Abstract
Barley yellow dwarf viruses (BYDVs) cause significant economic losses on barley, wheat, and oats worldwide. 17-kDa protein (17K) of BYDVs plays a key role in viral infection in plants, whereas the underlying regulation mechanism of 17K in virus infection remains elusive. In this study, we determined that 17K of BYDV-GAV, the most common species found in China in recent years, was involved in viral pathogenicity. To identify the host factors interacting with 17K, the full length coding sequence of 17K was cloned into pGBKT7 to generate the bait plasmid pGBKT7-17K. 114 positive clones were identified as possible host factors to interact with 17K through screening a tobacco cDNA library. Gene ontology enrichment analysis showed that they were classified into 35 functional groups, involving three main categories including biological processes (BP), cellular components (CC), and molecular functions (MF). Kyoto Encyclopedia of Genes and Genome (KEGG) analysis indicated the acquired genes were assigned to 49 KEGG pathways. The majority of these genes were involved in glyoxylate and dicarboxylate metabolism, carbon fixation in photosynthetic organisms, and glycolysis/gluconeogenesis. The interactions between 17K and the 27 proteins with well-documented annotations were verified by conducting yeast two-hybrid assays and 12 of the 27 proteins were verified to interact with 17K. To explore the putative function of the 12 proteins in BYDV-GAV infection, the subcellular localization and expression alterations in the presence of BYDV-GAV were monitored. The results showed that, under the condition of BYDV-GAV infection, RuBisCo, POR, and PPD5 were significantly up-regulated, whereas AEP and CAT1 were significantly down-regulated. Our findings provide insights into the 17K-mediated BYDV-GAV infection process.
Similar content being viewed by others
Introduction
Barley yellow dwarf viruses (BYDVs), a group of viral pathogens that belong to the genus Luteovirus or Polerovirus or are unassigned to a genus (family Luteoviridae), cause significant economic losses on barley, wheat, and oats worldwide1. BYDVs are transmitted by aphids in a persistent, circulative, and non-propagative manner and phloem-limited. BYDVs-diseased plants always exhibit typical disease symptoms such as yellowing of leaves and stunting. In China, BYDVs have been classified into four isolates (GPV, GAV, PAV, and RMV) based on the specificity of vectors and serological properties2,3,4,5 and BYDV-GAV is the most common species in recent years6.
The BYDV-GAV is about 5.7 kb in length and comprises seven open reading frames (ORFs), which encode P1, P2, P3, P3a, P4, P5, and P6, respectively. They are involved in viral replication (P1 and P2), virion assembly (P3, the coat protein [CP]), virus spread (P3, P3a, and P4), transmission by the aphid vector (P5), and suppression of RNA silencing (P4 and P6)2,7,8,9,10,11,12. Luteovirus P4 encodes a 17-kDa protein (17K) that functions in viral movement, suppression of host RNA silencing, and viral pathogenesis8,12,13. For instance, BYDV-GAV 17K disrupts mitosis by interrupting the function of Wee1-Cdc25-CDKA/Cdc2 via direct protein–protein interactions12. But, how 17K assists virus movement, suppresses host antiviral RNA silencing and promotes virus infection remains largely unknown.
In this study, we determined the function of 17K in viral pathogenicity. We identified the host factors interacting with 17K by screening a tobacco cDNA library, and the interactions between 12 host factors and 17K were confirmed by Y2H assays. We further detected the subcellular localization of the interacting host factors and the expression of the interacting host factors after BYDV-GAV infection. Our study may help to comprehensively understand the pathogenesis of BYDV-GAV infection.
Results
17K enhances viral pathogenicity in a heterologous virus expression system
To test whether 17K is associated with viral pathogenicity, the protein was expressed in Nicotiana benthamiana (N. benthamiana) using a PVX-based vector. At 5 days post-infiltration (dpi), the systemic leaves of PVX17K plants showed obvious mosaic symptoms, whereas those treated with PVX showed no symptoms (Fig. 1A). PVX accumulation in PVX17K plants at 5 dpi was indicated by performing Western blotting with PVX CP antibodies, whereas no signal was observed in the PVX control (Figs. 1B, S1). These results suggest that 17K enhances viral pathogenicity of PVX. Additionally, silencing suppressor activity of 17K was tested using a N. benthamiana 16c system, and strong fluorescence was observed at 3 dpi in the combinations GFP + P19 and GFP + 17K, indicating silencing suppressor activity of 17K (Fig. S2).
