Abstract
A major contributor to biomaterial associated infection (BAI) is Staphylococcus aureus. This pathogen produces a protective biofilm, making eradication difficult. Biofilms are composed of bacteria encapsulated in a matrix of extracellular polymeric substances (EPS) comprising polysaccharides, proteins and extracellular DNA (eDNA). S. aureus also produces micrococcal nuclease (MN), an endonuclease which contributes to biofilm composition and dispersion, mainly expressed by nuc1. MN expression can be modulated by sub-minimum inhibitory concentrations of antimicrobials. We investigated the relation between the biofilm and MN expression and the impact of the application of antimicrobial pressure on this relation. Planktonic and biofilm cultures of three S. aureus strains, including a nuc1 deficient strain, were cultured under antimicrobial pressure. Results do not confirm earlier findings that MN directly influences total biomass of the biofilm but indicated that nuc1 deletion stimulates the polysaccharide production per CFU in the biofilm in in vitro biofilms. Though antimicrobial pressure of certain antibiotics resulted in significantly increased quantities of polysaccharides per CFU, this did not coincide with significantly reduced MN activity. Erythromycin and resveratrol significantly reduced MN production per CFU but did not affect total biomass or biomass/CFU. Reduction of MN production may assist in the eradication of biofilms by the host immune system in clinical situations.
Similar content being viewed by others
Introduction
An increasing number of biomedical implants is being used in clinical practice. This is due to technical advancement, innovative therapies, increased patient demands and life expectancy. These implants come with the risk of biomaterial associated infection (BAI)1. Simultaneously the rate of BAI per implant is increasing due to increasing age and comorbidities of the patients receiving such implants2, 3. Treatment of BAI often entails revision of the implant and antibiotic therapy with a risk of secondary infection4. This results in increased morbidity and high healthcare costs1. Primary antibiotic therapy is usually ineffective since bacteria create a protective shelter when attached to the surface of the foreign material, also designated as a ‘biofilm’. Biofilms are composed of bacteria encapsulated in a matrix of extracellular polymeric substances (EPS) providing protection from antibiotics, the host immune system, and physical and mechanical stress5. The EPS content includes polysaccharide intercellular adhesin (PIA), proteins and extracellular DNA (eDNA), all playing a major role in biofilm maturation and structure6, 7. eDNA in particular has a glue-like function by keeping bacteria entangled within the biofilm by electrostatic and acid–base interactions with cell surfaces and polysaccharides7, 8.
One of the most prominent pathogens in BAI is Staphylococcus aureus, a strong biofilm producer responsible for 34% of all orthopedic BAI9. An important virulence factor of S. aureus is micrococcal nuclease (MN), a thermostable endonuclease that degrades eDNA as a constituent of the biofilm10, 11. Biofilm formation and maturation is expected to be intrinsically affected by the production of MN since it cleaves eDNA. MN production is largely regulated by the SaeRS gene system10, 12. SaeRS consists of a sensory SaeS part and a SaeR response regulator and is activated mainly in the post-exponential growth phase by phagocytosis-related signals, sub-minimum inhibitory concentrations (sub-MIC) of some antibiotics and certain chemical stimuli13. S. aureus produces two types of MN. The first is secreted by the bacterium and encoded by the nuc1 gene. The second is cell-wall bound and encoded by nuc210, 14. Nuc1 and nuc2 are expressed in different ratios depending on the growth phase, with nuc1 being expressed mainly in the post-exponential phase and nuc2 in the early-exponential phase10, 15. In post-exponential cultures of S. aureus nuc2 accounts for a minimal part of the DNA degrading capabilities of S. aureus cultures15. Our previous research estimated that about 1% of the nuclease activity at the post-exponential phase can be contributed to nuc2, as nuclease activity is decreased by 99% when comparing a S. aureus Newman and its nuc1 deficient mutant16. Controversy exists about the effect of MN on biofilm formation. A negative correlation was observed in vitro between biofilm biomass and the nuclease activity in biofilms of various S. aureus strains and nuc1 mutants17, whereas in an in vitro catheter model no effect was found on biofilm formation in a nuc1 and nuc2 mutant of S. aureus UAMS-1 strains18.
MN expression and in particular the SaeRS system can be modulated by sub-minimum inhibitory concentrations of antimicrobials (see also Table 1)19, 20. Vice versa, SaeRS may also regulate the AtlA autolysin gene, responsible for programmed cell lysis and eDNA release21, 22, which makes the production of nuclease and release of e-DNA delicately balanced processes.
Sub-MIC antimicrobial pressure exists in tissue surrounding biomaterial implants because treatment by antimicrobials is hindered by poor penetration into the biofilm or by pathological changes in the implant site, like the formation of a fibrous layer around the implant in case of older implants (> 1 month)26, or changes to the bone structure in joint prosthesis27.
