir, GA Ganjam, MU Muzzafarnagri, NL Nali, CH Chokla, KH Kheri, MA Marwari, JA Jaisalmeri, SO Sonadi, PA Patanwadi, DE Deccani, NE Nellore, and GR Garole. The MDS plot was generated using R program (www.r-project.org) and the final output was edited in Inkscape 1.0 (https://inkscape.org).
Median-joining network of Indian domestic sheep lineage A. The MJ network was constructed using 655 D-loop sequences (612 bp) belonging to 27 Indian sheep breeds. Size of the node is proportional to the number of sheep present in the node. The length of the line is proportional to the number of mutations (# indicates that the length of the line is not proportional to the number of mutations. There is only one mutation between the two haplotypes but length of the line has been adjusted for the convenient arrangement of the node). Sheep breeds are indicated by different colours. The small red rectangles represent the median vector. The network was constructed using Network 10.2.0.017 and edited in Inkscape 1.0 (https://inkscape.org).
Median-joining network of Indian domestic sheep Lineage B and C. (A) The MJ network of lineage B was constructed using 612 bp D-loop sequences. A total of 86 sequences belonging to 17 domestic sheep breeds were used. (B) The MJ network of lineage C was constructed using five D-loop (612 bp) sequences belonging to three sheep breeds. The sizes of nodes are proportional to the number of sheep present in the node. The length of the line is proportional to the number of mutations. Sheep breeds are indicated by different colours. The small red rectangles represent the median vector. The network was constructed using Network 10.2.0.017 and edited in Inkscape 1.0 (https://inkscape.org).
Median-joining network of Indian domestic sheep lineage A, B and C of CYTB gene sequences. (A) The MJ network of lineage A was constructed using 667 CYTB gene sequences (737 bp) of 27 Indian sheep breeds. (B) The MJ network of lineage B was constructed using 67 CYTB gene sequences (737 bp) of 17 sheep breeds. (C) The MJ network of lineage C was constructed using 4 CYTB gene (737 bp) sequences of two sheep breeds. Size of the node is proportional to the number of sheep present in the node. The length of the line is proportional to the number of mutations. Sheep breeds are indicated by different colours. The small red rectangles represent the median vector. The network was constructed using Network 10.2.0.017 and edited in Inkscape 1.0 (https://inkscape.org).
Origin and migration of lineage A and B
The ABC analysis was capable to reconstruct the most probable colonization scenarios for lineages of A and B of the domestic sheep using D-loop sequences (Table 2). For lineage A, the scenario with the highest posterior probability (0.9500; 95% CI 0.9375–0.9625) identified the Indian subcontinent as the source population, which first spread to the Middle East region and then expand to the Mongolian Plateau. It indicated that the domestication of sheep occurred in Indian subcontinent much earlier than previously reported. The posterior predictive error was considerably low (0.285). The posterior distributions of demographic parameters were inferred under the best colonization scenario (Supplementary Table S3). Most parameter estimates showed high RMAE values (> 0.2) and cannot be considered fully reliable. However, because our main goal was to understand the origin of domestic sheep mtDNA lineages and colonization routes, the high RMAE values did not affect our evaluation of the drawn and tested scenarios. For lineage B, ABC analysis identified the Arabian sea route as the most probable colonization route to the Indian subcontinent (posterior probability: 0.6801; 95% CI: 0.6442–0.7159). This scenario also showed low posterior predictive error (0.254), which represented high confidence in the chosen scenario. All estimated parameters of the best scenario and their RMAE can be also found in Supplementary Table S3.