17K promoted PVX infection in N. benthamiana. (A) Disease symptoms exhibited in N. benthamiana leaves inoculated with PVX or PVX17K. (B) PVX accumulation in the PVX- or PVX17K-inoculated N. benthamiana leaves were detected by conducting Western blotting analysis with anti-CP antibodies. Coomassie Brilliant Blue (CBB) staining of the large subunit of RuBisCo served as loading controls.
Subcellular localization pattern of BYDV-GAV 17K
Using PortII (https://psort.hgc.jp/form2.html) and seqNLS (http://mleg.cse.sc.edu/seqNLS/), 17K was predicted to be localized in the nucleus and would thus show a nuclear localization signal (Fig. 2A). To illustrate the subcellular localization of BYDV-GAV 17K, 17K-GFP was measured in epidermal cells of N. benthamiana plants. The results showed that the green fluorescent signal of 17K-GFP merged with the red fluorescent signal of H2B-RFP, indicating the nuclear localization of BYDV-GAV 17K (Fig. 2B). Our finding is consistent with a previous study12. Notably, we found that 17K was monitored mainly at the nuclear envelope (Fig. 2B).
Subcellular localization of 17K in N. benthamiana epidermal cells. (A) Online analysis of the 17K nuclear localization signal using PortII (https://psort.hgc.jp/form2.html) and seqNLS (http://mleg.cse.sc.edu/seqNLS). The predicted nuclear localization signal sequence is colored in red and underlined. (B) Localization of 17K in N. benthamiana epidermal cells. GFP-tagged 17K (17K-GFP) was expressed in planta. Confocal images were taken at 2 dpi. Bars represent 20 μm and 5 μm, respectively.
Identification of 17K-interacting host factors by screening a tobacco library
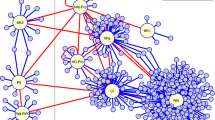
After screening, a total of 183 yeast clones were collected from the SD/-Trp/-Leu/-His/-Ade (QDO) agar plates. In total, 114 cDNA fragments were obtained by colony PCR amplification and sequentially sequenced. We used BLASTn search, gene ontology (GO), and the Kyoto Encyclopedia of Genes and Genome (KEGG) databases to identify the main functional groups of the acquired clones. Based on the results, a total of 90 genes were annotated. The full list of specific annotation results is shown in Supplementary Tables S1–S3. The identified genes were subjected to a GO enrichment analysis. The 90 acquired genes were classified into 35 functional groups, which belonged to three main categories: biological processes (BP), cellular components (CC), and molecular functions (MF). The main subcategories within BP were cellular processes, metabolic processes, and biological regulation. Within the MF category, catalytic activity and binding were predominant. The three main CC subcategories were cell, cell parts, and organelles (Fig. 3). In addition, 49 KEGG pathways for the acquired genes were identified (Table S3). The pathways with the highest numbers of screened genes were glyoxylate and dicarboxylate metabolism, carbon fixation in photosynthetic organisms, and glycolysis/gluconeogenesis (Fig. 4). Based on the analysis, we found that most screened genes were associated with signal transduction (KO04066, KO04068, KO04011, KO04016, KO02020, KO04152, KO04371, KO04020, KO04022, KO04151, KO04014, and KO04150), amino acid metabolism (KO00220, KO00250, KO00260, KO00380, and KO00290), carbohydrate metabolism (KO00630, KO00010, KO00030, KO00051, KO00520, KO00500, KO00562, and KO00040), and energy metabolism (KO00710, KO00680, and KO00910).
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses for the 17K-interacting host genes14.
Genes involved in signal transduction
A subset of genes were related to signal transduction pathways, including the HIF-1 signaling pathway (KO04066), forkhead box O (FOXO) signaling pathway (KP04068), MAPK signaling pathway (KO04011 and KO04016), two-component pathway (KO02020), AMPK signaling pathway (KO04152), Apelin signaling pathway (KO04371), calcium signaling pathway (KO04020), cGMP-PKG signaling pathway (KO04022), PI3K-Akt signaling pathway (KO04151), Ras signaling pathway (KO04014), and mTOR signaling pathway (KO04150). In this study, four genes were associated with the MAPK signaling pathway (XM_016653320.1, XM_016590541.1, NM_001325412.1, and NM_001326196.1) (Table S4). FOXOs are key transcription factors (TFs) that protect cells from various stresses. In Caenorhabditis elegans, heat shock factor 1 and FOXO together promote a long life span15. In plants, the function of the FOXO signaling pathway remains unclear. In total, six genes were associated with the FOXO signaling pathway (XM_016653320.1, XM_016584004.1, XM_016633060.1, XM_016595156.1, NM_001325412.1, and NM_001326196.1) (Table S4).