This study aimed at investigating the relation between biofilm formation and MN production and the impact of sub-MIC antimicrobial pressure on this relation. Therefore biofilm formation and MN activity in planktonic and biofilm cultures were studied while applying various sub-MICs of antimicrobials to three strains of S. aureus, the S. aureus ATCC12600, the bioluminescent S. aureus Newman lux, and the S. aureus Newman lux Δnuc1 mutant which is deficient in producing MN17. The ATCC12600 is often used as a reference strain. The Newman WT strain was chosen because of its constitutive expression of MN due to a point mutation SaeSp that constitutively activates the response regulator SaeR, even in the early-exponential growth phase28. Five antibiotics were involved with different working mechanisms (Table 1) and one antimicrobial which has been shown to inhibit SaeRS activity19. Sub-MIC antimicrobial pressure, in particular by aminoglycocides, is also known to affect the production of PIA, the other main important biofilm apart from e-DNA, that contributes to biofilm formation and immune evasion mechanisms29. Thus, in order to take into account biofilm characteristics that may be affected by sub-MICs of antimicrobials in relation to MN activity, both total biomass and polysaccharide quantity in the EPS were investigated as well as colony forming units (CFU’s) and bioluminescence as a measure of metabolic activity30.
Results
CFU, polysaccharides and biomass of biofilms in absence of antimicrobials
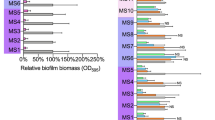
The total biomass, polysaccharide content and CFUs of the two MN proficient strains and one MN deficient S. aureus strain after 24 h of biofilm growth were not significantly different (Fig. 1A–C) due to relatively large standard deviations encountered over all cultures and experiments for all parameters. The analysis, of biomass and polysaccharides per CFU however, executed within each separate culture and experiment on one and the same day revealed that biomass per CFU was raised in the MN deficient strain, although not significantly (Fig. 1D). However, the amount of polysaccharides produced per CFU of the MN deficient S. aureus Newman lux Δnuc1 grown under biofilm conditions was significantly higher than the other strains (Fig. 1E), indicating that an equally amount of EPS was produced populated by fewer viable bacteria. Note that biomass and polysaccharide content do not need to be directly proportional to each other, since biomass includes both bacterial mass (live and dead) and biofilm matrix.
Biofilm characteristics of S. aureus ATCC12600, Newman lux, and Newman lux Δnuc1. (A) Biomass of biofilms measured by spectral absorption after crystal violet staining. (B) Polysaccharide content of biofilms measured by fluorescent intensity after calcofluor white staining (C) Colony forming units (CFU’s) per cm2 of biofilms grown for 24 h in a 24 wells plate. (D) Ratios of biomass/CFU and (E) polysaccharides/CFU of biofilms. All data shown are averages of triplicate measurements in three separate cultures, repeated on separate days. Error bars indicate standard error of the mean. Statistical differences were calculated using an ANOVA. *Significant difference (p < 0.05). In (D, E) the natural logarithm of the values was used for statistical analysis as the data are lognormally distributed (see also Fig. S3).
Changes in biofilm composition under antimicrobial pressure
As expected, most S. aureus biofilms required a higher concentration of antimicrobials to achieve inhibition of growth than planktonically grown bacteria resulting in MBIC > MIC (Table 2). The exceptions being doxycycline and erythromycin, for which no differences between planktonic and biofilm inhibition were observed for both Newman strains. It should be noted that the susceptibility to antimicrobial substances of the Newman nuc1-deficient strain was identical to its parent strain (Table 2).
Most sub-inhibitory concentrations of antimicrobials had no significant inhibiting effects on total biomass and polysaccharides (see Fig. 2). Ciprofloxacin had a stimulating effect on biomass at low doses (\({\raise0.7ex\hbox{$1$} \!\mathord{\left/ {\vphantom {1 8}}\right.\kern-\nulldelimiterspace} \!\lower0.7ex\hbox{$8$}}\) and \({\raise0.7ex\hbox{$1$} \!\mathord{\left/ {\vphantom {1 4}}\right.\kern-\nulldelimiterspace} \!\lower0.7ex\hbox{$4$}}\) MBIC) in both S. aureus Newman strains. Total MN activity of the biofilms was inhibited up to 80% by doxycycline, erythromycin and resveratrol at concentrations below the MBIC (See Fig. 2). At high concentrations (± 100 μg/mL) resveratrol gradually precipitated, binding crystal violet and causing a strong false positive signal. Precipitated resveratrol, however, had no effect on the quantification of MN activity and polysaccharides (data not shown).
Biomass, polysaccharides and micrococcal nuclease (MN) activity of S. aureus ATCC12600, Newman lux, and Newman lux Δnuc1 biofilms under antimicrobial pressure (at 0, 1/8, \({\raise0.7ex\hbox{$1$} \!\mathord{\left/ {\vphantom {1 4}}\right.\kern-\nulldelimiterspace} \!\lower0.7ex\hbox{$4$}}\), \({\raise0.7ex\hbox{$1$} \!\mathord{\left/ {\vphantom {1 2}}\right.\kern-\nulldelimiterspace} \!\lower0.7ex\hbox{$2$}}\) or 1 times MBIC). All data are normalized to a control culture without antimicrobials (= 100%; horizontal dotted line). No MBIC was found for resveratrol, therefore it is set at the highest attainable concentration, 512 μg/mL. Statistical differences were calculated using an ANOVA. *Significant difference for concentrations smaller than MBIC when compared to the culture without antibiotics. (*p < 0.05; **p < 0.01; ***p < 0.001).
No significant difference was found in biomass and polysaccharide between S. aureus Newman lux and the nuc1-deficient strain at any concentration of any antimicrobial (Fig. S1).
