Abstract
Simplicillium species are commonly found from soil, seawater, rock surface, decayed wood, air and as symbiotic, endophytic, entomopathogenic and mycoparasitic fungi. Minority insect-associated species was reported. Simplicillium coccinellidae, S. hymenopterorum, S. neolepidopterorum and S. scarabaeoidea were introduced as the newly insect-associated species. The phylogenetic analyses of two combined datasets (LSU + RPB1 + TEF and SSU + ITS + LSU) revealed that S. coccinellidae and S. hymenopterorum were both nested in an independent clade. S. neolepidopterorum and S. scarabaeoidea have a close relationship with S. formicidae and S. lepidopterorum, respectively. S. neolepidopterorum can be easily distinguished from S. formicidae by ellipsoidal to cylindrical, solitary conidia which occasionally gather in short imbricate chains. S. scarabaeoidea could be easily distinguished from S. lepodopterorum by having longer phialides and larger conidia. Based on the morphological and phylogenetic conclusion, we determine the four newly generated isolates as new species of Simplicillium and a new combination is proposed in the genus Leptobacillium.
Similar content being viewed by others
Introduction
The genus Simplicillium was established for the typical species S. lanosoniveum (J.F.H. Beyma) Zare & W. Gams and three other species S. obclavatum (W. Gams) Zare & W. Gams, S. lamellicola (F.E.V. Sm.) Zare & W. Gams and S. wallacei H.C. Evans1. The typical characteristic of Simplicillium is its solitary phialides, which could be easily distinguished from its closely genus Lecanicillium W. Gams & Zare. S. wallacei was transferred to the genus Lecanicillium based on the phylogenetic analysis by Zare & Gams2. Fourteen species were reported later. Okane et al.3 transferred S. chinense F. Liu & L. Cai and S. coffeanum A.A.M. Gomes & O.L. Pereira to the genus Leptobacillium and this transfer was confirmed by Wang et al.4.
Simplicillium species have diverse ecology, but most species are known from few strains impeding to define their habitat and ecology accurately. Species were found from soil (e.g., S. cylindrosporum, S. minatense, S. subtropicum, and S. sympodiophorum5), as plant endophyte (e.g. S. coffeanum and S. filiforme isolated from Coffea arabica6 and Citrullus lanatus7), from decaying wood or rock (S. calcicola8 and S. chinense9) or from multiple sources. Simplicillium obclavatum was isolated from air, soil, bark, human nail, and seawater1,10, whereas S. aogashimaense was isolated from soil, seawater, and as symbiotic fungi from Nilaparvata lugens Stål5,11,12. Simplicillium lamellicola was isolated as endophytic, entomopathogenic, and mycoparasitic fungi1,13,14,15. Simplicillium lanosoniveum was isolated as cyanobacterium-symbiotic, endophytic, entomopathogenic, and mycoparasitism fungi16,17,18,19. Among those Simplicillium species, six species viz. S. cicadellidae, S. formicae, S. formicidae, S. lamellicola, S. lanosoniveum and S. lepidopterorum, were found associated with insects.
In the present study, four novel insect-associated species viz. Simplicillium coccinellidae, S. hymenopterorum, S. neolepidopterorum and S. scarabaeoidea, were introduced based on morphological comparison and molecular phylogenetic analyses, and this may contribute to the control of insect pest and the discovery of useful novel compounds.
Result
Phylogenetic analyses
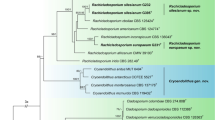
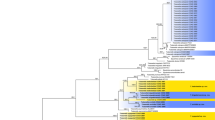
In the phylogenetic tree, Purpureocillium lilacinum (Thom) Luangsa-ard, Houbraken, Hywel-Jones & Samson (CBS 284.36 and CBS 431.87) and Pochonia chlamydosporia (Goddard) Zare & W. Gams (CBS 103.65) were used as the outgroup in analysis 1 and analysis 2, respectively. The concatenated sequences of analysis 1 and analysis 2 included 46 and 22 taxa, and consisted of 1,729 (LSU: 497, RPB1: 550 and TEF: 682) and 1,904 (SSU: 845, ITS: 541 and LSU: 518) characters with gaps, respectively.