Genes involved in stress responses
Infection by plant viruses induces the expression of a variety of genes that are usually regulated by TFs16. TFs are triggers for gene expression and play important roles throughout the lifetime of plants, especially in plant growth, development, and responses to abiotic and biotic stresses17. In this study, we identified four TFs after screening, including TF PosF21 (XM_016587072.1), nuclear TF Y subunit C-1-like (XM_009781056.1), zinc finger protein CONSTANS-LIKE 5-like (XM_016580159.1), and GATA TF 5-like (XM_016591336.1). PosF21 shows all the characteristics of a basic region/leucine zipper motif (bZIP) type of DNA-binding domain18. As one of the largest TFs, bZIP TFs play pivotal, life-long roles in plant growth17.
Genes involved in chloroplast-related functions
Photosynthesis, the most fundamental and complex physiological process in plants, played an important role in viral infection. In this study ten photosynthesis and chloroplast related proteins were identified through screening which are light-harvesting complex I chlorophyll a/b binding protein 3(XM_016643125.1), light-harvesting complex I chlorophyll a/b binding protein 4(XM_016594598.1), light-harvesting complex II chlorophyll a/b binding protein 1(XM_016632344.1), psbP domain-containing protein 5(XM_016648932.1), phosphomethylpyrimidine synthase (XM_016635286.1), glutamine synthetase(XM_016584731.1, XM_016625469.1), protochlorophyllide reductase-like(XM_016616425.1), fructose-bisphosphate aldolase 1(XM_016590854.1, XM_016650131.1), probable 1-deoxy-D-xylulose-5-phosphate synthase(NM_001325496.1), ribulose-phosphate 3-epimerase(XM_016659233.1), glyceraldehyde-3-phosphate dehydrogenase B(XM_016624443.1), PTST(XM_016593025.1), and ribulose bisphosphate carboxylase small chain S41(XM_016613904.1) (Table S5). Among them light-harvesting complex II chlorophyll a/b binding protein, psbP domain-containing protein 5, and glutamine synthetase were previously found to be markedly regulated by BYDV-GAV infection19.
Verification of the interactions between the identified host proteins and BYDV-GAV 17K by yeast two-hybrid assays
Based on the screened data, 27 clones involved in different pathways were selected for further research (Tables 1 and S6). We used Y2H method to verify the interactions between BYDV-GAV 17K and the 29 proteins. The cDNAs of the 27 proteins were individually amplified and constructed into the prey vector pGADT7 to generate pGADT7 fusion clones, followed by co-transformation with the pGBKT7-17K into a yeast strain Y2HGold and cultured on SD/-Leu/-Trp (DDO) and SD/-Leu/-Trp/-His/-Ade (QDO) culture media supplemented with X-α-Gal and Aba. Blue colonies were grown from yeast cell co-transformation of BD-17K in 12 of the 27 host proteins on the QDO/X-α-Gal and Aba culture media (Fig. 6). The 12 proteins are shown as follows: SRC2 homolog (SRC2), psbP domain-containing protein 5 (PPD5), catalase isozyme 1-like (CAT1), acidic-endochitinase P (AEP), 26S protease regulatory subunit 6B homolog, sucrose nonfermenting 1 (SNF1)-related protein kinase regulatory subunit beta-2-like (SnRK2), SNF1-related protein kinase regulatory subunit beta-1-like (SnRK1), nuclear TF Y subunit C-1-like (NF-YC1), protochlorophyllide reductase-like (POR), ribulose bisphosphate carboxylase small chain S41 (RuBisCo), zinc finger protein CONSTANS-LIKE 5-like (CO5), and BOI-related E3 ubiquitin-protein ligase 2 (BRG2) . Yeast cells co-transformed in pairs with the other 15 prey vectors, and pGBKT7-17K were able to grow on DDO media but showed no growth on QDO/X-α-Gal and Aba media (Fig. 5).