Effects of antimicrobials on EPS production per bacterium
Some antimicrobials induced increase of EPS production per CFU under antimicrobial pressure was observed. Ciprofloxacin, doxycycline, and erythromycin increased the biomass and polysaccharides per CFU significantly by up to five times that of the control (Fig. 3), whereas gentamicin, vancomycin and resveratrol had a moderate to no effect. This is due to the biomass and polysaccharide content staying largely unchanged while the number of CFU’s decreased (Figs. 2 and S2).
Change in biomass per CFU and polysaccharides per CFU ratios of S. aureus ATCC12600 and Newman lux biofilms. Biomass is quantified by crystal violet staining and subsequent measurement of spectral absorption at 590 nm. Polysaccharides are quantified by calcofluor white and subsequent measurement of fluorescent intensity at 355/490 nm (Ex/Em). Values shown is increase relative to the control biofilms without antibiotics (= 1; dotted horizontal line). Data shown are based on averages of triplicate measurements, repeated on separate days. Error bars indicate standard error of the mean. Statistical differences were calculated using an ANOVA comparing lognormal values (see “Materials and methods”). *Significant difference when compared to the culture without antibiotics (*p < 0.05; **p < 0.01; ***p < 0.001).
Effects of antimicrobials on nuclease activity per bacterium
CFU-counts in both biofilm and planktonic cultures (Fig. S2) were used to calculate the effect of antimicrobials on MN activity per viable bacterium. In contrast to the stimulating effect of some antimicrobials on biomass and polysaccharides, MN activity per CFU was unaffected or inhibited by antimicrobials (Fig. 4). Lognormal values of MN/CFU were used because this variable was lognormally distributed (see Fig. S3). In particular resveratrol inhibited MN per CFU significantly at concentrations as low as \({\raise0.7ex\hbox{$1$} \!\mathord{\left/ {\vphantom {1 8}}\right.\kern-\nulldelimiterspace} \!\lower0.7ex\hbox{$8$}}\) MIC and MBIC, except for S. aureus ATCC12600 biofilms in which MN per CFU was inhibited at \({\raise0.7ex\hbox{$1$} \!\mathord{\left/ {\vphantom {1 8}}\right.\kern-\nulldelimiterspace} \!\lower0.7ex\hbox{$8$}}\) and \({\raise0.7ex\hbox{$1$} \!\mathord{\left/ {\vphantom {1 4}}\right.\kern-\nulldelimiterspace} \!\lower0.7ex\hbox{$4$}}\), but not at \({\raise0.7ex\hbox{$1$} \!\mathord{\left/ {\vphantom {1 2}}\right.\kern-\nulldelimiterspace} \!\lower0.7ex\hbox{$2$}}\) MBIC. Erythromycin reduced MN activity of the Newman strain by almost 75% at \({\raise0.7ex\hbox{$1$} \!\mathord{\left/ {\vphantom {1 4}}\right.\kern-\nulldelimiterspace} \!\lower0.7ex\hbox{$4$}}\) MBIC.
Micrococcal nuclease (MN) production per CFU of S. aureus ATCC12600 and Newman lux, growing planktonically and in biofilm under antibiotic pressure. Data shown are averages of triplicate measurements, repeated on separate days. Error bars indicate standard error of the mean. Statistical differences were calculated using an ANOVA comparing lognormal values with control values where antimicrobial concentration = 0). *Significant difference between value and the culture without antibiotics (*p < 0.05; **p < 0.01).
Effect of different type of culturing
Bacteria were grown both in stationary biofilm cultures from adhering bacteria, and under rotational movement to stimulate planktonic growth and reduce biofilm formation (see also Fig. S4). MN production and luminescence per CFU was quantified and analyzed using a two-way ANOVA. Lognormal values of MN/CFU were used as this variable is lognormally distributed (see Fig. S3). Planktonically grown bacteria produced more MN per CFU than those growing in a biofilm (p = 0.0118) (Fig. 5A). Strain was not a significant determining factor, neither was interaction between strain and type of culturing. As expected, the nuc1-deficient strain produced less than 1% of the amount MN produced by the parent strain. Luminescence per CFU in strains grown under rotation was 150% higher than when grown stationary (p = 0.0003) (Fig. 5B). In case of luminescence the deletion of the nuc1 gene was a significant factor in increasing luminescence (p = 0.0028), with a significant interaction between type of culturing and nuc1 deletion (p = 0.0219). The luminescence was used as a marker for metabolic activity of the bacteria30.
(A) Micrococcal nuclease activity (U), per colony forming unit (CFU) of S. aureus ATCC12600 and S. aureus Newman lux from planktonic and biofilm cultures. The type of culture (biofilm or planktonic) significantly affects the nuclease production. (B) Bacterial luminescence per CFU (photons/second/CFU) of S. aureus Newman lux and Δnuc1 mutant. The type of culture (biofilm or planktonic) significantly affects the luminescence for both strains individually. Data shown are averages of triplicate measurements, repeated on separate days. Statistical analysis was done using an ANOVA accounting for strain and type of culture. Error bars indicate standard deviations. Asterisks indicate significant differences (*p < 0.05; ***p < 0.001).