Analysis 1: The P-value of PAUP4.0b10 using the command “hompart” is 0.01, and indicated the dataset LSU + RPB1 + TEF is not suitable for the combined analysis. The selected model for LSU, RPB1 and TEF were SYM + G4, SYM + G4 and GTR + F + I + G4, respectively. The final value of the highest scoring tree was –17,856.725706, which was obtained from the ML analysis of the dataset (LSU + RPB1 + TEF). The parameters of GTR model to analysis of the dataset were estimated base frequencies; A = 0.235757, C = 0.286704, G = 0.270379, T = 0.207160; substitution rates AC = 0.874437, AG = 2.344268, AT = 0.877112, CG = 0.872563, CT = 6.144163, GT = 1.000000; gamma distribution shape parameter α = 0.441982. In the phylogenetic tree (Fig. 1), both analyses of ML and BI trees were largely congruent, and strongly supported in most branches. All Simplicillium species were nested in an independent clade, which was the earliest diverging lineage in Cordycipitaceae. The four new species, S. coccinellidae, S. hymenopterorum, S. neolepidopterorum and S. scarabaeoidea were both formed an independent branch and clustered with S. cicadellidae, S. formicidae and S. lepidopterorum in a subclade.
Analysis 2: The P-value of PAUP4.0b10 using the command “hompart” is 0.99, and indicated the dataset SSU + ITS + LSU is suitable for the combined analysis. The selected model was JC for SSU and K2P + G4 for ITS + LSU. The final value of the highest scoring tree was –6,637.139922, which was obtained from the ML analysis of the dataset (SSU + ITS + LSU). The parameters of GTR model to analysis of the dataset were estimated base frequencies; A = 0.251177, C = 0.239762, G = 0.263036, T = 0.246025; substitution rates AC = 1.301732, AG = 2.440073, AT = 0.844382, CG = 1.306407, CT = 3.262235, GT = 1.000000; gamma distribution shape parameter α = 0.552466. In the phylogenetic tree (Fig. 2), both analyses of ML and BI trees were largely congruent, and strongly supported in most branches. Four well-supported clades representing four new novel species S. coccinellidae, S. hymenopterorum, S. neolepidopterorum and S. scarabaeoidea were obtained. These new species clustered with S. cicadellidae, S. formicidae and S. lepidopterorum in a well-supported subclade within the Simplicillium lineage. S. coccinellidae and S. hymenopterorum were both nested in an independent clade. S. neolepidopterorum and S. scarabaeoidea have a close relationship with S. formicidae and S. lepidopterorum, respectively.
Taxonomy
Simplicillium coccinellidae W.H. Chen, Y.F. Han, Z.Q. Liang sp. nov. (Fig. 3).
MycoBank No.: MB 835583.
Etymology: referring to its insect host, family Coccinellidae.
Description: The colonies were moderate-growing on PDA medium, reaching a diameter of 31–36 mm, in 14 days at 25 °C, convex, with white velutinate aerial mycelium, reverse yellowish to pale brown, especially in the middle, margin entire, soluble pigment not produced. Vegetative hyphae branched, hyaline, smooth-walled, septate, 1.1–1.9 μm wide. Phialides produced on aerial hyphae, always solitary, aseptate, hyaline, smooth-walled, relatively slender, and tapering toward the tip, 24.9–62.1 × 1.0–1.5 μm. Conidia in small subglobose slimy heads at the apex of the phialides, hyaline, cylindrical, ellipsoidal to globose, aseptate, smooth-walled, 1-celled, 2.0–3.4 × 1.6–2.0 μm, Octahedral crystals absent.
Material examined: CHINA, Guizhou, Guiyang, Duyun City (26°21′27.96″ N, 107°22′48.22″ E). On dead sacrab (Coccinellidae), 1 October 2019, Wanhao Chen, DY10179 (GZAC DY10179, holotype), was deposited at the Institute of Fungus Resources, Guizhou University (formally Herbarium of Guizhou Agricultural College; code, GZAC), Guiyang City, Guizhou, China; ex-type living cultures, DY101791, DY101792. Sequences from isolated strain DY101791 has been deposited in GenBank with accession numbers: ITS = MT453861, SSU = MT453863, LSU = MT453862 and TEF = MT471341.
Know distribution: China, Guizhou Province, Duyun City (26°21′27.96″ N, 107°22′48.22″ E).