Verification of the interactions between 17K and 27 screened host factors. BD17K was co-transformed with each of 27 host proteins into Y2HGold yeast cells and plated on low-(SD/–Trp/–Leu) and high-(SD/–Trp/–Leu/–His/–Ade/Aba/X-α-Gal) stringency selection media. ADT + BDP53 and ADT + BDLam served as positive and negative controls, respectively.
Subcellular localization patterns of the identified 17K-interacting host factors
To determine the subcellular localization of the identified 17K-interacting host factors, we transiently expressed 6 proteins as fusions to the N-terminus of YFP in N. benthamiana leaves. As shown in Fig. 7, POR-YFP and CAT1-YFP showed fluorescence in both the cell periphery and the nucleus. SnRK1-YFP and SnRK2-YFP showed fluorescence in the cell periphery, whereas, BRG2-YFP was perhaps only localized in the nuclear and PPD5 showed the localization at the chloroplast (Figs. 6, S3). The various distributions of identified host proteins suggested 17K protein is possibly involved in several distinct cellular pathways.
Subcellular localization of six 17K-interacting host proteins in N. benthamiana epidermal cells. YFP-tagged POR (POR-YFP), YFP-tagged SnRK1 (SnRK1-YFP), YFP-tagged SnRK2 (SnRK-YFP), YFP-tagged CAT1 (CAT1-YFP), YFP-tagged PPD5 (PPD5-YFP), and YFP-tagged BRG2 (BRG2-YFP) were expressed in N. benthamiana leaves through agrobacterium-mediated transient expression system. Confocal images were taken at 2 dpi using a ZeissLSM710 laser scanning microscope. The bar represents 20 μm.
Expression of interacting proteins after BYDV infection
To determine the role of the identified host factors in viral infection, 10 interacting host genes were selected for quantitative RT-PCR (qRT-PCR) analysis using BYDV-inoculated N. benthamiana leaves. The results showed that RuBisCo, POR, and PPD5 were significantly up-regulated by viral infection, whereas AEP and CAT1 were significantly down-regulated (Fig. 7). PPD5 was previously reported to be up-regulated in BYDV-infected wheat leaves at 35 dpi, consistent with our results19.
Detection of differentially expressed 17K-interacting host factors under BYDV-GAV infection by qRT-PCR. The relative expression levels of 17K-interacting host factors in mock- and BYDV infected N. benthamiana leaves at 5 dpi using qRT-PCR. Data were pooled across experiments and analyzed using t-tests. Bars represent the grand means ± standard deviation (SD). *, P < 0.05; **, P < 0.01.
Discussion
Infection by BYDVs causes leaf yellowing and plant dwarfism in wheat, leading to significant economic losses. To realize successful infection, viruses always have to manipulate or utilize many host factors through directly interacting with them. Multifunctional viral proteins such as γb encoded by Barley stripe mosaic virus (BSMV) which are suppressor of RNA silencing and also affects symptom development and seed transmission of BSMV. Previous studies have shown that multiple host factors interact with γb and affects the roles that γb plays during viral infection20,21,22. 17K protein is also a multifunctional protein and its roles in suppression of RNA silencing, virus movement and symptom development are unclear. Identification of interacting host factors will provide insights into the 17 mediated viral process. In this study, we identified 12 host proteins that interact with BYDV 17K. We also tested the possible interaction between 17K and the other BYDV encoded proteins. No interaction was found using Y2H assay.
Reactive oxygen species (ROS) such as hydrogen peroxide (H2O2) or superoxide anions (O2 −) are one of the earliest cellular responses against pathogen infections20. Two identified host factors, NbSRC2 and NbCAT1, were involved in ROS production. Arabidopsis SRC2 acts as a novel activator of NADPH oxidase AtRbohF-mediated ROS production and may play a role in cold stress23. The rapid production of ROS under biotic or abiotic challenges, known as an oxidative response, is an important signal for both local immune responses and cell-to-cell communication24. Catalase serves to protect cells from the toxic effects of hydrogen peroxide. The interaction between 17K and NbSRC2/ NbCAT1 may influence viral infection by regulating ROS production.
Two chloroplast proteins were identified: PsbP domain-containing protein 5 and ribulose bisphosphate carboxylase small-chain S41. Plants contain an extensive family of PsbP domain (PPD) proteins, which are localized in the thylakoid lumen. Members of the PsbP family have been shown to exhibit a variety of functions. Except for the function of photosynthetic electron transfer, the Arabidopsis ppd5 mutant showed striking morphological defects related to a deficiency in strigolactone biosynthesis25. RuBisCo is the most abundant enzyme in plants. It catalyzes the carboxylation of ribulose-1,5-biphosphate in chloroplasts and thus is responsible for fixing CO2 during photosynthesis. Hence, 17K may be involved in different pathways in chloroplasts and thus influence symptom development.