Discussion
The aim of this study was to investigate the relation between biofilm formation and MN activity and the impact on it by sub-MIC antimicrobial pressure.
Various earlier reports are not unambiguous on whether MN has an inhibitory effect on biofilm formation with results varying from increased to decreased biofilm formation, also depending on in vitro17 or in vivo setup11, 17, 18, 32. Our results revealed no significant differences between the two WT-strains and the MN-deficient strain with respect to the number of CFU, the total biomass and polysaccharides (Figs. 1A–C and S1). It might have been anticipated that S. aureus Newman strains do not produce biofilms due to a defect in the expressed fibronectin binding proteins (FnBPs) but it has been suggested earlier that the lack of proper fibronectin binding proteins is compensated by the enhanced production of extracellular adhesive proteins (Eap) in the Newman strain33. Forson et al. also suggested that the eDNA in the biofilm is stabilized, possibly via interactions with DNA-binding proteins like Eap and in particular SaeP, which is expressed as an auxiliary protein in the SaeRS two component system32. There are several possible explanations for the observation that biofilms in our experiments did not increase in mass upon deletion of the nuc1 gene as was observed in other research17, such as the lack of added glucose in the growth medium, longer incubation time (24 h), and the lower initial bacterial load that may alter the response of the biofilm to MN11, 34, 35.
When the present results, however, were normalized with respect to the number of CFU’s in the assays, the results revealed, for the first time, a significant, 20% enhancement of biomass and polysaccharides production per CFU of the nuc1 deficient S. aureus Newman lux strain compared to both other S. aureus strains. This suggests an inverse relationship between EPS production per CFU and MN activity per CFU. This is in line with earlier findings32, based on bacterial counts and optical coherence tomography observations, that the bacterial density in biofilms is reduced in the nuclease deficient S. aureus Newman strain. Uncleaved eDNA in the nuclease deficient strain tends to expand the biofilm and may keep bacteria at relatively long distances from each other, also pointing to an increased biomass production per CFU in MN deficient strains32. It is very likely that the differences found between both Newman strains are due to the missing nuc1 gene alone and not to other secondary mutations in regulatory genes. Both Newman strains showed very similar adhesion characteristics on both hydrophobic and hydrophilic substrates and were also highly similar in biofilm structure32. Moreover, both strains after inoculation in mice to induce a local infection in the murine thigh, resulted in similar infections as observed by in vivo imaging36.
In this study we anticipated that some antibiotics may modulate MN production, the most significantly in the S.aureus ATCC12600 strain, since the constitutive character of the SaeSp induced kinase activity. The point mutation in the Newman strain expression, however, allows some substances (such as a biocide “perform” and Sodium dodecyl sulfate) to raise the transcription level of SaeS even further33 indicating that environmental factors may still modulate the Sae system and potentially modulate nuclease production as remarkably found in our study in both S. aureus strains. We observed significant reductions, up to 80% in MN production for both S. aureus strains by doxycyclin, erythromycin and resveratrol by (Fig. 2). The inhibition of MN activity by erythromycin is in line with reports of clindamycin, which has a similar working mechanism by binding to the ribosomal 50 s subunit and inhibits the SaeRS system (Table 1). The reduction by resveratrol is attributed to the direct inhibition of the SaeRS system as reported earlier19. Since it was concluded from Fig. 1E that MN reduction had effect on the EPS production per CFU rather than on the total CFU count (Fig. 1C), biomass (Fig. 1A) and polysaccharides (Fig. 1B), it was expected that EPS production per CFU subsequently would be affected by sub-MIC pressure of antimicrobials as well. Indeed, significant higher polysaccharide content per CFU was found in the presence of sub-MIC doxycyclin and erythromycin, up to a factor of 5 with respect to the control (Fig. 3), but only in case of erythromycin this coincides with significant reduction in MN per CFU (Fig. 4) and not in case of sub-MIC doxycyclin. These results strongly suggest that sub-MIC doxycyclin stimulates polysaccharide production, an effect that has been reported earlier for aminoglycosides in S. epidermidis29. The aminoglycoside in this study (gentamycin), however, did not show polysaccharide stimulation.
It should be noted that as a result of the highly increased histidine kinase activity in the Newman strain, we expected that S. aureus ATCC12600 would show a lower nuclease productivity. Instead, we found relatively high amounts of nuclease produced by the S. aureus ATCC12600, in the same range as the Newman strain (see Fig. 5A). We earlier identified S. lugdunensis as a additional strain producing an equal amount of nuclease, and S. aureus RN 4220 hardly showed lower nuclease production as compared to the Newman strain16. Moreover it should be noticed that the Newman SaeSp activates SaeR in the early exponential phase, resulting in an early nuc1 mediated reaction28, 37,38,39. The ‘normal’ SaeS found in ATCC12600 activates SaeR only after 6 h of growth. Since our measurements have all been made after 24 h of growth this difference may have disappeared entirely, since earlier experiments have shown that nuclease production of strains usually levels to a plateau value after 12 h of growth16. As was observed earlier32, MN activity appeared significantly inhibited in the biofilm mode of growth compared to planktonic growth (Fig. 5A). The reduction of MN in biofilms compared to planktonic cultures, is similar to the reduction of bioluminescence in biofilms as related to planktonic cultures (Fig. 5B). The reduction in bioluminescence most likely results from a limited availability of oxygen and lower metabolic activity of bacteria located deeper in the biofilm30, 40. Decreased oxygenation and metabolic activity is probably also the dominant factor reducing MN production in biofilms.