Notes: S. coccinellidae share similar conidial and phialide morphologies with the related species (Table 1). However, the pairwise dissimilarities of ITS sequences show 30, 127, 31, 29, 45, 33 bp difference within 584 bp between S. coccinellidae and S. cicadellidae, S. formicidae, S. lepodopterorum, S. hymenopterorum, S. neolepidopterorum, S. scarabaeoidea respectively. Jeewon & Hyde39 recommended that a minimum of > 1.5% nucleotide differences in the ITS regions may be indicative of a new species. Besides, based on the analysis of the combined dataset LSU + RPB1 + TEF and SSU + ITS + LSU, S. coccinellidae was nested in a separate group in both phylogenetic trees. Thus, the molecular phylogenetic results supported that S. coccinellidae was a new species in the genus Simplicillium.
Simplicillium hymenopterorum W.H. Chen, Y.F. Han, Z.Q. Liang sp. nov. (Fig. 4).
MycoBank No. : MB 835581.
Etymology: referring to its insect host, order Hymenoptera.
Description: The colonies were rapid-growing on PDA medium, reaching a diameter of 40–42 mm, in 14 days at 25 ℃, convex, with white velutinate aerial mycelium, reverse pale yellow, especially in the middle, margin entire, soluble pigment not produced. Phialides produced on prostrate aerial hyphae, mainly solitary, aseptate, hyaline, smooth-walled, relatively slender, and tapering toward the tip, 19.3–46.2 × 1.1–2.3 μm. Conidia in small subglobose heads at the apex of the phialides, hyaline, cylindrical to subellipsoidal, aseptate, smooth-walled, 1-celled, 2.1–2.8 × 1.3–1.9 μm, Octahedral crystals absent.
Material examined: CHINA, Guizhou, Guiyang, Duyun City (26°21′27.96″ N, 107°22′48.22″ E). On dead ant (Hymenoptera), 1 October 2019, Wanhao Chen, DY10169 (GZAC DY10169, holotype), was deposited at the Institute of Fungus Resources, Guizhou University (formally Herbarium of Guizhou Agricultural College; code, GZAC), Guiyang City, Guizhou, China; ex-type living cultures, DY101691, DY101692. Sequences from isolated strain DY101691 has been deposited in GenBank with accession numbers: ITS = MT453848, SSU = MT453849, LSU = MT453850, RPB1 = MT471344 and TEF = MT471337.
Notes: Based on the analysis of the combined dataset LSU + RPB1 + TEF and SSU + ITS + LSU, S. hymenopterorum was nested in a separate group in two phylogenetic trees. The pairwise dissimilarities of ITS sequences show 105, 24, 31, 17 bp difference within 582 bp between S. hymenopterorum and S. formicidae, S. lepodopterorum, S. coccinellidae, S. neolepidopterorum, respectively. The pairwise dissimilarities of RPB1 sequences show 25, 16 bp difference within 737 bp between S. hymenopterorum and S. cicadellidae, S. scarabaeoidea respectively. When compared with the typical characteristics of S. cicadellidae and S. scarabaeoidea (Table 1), S. hymenopterorum could be easily distinguished from S. cicadellidae and S. scarabaeoidea by having subglobose slimy heads of conidia, cylindrical to subellipsoidal conidia, 2.1–2.8 × 1.3–1.9 μm and phialides, 19.3–46.2 × 1.1–2.3 μm. Thus, morphologically based conclusion supported the molecular phylogenetic results that S. hymenopterorum was a new species in the genus Simplicillium.
Simplicillium neolepidopterorum W.H. Chen,Y.F. Han, Z.Q. Liang sp. nov. (Fig. 5).
MycoBank No. : MB 835582.
Etymology: referring to its insect host, order Lepidoptera.
Description: Insect host was completely covered by white to yellowish, loosely mycelium. Conidiophore mononematous. The colonies were slow-growing on PDA medium, reaching a diameter of 28–31 mm, in 14 days at 25 ℃, convex, with white velutinate aerial mycelium, reverse yellowish to pale brown, especially in the middle, margin entire, soluble pigment not produced. Vegetative hyphae branched, hyaline, septate, smooth-walled, 1.3–1.4 μm wide. Phialides produced on aerial hyphae, always solitary and rather long and narrow, aseptate, hyaline, smooth-walled, relatively slender, and tapering toward the tip, 34.1–44.3 × 1.0–1.7 μm. Conidia solitary, occasionally in short imbricate chains, hyaline, ellipsoidal to cylindrical, aseptate, smooth-walled, 1-celled, 2.5–3.8 × 1.5–2.1 μm, Octahedral crystals absent.