Discussion
Sheep domestication and phylogeography have been subject of long stretched discussion worldwide during the past a few decades. To have a comprehensive view on the origin and phylogeography of Indian sheep breeds, we analysed the mtDNA D-loop and CYTB gene sequences of 27 Indian sheep breeds. The lineage A was predominant whereas lineage B was observed at a low frequency among Indian sheep as was the scenario in Chinese sheep breeds22,23,24,25,26. Surprisingly, 79% of samples of Mandya breed from south India were of lineage B. It may be recalled that Sonadi breed from northwestern India also had relatively high frequency (48%) of lineage B8 (Fig. 1). Lv et al.11 suggested that the lineage B entered into India through Mongolian Plateau. In such a scenario, one would expect a gradient of lineage B from north to south India. But the high frequencies of lineage B in Mandya breed from south India and in Sonadi breed from northwestern India would argue against the proposal of Lv et al.11. It may also be noted that the native tract of these two breeds are close to the Arabian Sea. The ABC analysis strongly supported the migration of lineage B type sheep from the Middle East into the Indian subcontinent through Arabian Sea route. Therefore, we support the earlier suggestion of Singh et al.8 that the lineage B might have arrived into India through sea route rather from the Mongolian Plateau as argued by Lv et al.11. The sea route has been used to export elephant and buffalo from India to other countries in the ancient times27,28 and this further strengthens our findings. Lineage C was only observed among sheep breeds of north and east India at a very low frequency (< 1%) due probably to its recent introduction into the Indian subcontinent as mentioned in the previous study11.
Observing the immense diversity of lineage A in Indian sheep breeds, we constructed a MJ network for lineage A using D-loop sequences of Indian sheep and sheep from other regions of the world to understand the diversity and relationship of lineage A haplotypes. The MJ network was extremely complex and the haplotype diversity was higher in Indian sheep as compared to that of sheep breeds of other regions (Fig. 8). Only in Indian sheep, all the five major haplogroups of A1, A2, A3, A4 and A5 were found in contrast to lineage A sheep from any other country. Specifically, the Indian sheep had higher frequency of haplogroups of A2, A4 and A5. In addition, there were several singleton haplotypes in Indian sheep. The presence of multiple expanding haplogroups in Indian sheep breeds suggested a population expansion of lineage A in India. The same was also reflected in the mismatch distribution curve (Supplementary Fig. S6).
Median-joining network of domestic sheep lineage A. MJ network was constructed using 1303 D-loop sequences (482 bp) of domestic sheep belonging to India, China, Central Asia, Caucasus and West Russia, Middle East, Mongolia and Europe. GenBank accession numbers of the domestic sheep sequences are provided in the Supplementary Table S1. The sizes of nodes are proportional to the number of sheep present in the node. The length of the line is proportional to the number of mutations (# indicates that the length of the line is not proportional to the number of mutations. There is only one mutation between the two haplotypes, but length of the line has been adjusted for the convenient arrangement of the node). The colour of the node indicates the sampling location of sheep. The small red rectangles represent the median vector. The network was constructed using Network 10.2.0.017 and edited in Inkscape 1.0 (https://inkscape.org).
The wild sheep Ovis orientalis (O. gmelini as per recent nomenclature) is comprised of a number of subspecies which are mostly distributed in the Middle East and Southwest Asia, including the Indian subcontinent29. It has been suggested to be the wild ancestor of the modern-day domestic sheep3,11,30,31. Demirci et al.31 suggested that, the present-day domestic sheep might have originated from two maternally distinct ancestral O. gmelini populations. Lv et al.11 observed the close genetic relationship of O. musimon and O. orientalis with domestic sheep lineage B based on the whole mitogenome. However, the D-loop and CYTB gene based phylogenies were different from the whole mitogenome wherein the O. musimon and O. orientalis clustered with lineages A, B, and C of domestic sheep. This leaves open copious questions corresponding to the wild ancestors and their contribution to the gene pool of modern domestic sheep. Therefore, we constructed NJ trees using mtDNA D-loop and CYTB gene sequences of Indian domestic sheep and various wild sheep in order to identify the wild ancestor of Indian domestic sheep. The NJ tree of D-loop showed that Indian domestic sheep lineage A shared ancestry with wild species O. orientalis anatolica whereas lineage B shared ancestry with O. musimon and O. Orientalis (Supplementary Fig. S7) which is in accordance with previous studies11. In case of CYTB gene, the O. orientalis subspecies (O. orientalis gmelini, O. orientalis anatolica and O. orientalis isphahanica) clustered with lineage A while O. aries musimon and O. orientalis gmelini clustered with lineage B as has been reported previously11. We found that one of the wild O. vignei blanfordi sample (collected from Pakistan) clustered with lineage A of Indian domestic sheep (Fig. 9). Its close phylogenetic relationship with lineage A raises speculations that O. vignei blanfordi could be a possible wild ancestor of the lineage A of Indian domestic sheep. The distribution of O. vignei blanfordi in the Balochistan and Sindh (Pakistan) helps further in strengthening the argument that lineage A might have been domesticated in the Indian subcontinent.