We recognize a wide debate on the various mechanisms involved in biofilm formation and we in particular elaborated on the understudied aspects of MN production and its potential impact on biofilm characteristics. In summary our results do not confirm earlier findings that MN directly influences total biomass of the biofilm, but rather that deletion of nuc1 stimulates the EPS production per CFU, as was found in the S. aureus Newman strain. MN production can be further modulated by antimicrobial pressure, and did in some cases coincide with significantly increased production of polysaccharides, but this was not a consistent relationship and might be a result of other more complex regulatory mechanisms. It is suggested that various antibiotics differentially impact the delicate balance of eDNA release and nuclease production. Unraveling the underlying mechanisms, however, was not the primary objective of this study, rather investigating the effect of antibiotics on nuclease and biofilm formation as it has clinical relevance by various reasons. First sub-MIC antibiotic pressure develops around biomaterial implants during antibiotic treatment. Second, modulation of MN production of S. aureus may result in clinically relevant strategies of preventing and treating S. aureus infections by arresting biofilm dispersal. Finally, S. aureus bacteria possess escape mechanisms from the host immune system in particular by MN that disassembles neutrophil extracellular traps, rendering the bacteria unaffected by this particular defense mechanism of the immune system41, 42. Reduction of MN by sub-MIC antimicrobials may thus assist in the eradication of biofilms by the host immune system, even in case of clinical resistance to these antimicrobials.
Materials and methods
Antimicrobials
The antimicrobials used are mentioned in Table 1 together with their working mechanism. They were purchased from Merck (Darmstadt, Germany). All antimicrobials were prepared from powder-form in ultrapure water and sterilized by filtering through a 0.22 μm pore filter according to the manufacturers protocol. Resveratrol was dissolved in DMSO (dimethylsulfoxide) to a concentration of 52 mg/ml before filtration. Due to the poor aqueous solubility of resveratrol no concentrations of resveratrol higher than 512 µg/ml in ultrapure water containing 1% DMSO were used. All cultures containing resveratrol including the control for these cultures without antibiotics, contain 1% DMSO.
Nuclease probe
In order to measure MN production a nuclease activatable fluorescence probe was applied16, 36. The probe (Merck) consists of a 11-thymine base oligonucleotide flanked by a fluorescein amidine fluorophore and both a ZEN and Black hole quencher (5′-/FAM/TTTTTTTTTTT/ZEN/BhQsp/-3′). Before use the lyophilized probe was diluted in 10 mM Tris–HCl and 10 mM CaCl2, pH 8.0 to a working stock concentration of 2 µM.
Bacterial strains, culturing and harvesting
Experiments were conducted with S. aureus ATCC12600 and a luminescent S. aureus Newman lux strain (AH2600) in which LuxABCDE genes and kanamycin resistance were transduced from Photorhabdus luminescens using bacteriophage 1143. The bioluminescence is the result of a LuxABCDE gene, regarded as a global marker for cellular activity. The S. aureus Newman lux Δnuc1 mutant strain (AH2627) was obtained by deletion of nuc1 using the Targetron Gene Knockout System (Merck)17. Both S. aureus Newman strains were constructed previously17, 36.
All media were prepared according to manufacturer’s protocol. S. aureus ATCC12600, S. aureus Newman lux and S. aureus Newman lux Δnuc1 were cultured from cryopreservative beads onto Tryptic Soy Broth Agar (TSA) (Oxoid, Basingstoke, UK). Kanamycin (200 μg/ml) was added to the agar plates, pre-, and main-cultures of the S. aureus Newman lux strains. After inoculation the agar plates were incubated for 24 h at 37 °C in ambient air.
A pre-culture was made by inoculating one colony in tryptic soy broth (TSB) (10 ml) (Oxoid) and cultured for 24 h at 37 °C, 150 RPM. The main culture was made by inoculating 40 ml TSB with 2 ml of the pre-culture and cultured for 16 h at 37 °C, 150 RPM.
The bacteria were harvested by centrifugation for 5 min at 10 °C, 5000g (Avanti J-E centrifuge, JLA-16.259 rotor, Beckman-Coulter) and subsequently washed three times with phosphate buffered saline (PBS). All cultures were sonicated three times for 10 s at 30 W on ice, to remove aggregates. The number of bacteria was determined from a 1:200 diluted sample in a Bürker-Türk counting chamber in order to establish the required concentration of bacteria to start the experiments.
Minimum inhibitory concentration and minimum biofilm inhibitory concentration
The MIC of all antimicrobials were determined by incubating all precultured strains for 24 h at 37 °C in a 1:1 serially diluted antimicrobial concentration under rotating conditions (150 RPM) starting from 512 µg/ml. To this end all wells but the first were filled with 100 μl of ultrapure water. Then an antimicrobial solution (200 μl) at double the final concentration (1024 µg/ml in this case) was added to the first well. Subsequently 100 μl was transferred from the first well to the next, and then from that one to the next etc. Then a suspension of double the final concentration of bacteria (2 × 105 cells/mL in this case) in double concentrated TSB was prepared, and 100 μl was added to all wells, resulting in all wells containing the final concentration of bacterial inoculum in TSB, with an antimicrobial gradient. Wells with sterile TSB were included as negative controls. After 24 h the plates were assessed for the lowest concentration that resulted in no visible growth (MIC).