Material examined: CHINA, Guizhou, Guiyang, Duyun City (26°21′27.96″ N, 107°22′48.22″ E). On dead insect (Lepidoptera), 1 October 2019, Wanhao Chen, DY10175 (GZAC DY10175, holotype), was deposited at the Institute of Fungus Resources, Guizhou University (formally Herbarium of Guizhou Agricultural College; code, GZAC), Guiyang City, Guizhou, China; ex-type living cultures, DY101751, DY101752. Sequences from isolated strain DY101751 has been deposited in GenBank with accession numbers: ITS = MT453854, SSU = MT453856, LSU = MT453855 and TEF = MT471339.
Notes: Based on the analysis of the combined dataset SSU + ITS + LSU, S. neolepidopterorum is phylogenetically close to S. formicidae. Besides, the pairwise dissimilarities of ITS sequences show 153 bp difference within 580 bp between S. neolepidopterorum and S. formicidae. When compared with the typical characteristics of S. formicidae (Table 1), S. neolepidopterorum could easily distinguished from S. formicidae by having solitary conidia, occasionally in short imbricate chains, and ellipsoidal to cylindrical conidia. Thus, molecular phylogenetic results and morphologically based conclusion were supported S. neolepidopterorum was a new species in the genus Simplicillium.
Simplicillium scarabaeoidea W.H. Chen, Y.F. Han, Z.Q. Liang sp. nov. (Fig. 6).
MycoBank No. : MB 835580.
Etymology: referring to its insect host, family Scarabaeoidea.
Distribution: Insect host was completely covered by white, yellowish to pinkish, densely mycelium. Conidiophore mononematous. The colonies were rapid-growing on PDA medium, reaching a diameter of 44–47 mm, in 14 days at 25 ℃, convex, with white velutinate aerial mycelium; reverse pale yellow, margin entire, soluble pigment not produced. Phialides produced on prostrate aerial hyphae, mainly solitary, aseptate, hyaline, smooth-walled, relatively slender, and tapering toward the tip, 18.5–63.4 × 1.1–1.4 μm. Conidia in small globose heads at the apex of the phialides, hyaline, ellipsoidal, aseptate, smooth-walled, 1-celled, 1.9–2.9 × 1.4–2.0 μm. Octahedral crystals absent.
Material examined: CHINA, Guizhou, Guiyang, Duyun City (26°21′27.96″ N, 107°22′48.22″ E). On dead insect (Lepidoptera), 1 October 2019, Wanhao Chen, DY10139 (GZAC DY10139, holotype), was deposited at the Institute of Fungus Resources, Guizhou University (formally Herbarium of Guizhou Agricultural College; code, GZAC), Guiyang City, Guizhou, China; ex-type living cultures, DY101391, DY101392. Sequences from isolated strain DY101391 has been deposited in GenBank with accession numbers: ITS = MT453842, SSU = MT453843, LSU = MT453844, RPB1 = MT471343 and TEF = MT471335.
Notes: Based on the analysis of the combined dataset SSU + ITS + LSU, S. scarabaeoidea is phylogenetically close to S. lepodopterorum. However, the pairwise dissimilarities of RPB1 sequences show 31 bp difference within 760 bp between S. scarabaeoidea and S. lepodopterorum. When comparing with the typical characteristics of S. lepodopterorum (Table 1), S. scarabaeoidea could be easily distinguished from S. lepodopterorum by having longer phialides and larger conidia. Thus, molecular phylogenetic results and morphologically based conclusion were supported S. scarabaeoidea was a new species in the genus Simplicillium.
Leptobacillium filiform (R.M.F. Silva, R.J.V. Oliveira, Souza-Motta, J.L. Bezerra & G.A. Silva) W.H. Chen, Y.F. Han J.D. Liang & Z.Q. Liang, comb. nov.
Mycobank No.: MB839923.