Neighbour-joining tree of Indian domestic sheep and wild sheep. The NJ tree was constructed using 737 bp CYTB gene sequence of domestic and wild sheep. The bootstrap values are given above/below the branches. GenBank accession numbers of the wild sheep sequences are provided in the Supplementary Table S1. The NJ tree was constructed using the software MEGA (version 7)14 and the final output was edited in Inkscape 1.0 (https://inkscape.org).
Chen et al.22 have argued that sheep might have been domesticated independently in multiple places and not only confined to the Near East as proposed elsewhere. Similarly, Singh et al.8 proposed that the Indian subcontinent as the one of domestication centres for the lineage A sheep. The Indus valley region of the Indian subcontinent is regarded to be one of the earliest centres of domestication for several plant and animal species32,33,34,35. The presence of wild cattle, wild goat and wild sheep has been reported in Baluchistan in the sixth millennium B.C. through archaeological evidences36. The horncores of wild sheep have also been excavated from Mehrgarh (Neolithic period) and Nausharo (Harapan period)37,38. Further, sheep and goat remains have been recovered from the Central India (Indo-Gangetic Plain and Belan river valley) dated to 12,000–8000 BC39. Since animal domestication takes place locally, Jarrige and Meadow36 opined that inhabitant of Mehrgarh would have domesticated the sheep. The higher genetic diversity of Indian domestic sheep breeds gives strength to this proposition as it is expected that the domestication centres would have higher genetic variability8,40,41. Further, the ABC analysis also provides strength to the argument of Indian subcontinent as one of the domestication centres for lineage A by predicting independent origin of Indian sheep from the Middle East sheep. Therefore, based on our findings, previous molecular studies and archaeological evidences, we suggest that lineage A might have been domesticated in the Indian subcontinent.
In conclusion, our study suggests Indian subcontinent as one of the domestication centres for the lineage A sheep. The study also supports the earlier conclusion that the lineage B might have entered into India through sea route. Further investigation on O. vignei blanfordi from Pakistan is likely to extend understanding of the domestication history of sheep in Indian subcontinent.
References
Sharma, R. et al. Microsatellite and mitochondrial DNA analyses unveil the genetic structure of native sheep breeds from three major agro-ecological regions of India. Sci. Rep. 10, 20422. https://doi.org/10.1038/s41598-020-77480-6 (2020).
Wood, N. J. & Phua, S. H. Variation in the control region sequence of the sheep mitochondrial genome. Anim. Genet. 27, 25–33 (1996).
Hiendleder, S., Mainz, K., Plante, Y. & Lewalski, H. Analysis of mitochondrial DNA indicates that domestic sheep are derived from two different ancestral maternal sources: No evidence for contributions from urial and argali sheep. J. Hered. 89, 113–120 (1998).
Guo, J. et al. A novel maternal lineage revealed in sheep (Ovis aries). Anim. Genet. 36, 331–336 (2005).