To determine the minimum biofilm inhibitory concentration (MBIC), wells in a flat-bottom 96 wells plate (Greiner Bio-One) were filled with 200 μl of 5 × 108 bacteria/ml in TSB. Bacteria were allowed to adhere for 1 h at 37 °C under stationary conditions. Non-adhering bacteria were removed by washing three times with PBS before addition of an antimicrobial gradient as described above, followed by stationary incubation for 24 h at 37 °C. Biomass was then assessed by staining with crystal violet and spectral absorption. To assess the biomass, the biofilms were washed gently three times with PBS and dried at 60 °C to fix the biofilms. Biofilms were stained for 5 min with crystal violet (0.06%) and washed three times with demineralized water44. Subsequently, crystal violet was resolubilized in 30% acetic acid for at least 15 min. Finally, the crystal violet solution was diluted four times and the absorbance was measured at 590 nm in a Fluostar Optima plate reader (BMG labtech, Ortenberg, Germany). This device is able to measure absorbance up to a value of 4.5. Values measured across all experiments did not exceed 50% of this maximum. MBIC was defined as a ≥ 90% inhibition of biofilm formation45.
Effect of antimicrobials on planktonic cultures
All planktonic cultures were grown in a 24 (polystyrene) well plate (Greiner Bio-One, Frickenhausen, Germany) with a final volume of 1120 μl per well with decreasing antimicrobial concentrations, established by a 1:1 serial dilution ranging from 4 times to 1/16 times the MIC as described above with adjusted volumes. Wells with sterile TSB were included as negative controls. The final volume of 1120 μl for each well was chosen to preserve the volume-to-surface ratio across all experiments. 1% DMSO was added to the wells that served as a control for the strains grown with resveratrol. The concentration of bacteria at T = 0 was 1 × 105 bacteria/ml. All cultures were grown as described under MIC and MBIC.
For each of the concentrations of antimicrobials the number of CFU’s and the production of MN were determined in the same bacterial culture in order to allow the calculation of MN production per CFU.
Effect of antimicrobials on biofilm cultures
Biofilms were grown as described above. The differential concentrations of antibiotics were established by a 1:1 serial dilution that ranged from 1 time to 1/8 times MBIC. 1% of DMSO was added to the wells that served as a control for the strains grown with resveratrol. Wells with sterile TSB were included as negative controls. Separate biofilm cultures were prepared for measuring biofilm biomass, polysaccharide quantification, and CFU count/MN production. The former two were quantified from a single biofilm culture in order to allow the calculation of nuclease production per CFU. In order to relate biofilm formation at various antimicrobial concentrations, biofilms of all strains were grown simultaneously on the same day for all concentrations.
Determination of biomass and polysaccharides
To assess the biomass of the biofilm the same crystal violet procedure was followed as described for the determination of the MIC and MBIC. To quantify the polysaccharides 100 μl medium was carefully removed from each well containing a biofilm and replaced by 100 μl calcofluor white (Sigma Aldrich) solution (40 μg/ml) to yield a final concentration of 20 μg/ml calcofluor white per well which binds to polysaccharides44. After 1 min all wells were gently washed three times with PBS. Biofilms were resuspended by pipetting forcefully up and down and 5 min of sonication of the plate in a sonication bath. Fluorescence was measured in a Fluostar Optima plate reader (Excitation: 355 nm/Emission: 490 nm, setting: bottom).
Determination of CFU’s
All biofilms were resuspended by pipetting forcefully up and down and 5 min of sonication of the plate in a sonication bath. From the 24-h planktonic cultures a sample was taken directly from the suspension. The samples (20 μl) were serially tenfold diluted in PBS (180 μl). Three 10 μl aliquots of every dilution (10–107 times diluted) were put on a TSA plate and grown for 18 h at 37 °C. Then the number of colonies were counted, and the number of CFU per cm2 (biofilms) or per mL (planktonic) was calculated.
Nuclease activity
Bacterial suspensions of resuspended biofilm and planktonic cultures were obtained from the same cultures as from which the samples for CFU counting were taken. To measure nuclease activity samples were diluted 1000 times with 10 mM Tris–HCl, 10 mM CaCl2, pH 8.0 buffer. Twenty-five µl of diluted bacterial suspension was combined with 25 µl of probe working stock (2 µM) and 10 mM Tris–HCl, 10 mM CaCl2, pH 8.0 (150 µl), buffer in a 96 wells plate. As a negative control 1:1000 diluted sterile TSB was used instead of a sample from the culture. Fluorescence intensity was measured with a Fluostar Optima plate reader (Excitation: 490 nm; Emission: 520 nm) at 1-min intervals for 5 min at 37 °C. Nuclease activity was determined by the rate of fluorescence change per min. We used a known amount of purified MN (Merck) to calibrate the nuclease probe. The rate of fluorescence per min was shown to be linear with MN concentration (Fig. S5). One unit (U) is defined as the amount of enzyme required to release acid soluble oligonucleotides that produce an absorbance increase of O.D. 1.0 at 260 nm in 30 min at 37 °C, pH 8.846.