Basionym: Simplicillium filiform R.M.F. Silva, R.J.V. Oliveira, Souza-Motta, J.L. Bezerra & G.A. Silva, Persoonia 41: 403 (2018).
Notes: Okane et al.3 transferred Simplicillium chinense and S. coffeanum to the genus Leptobacillium. In the present study, S. chinense, S. coffeanum and S. filiform were clustered into an independent clade (Fig. 2), and supported by Crous et al.7, Chen et al.20 and Wei et al.28. Thus, L. filiform is proposed as a new combination.
Discussion
Sung et al.40 refined the classification of Cordyceps and the Clavicipitaceae; the genus Simplicillium thus belongs to the Cordycipitaceae sensu stricto. The result of phylogenetic analysis of the combined dataset (SSU, LSU, RPB1, RPB2 and TEF) showed that Simplicillium species were all clustered in an independent group and as the most ancient lineage in the phylogenetic tree41. In this study, all Simplicillium species were also clustered into a clade at the end of the tree (Fig. 1) based on the analysis of the concentrated dataset (LSU, RPB1 and TEF). The four newly identified species, S. scarabaeoidea, S. hymenopterorum, S. neolepidopterorum and S. coccinellidae, were all clustered in a separate subclade. Liu & Cai9 reported a new species based on the morphological comparison and phylogenetic analysis of ITS and LSU sequences, which was the earliest application for the identification of Simplicillium species. Kondo et al.29 added the loci SSU in the analysis of Simplicillum species. Thus, three loci (ITS, LSU and SSU) were applied in the analysis of the relationship among Simplicillium species in this study.
The nutritional mode from plant to animals and fungi is the evolutionary characteristics of Hypocreales42. Plants associated fungi, which including living plants and plant residues were the common ancestor in the families Hypocreaceae and Clavicipitaceae41. The animal pathogenic fungi are likely inherited from the plant associated fungi by a series of interkingdom host jumps42. In the phylogenetic tree of analysis 2 (Fig. 2), S. chinense, S. filiforme and S. coffeanum were nested in a clade and at the end of the tree. The substrates of S. chinense, S. coffeanum and S. filiforme were decaying wood, branches of Coffea arabica and leaves of Citrullus lanatus6,7,9. All of them were belongs to plants associated fungi, and might reflect the initial state of Simplicillium species, which then underwent a host jump or transferred their nutritional preference. Simplicillium species have rich diversity in substrates and life modes, such as soil, seawater, air, and isolated as symbiotic, endophytic, entomopathogenic and mycoparasitic fungi. Simplicillium species associated with predatory insects or animals, like spiders, will likely soon be reported. Thus, the genus Simplicillium will completely fit with the nutritional model of Hypocreales fungi and could be used as a model to study the evolutionary relationship.
Numerous new secondary metabolites were found from Simplicillium species, such as alkaloids43, diketopiperazine44 and anthraquinones45, especially aogacillin A, B and Simpotentin, which have antibacterial and antifungal activities and shown great potential applications in medicine46,47. In addition, some Simplicillium species were isolated as symbiotic, entomopathogenic and mycoparasitic fungi, and could be used to biocontrol of insect pest, nematode and microbial diseases48,49,50. Thus, it is expected that useful novel compounds will be discovered from the newly-reported Simplicillium species described here and be a natural resource for the application in biocontrol, medicine and health.
Materials and methods
Specimen collection and identification
Four infected insect specimens (DY10139, DY10169, DY10175 and DY10179) were collected from Duyun City (26°21′24.71″ N, 107°22′48.22″ E), Guizhou Province, on 1 October, 2019. Isolation of strains was conducted as described by Chen et al.20. Fungal colonies emerging from specimens were isolated and cultured at 25 °C for 14 days under 12 h light/12 h dark conditions following protocols described by Zou et al.21. Accordingly, strains were obtained. The specimens and the isolated strains were deposited in the Institute of Fungus Resources, Guizhou University (formally Herbarium of Guizhou Agricultural College; code, GZAC), Guiyang City, Guizhou, China.
Macroscopic and microscopic morphological characteristics of the fungi were examined and the growth rates were determined from PDA cultures incubated at 25 °C for 14 days. Hyphae and conidiogenous structures were mounted in lactophenol cotton blue or 20% lactate solution and observed with an optical microscope (OM, DM4 B, Leica, Germany).