Pedrosa, S. et al. Evidence of three maternal lineages in Near Eastern sheep supporting multiple domestication events. Proc. R. Soc. Lond. B. Biol. Sci. 272, 2211–2217 (2005).
Tapio, M. et al. Sheep mitochondrial DNA variation in European, Caucasian, and Central Asian areas. Mol. Biol. Evol. 23, 1776–1783 (2006).
Meadows, J. R., Cemal, I., Karaca, O., Gootwine, E. & Kijas, J. W. Five ovine mitochondrial lineages identified from sheep breeds of the Near East. Genetics 175, 1371–1379 (2007).
Singh, S., Kumar, S., Kolte, A. P. & Kumar, S. Extensive variation and sub-structuring in lineage A mtDNA in Indian sheep: Genetic evidence for domestication of sheep in India. PLoS ONE 8, e77858 (2013).
Pardeshi, V. C. et al. Mitochondrial haplotypes reveal a strong genetic structure for three Indian sheep breeds. Anim. Genet. 38, 460–466 (2007).
Arora, R., Yadav, H. S. & Mishra, B. P. Mitochondrial DNA diversity in Indian sheep. Livest. Sci. 153, 50–55 (2013).
Lv, F. H. et al. Mitogenomic meta-analysis identifies two phases of migration in the history of eastern Eurasian sheep. Mol. Biol. Evol. 32, 2515–2533 (2015).
Bhatia, S. & Arora, R. Biodiversity and conservation of Indian sheep genetic resources—An overview. Asian-Austral. J. Anim. Sci. 18, 1387–1402 (2005).
Kamalakkannan, R. et al. The complete mitochondrial genome of Indian gaur, Bos gaurus and its phylogenetic implications. Sci. Rep. 10, 11936 (2020).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2015).
Excoffier, L. & Lischer, H. E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 10, 564–567 (2010).
Meadows, J. R. S., Hiendleder, S. & Kijas, J. W. Haplogroup relationships between domestic and wild sheep resolved using a mitogenome panel. Heredity 106, 700–706 (2011).
Bandelt, H. J., Forster, P. & Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999).
Cornuet, J. M. et al. DIYABC v2.0: A software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30, 1187–1189 (2014).
Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256 (2008).
Cornuet, J. M., Ravigné, V. & Estoup, A. Inference on population history and model checking using DNA sequence and microsatellite data with the software DIYABC (v1.0). BMC Bioinform. 11, 1–11 (2010).
Beaumont, M. A., Zhang, W. & Balding, D. J. Approximate Bayesian computation in population genetics. Genetics 162, 2025–2035 (2002).
Chen, S. Y. et al. Origin, genetic diversity, and population structure of Chinese domestic sheep. Gene 376, 216–223 (2006).
Zhao, Y., Zhao, E., Zhang, N. & Duan, C. Mitochondrial DNA diversity, origin, and phylogenic relationships of three Chinese large-fat-tailed sheep breeds. Trop. Anim. Health Prod. 43, 1405 (2011).
Zhao, E., Yu, Q., Zhang, N., Kong, D. & Zhao, Y. Mitochondrial DNA diversity and the origin of Chinese indigenous sheep. Trop. Anim. Health Prod. 45, 1715–1722 (2013).
Liu, J. et al. Genetic diversity and phylogenetic evolution of Tibetan sheep based on mtDNA D-Loop sequences. PLoS ONE 11, e0159308 (2016).
Liu, J., Lu, Z., Yuan, C., Wang, F. & Yang, B. Phylogeography and phylogenetic evolution in Tibetan sheep based on MT-CYB sequences. Animals 10, 1177 (2020).
Bertman, S. Handbook to Life in Ancient Mesopotamia (Oxford University Press, 2003).
Nagarajan, M., Nimisha, K. & Kumar, S. Mitochondrial DNA variability of domestic river buffalo (Bubalus bubalis) populations: Genetic evidence for domestication of river buffalo in Indian subcontinent. Genome Biol. Evol. 7, 1252–1259 (2015).