Bioluminescence
To quantify bioluminescence, plates containing the biofilm or planktonic cultures were placed in an IVIS Lumina 2 system (PerkinElmer, Waltham Massachusetts US) and imaged for one minute (excitation filter: blocked, emission filter: open). Data was analyzed using the LivingImages 4.7.2 software (PerkinElmer). Reported units are in photons per second leaving the entire volume of the well.
Statistical analysis
All data were analyzed using Graphpad Prism 9 (Graphpad, San Diego, United States). Difference between 2 groups was calculated using a T-test, while differences between more groups were calculated using an ANOVA where appropriate. All statistics concerning MN/CFU, biomass/CFU, and polysaccharides/CFU were based on log-normal values, as these values are lognormally distributed and its log-values fit a normal distribution (Fig. S3). All experiments were performed in triplicate on each of three separate days with different cultures. Reported values are average values over nine measurements, standard deviations, standard errors of the mean and statistical tests are based on the three average values obtained, one for each culture.
References
Busscher, H. J. et al. Biomaterial-associated infection: Locating the finish line in the race for the surface. Sci. Transl. Med. 4, 153rv10 (2012).
O’Toole, P., Maltenfort, M. G., Chen, A. F. & Parvizi, J. Projected increase in periprosthetic joint infections secondary to rise in diabetes and obesity. J. Arthropl. 31, 7–10 (2016).
Voigt, A., Shalaby, A. & Saba, S. Continued rise in rates of cardiovascular implantable electronic device infections in the United States: Temporal trends and causative insights. PACE Pacing Clin. Electrophysiol. 33, 414–419 (2010).
Conlon, B. P. Staphylococcus aureus chronic and relapsing infections: Evidence of a role for persister cells: An investigation of persister cells, their formation and their role in S. aureus disease. BioEssays 36, 991–996 (2014).
Flemming, H. C. et al. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575 (2016).
Moormeier, D. E., Bose, J. L., Horswill, A. R. & Bayles, K. W. Temporal and stochastic control of Staphylococcus aureus biofilm development. MBio 5, e01341-e1414 (2014).
Peterson, B. W., Van der Mei, H. C., Sjollema, J., Busscher, H. J. & Sharma, P. K. A distinguishable role of eDNA in the viscoelastic relaxation of biofilms. MBio 4, e00497-13 (2013).
Dengler, V., Foulston, L., DeFrancesco, A. S. & Losick, R. An electrostatic net model for the role of extracellular DNA in biofilm formation by Staphylococcus aureus. J. Bacteriol. 197, 3779–3787 (2015).
Arciola, C. R., An, Y. H., Campoccia, D., Donati, M. E. & Montanaro, L. Etiology of implant orthopedic infections: A survey on 1027 clinical isolates. Int. J. Artif. Organs 28, 1091–1100 (2005).
Hu, Y., Xie, Y., Tang, J. & Shi, X. Comparative expression analysis of two thermostable nuclease genes in Staphylococcus aureus. Foodborne Pathog. Dis. 9, 265–271 (2012).
Mann, E. E. et al. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS ONE 4, e5822 (2009).
Kavanaugh, J. S. et al. Identification of extracellular DNA-binding proteins in the biofilm matrix. MBio 10, 1–30 (2019).
Geiger, T., Goerke, C., Mainiero, M., Kraus, D. & Wolz, C. The virulence regulator sae of Staphylococcus aureus: Promoter activities and response to phagocytosis-related signals. J. Bacteriol. 190, 3419–3428 (2008).
Davis, A., Moore, I. B., Parker, D. S. & Taniuchi, H. Nuclease B. A possible precursor of nuclease A, an extracellular nuclease of Staphylococcus aureus. J. Biol. Chem. 252, 6544–6553 (1977).
Kiedrowski, M. R. et al. Staphylococcus aureus Nuc2 is a functional, surface-attached extracellular nuclease. PLoS One 9, e95574 (2014).
Rosman, C. W. K. et al. ex vivo tracer efficacy in optical imaging of Staphylococcus aureus nuclease activity. Sci. Rep. 8, 1305 (2018).
Kiedrowski, M. R. et al. Nuclease modulates biofilm formation in community-associated methicillin-resistant staphylococcus aureus. PLoS ONE 6, e26714 (2011).
Beenken, K. E., Spencer, H., Griffin, L. M. & Smeltzer, M. S. Impact of extracellular nuclease production on the biofilm phenotype of Staphylococcus aureus under In vitro and In vivo conditions. Infect. Immun. 80, 1634–1638 (2012).
Duan, J. et al. Subinhibitory concentrations of resveratrol reduce alpha-hemolysin production in Staphylococcus aureus isolates by downregulating saeRS. Emerg. Microbes Infect. 7, 136 (2018).
Hodille, E. et al. The role of antibiotics in modulating virulence in Staphylococcus aureus. Clin. Microbiol. Rev. 30, 887–917 (2017).
Kaplan, J. B. et al. Low levels of β-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. MBio 3, 2–9 (2012).