DNA extraction, polymerase chain reaction amplification and nucleotide sequencing.
DNA extraction was carried out by Fungal genomic DNA Extraction Kit (DP2033, BioTeke Corporation) in accordance with Liang et al.22. The extracted DNA was stored at − 20 °C. The amplification of internal transcribed spacer (ITS) region, small subunit ribosomal RNA (SSU), large subunit ribosomal RNA (LSU) gene, RNA polymerase II largest subunit 1 (RPB1) and translation elongation factor 1 alpha (TEF) were amplified by PCR as described by White et al.23, Rakotonirainy et al.24, Castlebury et al.25 and van den Brink et al.26, respectively. PCR products were purified and sequenced at Sangon Biotech (Shanghai) Co. The generated sequences were submitted to GenBank.
Sequence alignment and phylogenetic analyses
Lasergene software (version 6.0, DNASTAR) was applied for the assembling and editing of DNA sequence in this study. The ITS, LSU, SSU, RPB1 and TEF sequences were downloaded from GenBank, based on Nonaka et al.5, Zhang et al.8, Gomes et al.6, Crous et al.7, Mongkolsamrit et al.27, Chen et al.20, Wei et al.28, Kondo et al.29 and others selected on the basis of BLAST algorithm-based searches in GenBank (Table 2). The Multiple datasets of ITS, LSU, SSU, RPB1 and TEF were aligned and edited by MAFFT v7.037b30 and MEGA631. Assembling of the combined datasets (LSU + RPB1 + TEF and SSU + ITS + LSU) were performed by SequenceMatrix v.1.7.832. The partition homogeneity test was conducted in PAUP4.0b1033 by using the command “hompart”.
The datasets (LSU + RPB1 + TEF and SSU + ITS + LSU) were analysis by Bayesian inference (BI) and maximum likelihood (ML) methods and aimed to analysis of the relationship among Simplicillium species and its related species in the family Cordycipitaceae (analysis 1) and the relationship among Simplicillium spp. (analysis 2), respectively. For BI, a Markov Chain Monte Carlo (MCMC) algorithm was used to generate phylogenetic trees with Bayesian probabilities using MrBayes v.3.234 for the combined sequence datasets. The model for BI analysis was selected by ModelFinder35 in the software PhyloSuite36. The Bayesian analysis resulted in 20,001 trees after 10,000,000 generations. The first 4000 trees, representing the burn-in phase of the analyses, were discarded, while the remaining 16,001 trees were used for calculating posterior probabilities in the majority rule consensus tree. After the analysis was finished, each run was examined using the program Tracer v1.537 to determine burn-in and confirm that both runs had converged. ML analyses were constructed with RAxMLGUI38. The GTRGAMMA model was used for all partitions, in accordance with recommendations in the RAxML manual against the use of invariant sites. The final alignment is available from TreeBASE under submission ID: 26290 (http://www.treebase.org).
References
Zare, R. & Gams, W. A revision of Verticillium section Prostrata. IV. The genera Lecanicillium and Simplicillium gen. nov. Nova Hedwigia 73, 1–50 (2001).
Zare, R. & Gams, W. A revision of the Verticillium fungicola species complex and its affinity with the genus Lecanicillium. Mycol. Res. 112, 811–824 (2008).
Okane, I., Nonaka, K., Kurihara, Y., Abe, J. P. & Yamaoka, Y. A new species of Leptobacillim, L. symbioticum, isolated from mites and sori of soybean rust. Mycoscience 61, 165–171 (2020).
Wang, Y. B. et al. Multigene phylogeny of the family Cordycipitaceae (Hypocreales): New taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Divers. 103, 1–46 (2020).
Nonaka, K., Kaifuchi, S., Ōmura, S. & Masuma, R. Five new Simplicillium species (Cordycipitaceae) from soils in Tokyo, Japan. Mycoscience 54, 42–53 (2013).
Gomes, A. A. et al. Simplicillium coffeanum, a new endophytic species from Brazilian coffee plants, emitting antimicrobial volatiles. Phytotaxa. 333, 188–198 (2018).
Crous, P. W. et al. Fungal Planet description sheets: 785–867. Persoonia 41, 238 (2018).