Ziaie, H. Middle East. In Wild Sheep and Goats and Their Relatives Status Survey and Conservation Action Plan for Caprinae (ed. Shackleton, D. M.) 49–55 (IUCN, 1997).
Reed, C. A review of the archaeological evidence on animal domestication in the prehistoric Near East. In Prehistoric Investigations in Iraqi Kurdistan, Studies in Ancient Oriental Civilization (eds Braidwood, R. J. & Howe, B.) 119–145 (The Oriental Institute of the University of Chicago, University of Chicago Press, 1960).
Demirci, S. et al. Mitochondrial DNA diversity of modern, ancient and wild sheep (Ovis gmelinii anatolica) from Turkey: New insights on the evolutionary history of sheep. PLoS ONE 8, e81952 (2013).
Gupta, A. K. Origin of agriculture and domestication of plants and animals linked to early Holocene climate amelioration. Curr. Sci. 87, 54–59 (2004).
Baig, M. et al. Phylogeography and origin of Indian domestic cattle. Curr. Sci. 89, 38–40 (2005).
Kumar, S., Nagarajan, M., Sandhu, J. S., Kumar, N. & Behl, V. Phylogeography and domestication of Indian river Buffalo. BMC Evol. Biol. 7, 186 (2007).
Fuller, D. Q. Finding plant domestication in the Indian subcontinent. Curr. Anthropol. 52, S347–S362 (2011).
Jarrige, J. & Meadow, R. H. The antecedents of civilization in the Indus Valley. Sci. Am. 243, 102–110 (1980).
Meadow, R. H. Early animal domestication in South Asia: A first report of the faunal remains from Merhgarh, Pakistan. In South Asian Archaeology 1979 (ed. Hertel, H.) 143–179 (Dietrich Reimer, 1981).
Meadow, R. H. Prehistoric wild sheep and sheep domestication on the eastern margin of the Middle East. In Animal Domestication and Its Cultural Context (eds Crabtree, P. J. et al.) 24–36 (University Museum, University of Pennsylvania, 1989).
Alur, K. R. Faunal remains from the Vindhyas and the Ganga Valley. In Beginning of Agriculture (eds Sharma, G. R. et al.) 201–227 (Abhinash Prakashan, 1980).
Troy, C. S. et al. Genetic evidence for Near-Eastern origins of European cattle. Nature 410, 1088–1091 (2001).
Beja-Pereira, A. et al. African origins of the domestic donkey. Science 304, 1781 (2004).
Acknowledgements
The study was partly supported by the Science and Engineering Research Board (SERB), Department of Science and Technology, (EMR/2015/000937) and the Department of Biotechnology (BT/PR14584/AAQ/1/647/2015), Government of India, New Delhi. Karippadakam Bhavana and Vandana R. Prabhu were supported by UGC (365844) and DST-INSPIRE (IF160266) through Ph.D. fellowship, respectively. We thank the anonymous reviewers for their critical comments and suggestions.
Author information
Authors and Affiliations
Contributions
M.N. conceived and designed the study. R.K. collected the samples. R.K., K.B., V.R.P., H.S.S. and D.S. performed the experiments. R.K., M.N. and C.B.M. performed the data analysis. M.N., S.K., R.K., V.R.P., C.B.M. and V.V. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kamalakkannan, R., Kumar, S., Bhavana, K. et al. Evidence for independent domestication of sheep mtDNA lineage A in India and introduction of lineage B through Arabian sea route. Sci Rep 11, 19733 (2021). https://doi.org/10.1038/s41598-021-97761-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-97761-y
This article is cited by
-
Genetic diversity and relationship among Indonesian local sheep breeds on Java Island based on mitochondrial cytochrome b gene sequences
Journal of Genetic Engineering and Biotechnology (2023)
-
Tracing the domestication of sheep in India
Nature India (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.