Mashruwala, A. A., Gries, C. M., Scherr, T. D., Kielian, T. & Boyd, J. M. SaeRS is responsive to cellular respiratory status and regulates Staphylococcus aureus. Infect. Immun. 85, 1–18 (2017).
Bisognano, C. et al. A RecA-LexA-dependent pathway mediates ciprofloxacin-induced fibronectin binding in Staphylococcus aureus. J. Biol. Chem. 279, 9064–9071 (2004).
Chopra, I. & Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260 (2001).
Ferrer, M. D. et al. Effect of antibiotics on biofilm inhibition and induction measured by real-time cell analysis. J. Appl. Microbiol. 122, 640–650 (2017).
Bergan, T. Pharmacokinetics of tissue penetration of antibiotics. Rev. Infect. Dis. 3, 45–66 (1981).
Jensen, L. K. et al. Suppurative inflammation and local tissue destruction reduce the penetration of cefuroxime to infected bone implant cavities. J. Comp. Pathol. 157, 308–316 (2017).
Cue, D. et al. SaeRS-dependent inhibition of biofilm formation in Staphylococcus aureus Newman. PLoS ONE 10, e0123027 (2015).
Nguyen, H. T. T., Nguyen, T. H. & Otto, M. The staphylococcal exopolysaccharide PIA—Biosynthesis and role in biofilm formation, colonization, and infection. Comput. Struct. Biotechnol. J. 18, 3324–3334 (2020).
Daghighi, S. et al. Influence of antibiotic pressure on bacterial bioluminescence, with emphasis on Staphylococcus aureus. Int. J. Antimicrob. Agents 46, 713–717 (2015).
The Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing CLSI supplement M100S (Clinical and Laboratory Standards Institute, 2016).
Forson, A. M., van der Mei, H. C. & Sjollema, J. Impact of solid surface hydrophobicity and micrococcal nuclease production on Staphylococcus aureus Newman biofilms. Sci. Rep. 10, 1–10 (2020).
Schäfer, D. et al. A point mutation in the sensor histidine kinase saeS of Staphylococcus aureus strain Newman alters the response to biocide exposure. J. Bacteriol. 191, 7306–7314 (2009).
Lovati, A. B., Bottagisio, M., Vecchi, E. De, Gallazzi, E. & Drago, L. Animal models of implant-related low-grade infections. A twenty-year review. Adv. Exp. Med. Biol. 971, 29–50 (2017).
Vidlak, D. & Kielian, T. Infectious dose dictates the host response during Staphylococcus aureus orthopedic-implant biofilm infection. Infect. Immun. 84, 1957–1965 (2016).
Hernandez, F. J. et al. Noninvasive imaging of Staphylococcus aureus infections with a nuclease-activated probe. Nat. Med. 20, 301–306 (2014).
Mainiero, M. et al. Differential target gene activation by the Staphylococcus aureus two-component system saeRS. J. Bacteriol. 192, 613–623 (2010).
Jeong, D. W. et al. Identification of the P3 promoter and distinct roles of the two promoters of the SaeRS two-component system in Staphylococcus aureus. J. Bacteriol. 193, 4672–4684 (2011).
Beenken, K. E. et al. Impact of the functional status of saeRS on in vivo phenotypes of Staphylococcus aureus sarA mutants. Mol. Microbiol. 92, 1299–1312 (2014).
Sjollema, J. et al. The potential for bio-optical imaging of biomaterial-associated infection in vivo. Biomaterials 31, 1984–1995 (2010).
Schilcher, K. et al. Increased neutrophil extracellular trap-mediated Staphylococcus aureus clearance through inhibition of nuclease activity by clindamycin and immunoglobulin. J. Infect. Dis. 210, 473–482 (2014).
Berends, E. T. M. et al. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J. Innate Immun. 2, 576–586 (2010).
Novick, R. P. Genetic systems in Staphylococci. Methods Enzymol. 204, 587–636 (1991).
Stiefel, P. et al. Is biofilm removal properly assessed? Comparison of different quantification methods in a 96-well plate system. Appl. Microbiol. Biotechnol. 100, 4135–4145 (2016).
Moskowitz, S. M., Foster, J. M., Emerson, J. & Burns, J. L. Clinically feasible biofilm susceptibility assay for isolates of Pseudomonas aeruginosa from patients with cystic fibrosis. J. Clin. Microbiol. 42, 1915–1922 (2004).
Heins, J. N., Suriano, J. R. & Anfinsen, B. Characterization of a nuclease produced by Staphylococcus aureus. J. Biol. Chem. 5, 1016–1020 (1967).
Author information
Authors and Affiliations
Contributions
H.C.v.d.M., J.S. and C.W.K.R. planned the study and wrote the manuscript. C.W.K.R. performed the experiments, analysed the data and prepared the figures. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rosman, C.W.K., van der Mei, H.C. & Sjollema, J. Influence of sub-inhibitory concentrations of antimicrobials on micrococcal nuclease and biofilm formation in Staphylococcus aureus. Sci Rep 11, 13241 (2021). https://doi.org/10.1038/s41598-021-92619-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-92619-9
This article is cited by
-
Strategy to combat biofilms: a focus on biofilm dispersal enzymes
npj Biofilms and Microbiomes (2023)
-
The biofilm matrix: multitasking in a shared space
Nature Reviews Microbiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.