Zhang, Z. F. et al. Culturable mycobiota from Karst caves in China, with descriptions of 20 new species. Persoonia 39, 1 (2017).
Liu, F. & Cai, L. Morphological and molecular characterization of a novel species of Simplicillium from China. Cryptogam Mycol. 33, 137–145 (2012).
Liang, X. et al. Eight linear peptides from the deep-sea-derived fungus Simplicillium obclavatum EIODSF 020. Tetrahedron 72, 3092–3097 (2016).
Gonçalves, V. N. et al. Taxonomy, phylogeny and ecology of cultivable fungi present in seawater gradients across the Northern Antarctica Peninsula. Extremophiles 21, 1005–1015 (2017).
Shentu, X. P., Xiao, Y., Cao, Z. Y., Fan, J. X. & Yu, X. P. Comparative analysis of the diversity of the microbial communities between non-fertilized and fertilized eggs of brown planthopper, Nilaparvata lugens Stål. Insects 11, 49 (2020).
Zhang, Q. et al. Diversity and biocontrol potential of endophytic fungi in Brassica napus. Biol. Control 72, 98–108 (2014).
Erper, İ., Saruhan, İ., Akça, İ., Aksoy, H. M. & Tuncer, C. Evaluation of some entomopathogenic fungi for controlling the green shield bug, Palomena prasina L. (Heteroptera: Pentatomidae). Egypt J. Biol. Pest Control 26, 573–578 (2016).
Shinn, T. S. et al. Development of a biofungicide using a mycoparasitic fungus Simplicillium lamellicola BCP and its control efficacy against gray mold diseases of tomato and ginseng. Plant Pathol. J. 33, 337–344 (2017).
Dong, Q. L., Dong, R. Z., Xing, X. Y. & Li, Y. K. A new antibiotic produced by the cyanobacterium-symbiotic fungus Simplicillium lanosoniveum. Nat. Prod. Res. 32, 1–5 (2017).
Yu, Y. T., He, S. H. & Zhao, Q. M. Isolation and identification of matrine-producing fungal endophytes from Sophora alopecuroides in Ningxia. Sci. Agric. Sin. 46, 2643–2654 (2013).
Wang, N., Xie, Y. P. & Fan, J. H. Pathogenicity of Simplicillium lanosoniveum TYL001 isolated from Pseudaulacaspis pentagona. Mycosystema 35, 559–568 (2016).
Gauthier, N. W. et al. Mycoparasitism of Phakopsora pachyrhizi, the soybean rust pathogen, by Simplicillium lanosoniveum. Biol. Control 76, 87–94 (2014).
Chen, W. H., Liu, C., Han, Y. F., Liang, J. D. & Liang, Z. Q. Three novel insect-associated species of Simplicillium (Cordycipitaceae, Hypocreales) from southwest china. Mycokeys 58, 83–102 (2019).
Zou, X., Liu, A. Y., Liang, Z. Q., Han, Y. F. & Yang, M. Hirsutella liboensis, a new entomopathogenic species affecting Cossidae (Lepidoptera) in China. Mycotaxon 111, 39–44 (2010).
Liang, J. D. et al. Optimal culture conditions for keratinase production by a novel thermophilic Myceliophthora thermophila strain GZUIFR-H49-1. J. Appl. Microbiol. 110, 871–880 (2011).
White, T. J., Bruns, T., Lee, S. & Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications (eds Innis, M. A. et al.) 315–322 (Academic Press, 1990).
Rakotonirainy, M. S., Cariou, M. L., Brygoo, Y. & Riba, G. Phylogenetic relationships within the genus Metarhizium based on 28S rRNA sequences and isozyme comparison. Mycol. Res. 98, 225–230 (1994).
Castlebury, L. A., Rossman, A. Y., Sung, G. H., Hyten, A. S. & Spatafora, J. W. Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycol. Res. 108, 864–872 (2004).
van den Brink, J., Samson, R. A., Hagen, F., Boekhout, T. & de Vries, R. P. Phylogeny of the industrial relevant, thermophilic genera Myceliophthora and Corynascus. Fungal Divers. 52, 197–207 (2012).
Mongkolsamrit, S. et al. Disentangling cryptic species with Isaria-like morphs in Cordycipitaceae. Mycologia 110, 230–257 (2018).
Wei, D. P., Wanasinghe, D. N., Hyde, K. D., Mortimer, P. E. & To-Anun, C. The genus Simplicillium. Mycokeys 60, 69–92 (2019).
Kondo, N., Iwasaki, H., Tokiwa, T., Ōmura, S. & Nonaka, K. Simplicillium spumae (Cordycipitaceae, Hypocreales), a new hyphomycetes from aquarium foam in Japan. Mycoscience 61, 116–121 (2020).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Vaidya, G., Lohman, D. J. & Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180 (2011).
Swofford, D. L. PAUP* 4.0b10: Phylogenetic Analysis Using Parsimony (*and Other Methods). (Sinauer, 2002).
Ronquist, F. et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K., Von Haeseler, A. & Jermiin, L. S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
Zhang, D. et al. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355 (2020).
Drummond, A. & Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007).
Silvestro, D. & Michalak, I. raxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 12, 335–337 (2012).
Jeewon, R. & Hyde, K. D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 7, 1669–1677 (2016).
Sung, G. H. et al. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 57, 1–64 (2007).
Kepler, R. M. et al. A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus. 8, 335–353 (2017).
Spatafora, J. W., Sung, G. H., Sung, J. M., Hywel-Jones, N. L. & White, J. F. Phylogenetic evidence for an animal pathogen origin of ergot and the grass endophytes. Mol. Ecol. 16, 1701–1711 (2007).
Fukuda, T., Sudoh, Y., Tsuchiya, Y., Okuda, T. & Igarashi, Y. Isolation and biosynthesis of preussin B, a pyrrolidine alkaloid from Simplicillium lanosoniveum. J. Nat. Prod. 77, 813–817 (2014).
Yan, B. et al. A new minor diketopiperazine from the sponge-derived fungus Simplicillium sp. YZ-11. Nat. Prod. Res. 29, 2013–2017 (2015).
Huang, Z., Yan, S. Z. & Chen, S. L. Optimization on fermentation conditions of Simplicillium obclavatum YX016 for the production of anthraquinones. Food Sci. Technol. 40, 12–17 (2015).
Takata, K. et al. Aogacillins A and B produced by Simplicillium sp. FKI-5985: New circumventors of Arbekacin resistance in MRSA. Org. Lett. 15, 4678–4681 (2013).
Uchida, R. et al. Simpotentin, a new potentiator of amphotericin B activity against Candida albicans, produced by Simplicillium minatense FKI-4981. J. Antibiot. 72, 134 (2019).
Ward, N. A., Robertson, C. L., Chanda, A. K. & Schneider, R. W. Effects of Simplicillium lanosoniveum on Phakopsora pachyrhizi, the soybean rust pathogen, and its use as a biological control agent. Phytopathology 102, 749–760 (2012).
Zhao, D. et al. Simplicillium chinense: A biological control agent against plant parasitic nematodes. Biocontrol Sci. Technol. 23, 980–986 (2013).
Chen, R. S., Huang, C. C., Li, J. C. & Tsay, J. G. Evaluation of characteristics of Simplicillium lanosoniveum on pathogenicity to aphids and in vitro antifungal potency against plant pathogenic fungi. Int. J. Environ. Agric. Res. 3, 55–61 (2017).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 31860002), High-level Innovative Talents Training Object in Guizhou Province (No. Qiankehepingtairencai [2020]6005), Science and Technology Foundation of Guizhou Province (No. Qiankehejichu [2020]1Y060), Program of Innovative Scientific and technological Talent Team of Guizhou Province(2020-5010) and Construction Program of Guizhou Engineering Research Center (Qian Fa Gai Gao Ji 2020-896) and National Survey of Traditional Chinese Medicine Resources (No. Caishe [2017]66, 216). We thank Catherine Dandie, PhD, from Liwen Bianji, Edanz Editing China (https://www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Author information
Authors and Affiliations
Contributions
W.C.: resources, data curation, writing-review & editing. Y.H.: writing-review & editing. J.L.: resources, review & editing. Z.L.: review & editing. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, WH., Han, YF., Liang, JD. et al. Taxonomic and phylogenetic characterizations reveal four new species of Simplicillium (Cordycipitaceae, Hypocreales) from Guizhou, China. Sci Rep 11, 15300 (2021). https://doi.org/10.1038/s41598-021-94893-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-94893-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.