TFs are important players in the response to biotic and abiotic stresses. In this study, two TFs were identified: nuclear TF Y subunit C-1-like (NF-YC1) and zinc finger protein CONSTANS-LIKE 5-like (CO5). Nuclear factor Y (NF-Y) is an evolutionarily conserved trimeric TF complex consisting of three subunits: NF-YA, NF-YB, and NF-YC. Recent studies have shown that the NF-Y complex plays multiple essential roles in plant growth, development, and stress responses26. Three NF-YC proteins (NF-YC3, NF-YC4, and NF-YC9) are positive regulators of photomorphogenesis27. CONSTANS/CONSTANS‐like (CO/COL) belongs to a family of zinc finger TFs and contains one or two B‐box zinc finger regions at the N‐terminus and a CCT (CO, COL, TOC1) domain at the C-terminus28. The CO genes have been reported to be involved in many molecular and genetic processes, including photoperiodically regulated developmental processes and abiotic stresses29,30,31. The function of NF-YC1 and CO5 in biotic stress remains unclear. The interaction between 17K and the two TFs may influence the regulation of gene expression by NF-YC1 and CO5, thereby influencing viral accumulation.
Three biotic stress-related genes that interact with 17K were identified, including SNF1-related protein kinase regulatory subunit beta-1-like, SNF1-related protein kinase regulatory subunit beta-2-like, and BOI-related E3 ubiquitin-protein ligase 2.SNF1-related protein kinase 1 (SnRK1) plays a central role in regulating energy and metabolism in plants and has been implicated in responses to abiotic and biotic stresses32,33,34,35. It is the best-characterized host protein kinase known to be involved in geminivirus infection34,36,37. SnRK1 phosphorylation of Rep interferes with viral replication. By contrast, the host defense response is enhanced by SnRK1 phosphorylation of AL2/C237 and β-satellite-encoded βC1 protein36,38. Protochlorophyllide reductase is a key enzyme in chlorophyll biosynthesis. BOI-related E3 ubiquitin-protein ligases represent a subclass of RING E3 ligases that contribute to plant disease resistance and abiotic stress tolerance through the suppression of pathogen-induced and stress-induced cell death39.
The screening and identification of 17K interacting host factor enhances the understanding of the molecular mechanism of BYDV-GAV infection. Since BYDV has different strains, our study will increase the knowledge on the mechanism of infection by BYDV-GAV and other strains.
Materials and methods
Plasmid construction
Primers used for plasmid construction are listed in Table S7. All the available constructs were sequenced.
To construct vectors for yeast two-hybrid analysis, the coding sequence of corresponding genes were amplified and inserted into EcoRI/BamHI digested pGADT7 vectors via homologous recombination.
PVX17K was constructed by introducing 17K into Potato virus X (PVX) vector pGR106 via ClaI and SalI digestion, followed by ligation with T4 DNA ligase (NEB). The fragments used were amplified using primer pairs PVX17KF/PVX17KR.
YFP fusion constructs were constructed using gateway strategy. The corresponding genes were cloned into entry vector pDONR221 (Invitrogen) using primer pairs to generate recombination vector. The resultant clone was used to construct the gateway vector 17K-YFP.
Yeast two hybrid assays
The N. tabacum cDNA library was screened according to the protocol handbook provided by the Matchmaker Gold Yeast Two-Hybrid System (Clontech Laboratories, Mountain View, CA, USA). The full-length BYDV-GAV 17K protein was amplified and cloned into yeast vector pGBKT7 to generate the bait vector BD17K. The cDNA library screening and interaction assay were performed as described previously40.
Plant materials and virus inoculation
N. benthamiana plants were grown in pots in a growth room under a 16 h light/8 h dark photoperiod at 25 °C with 60% humidity. For agroinfiltration, Agrobacteria strain GV3101 carrying infectious viral clones were suspended in infiltration buffer (10 mM MgCl2, 10 mM MES, and 200 μM acetosyringone, pH 5.6) at an OD600 of 1, kept at room temperature for 2 to 4 h and infiltrated into N. benthamiana leaves using a 1-mL needleless syringe.
Confocal laser scanning microscopy
For the subcellular localization assays, the corresponding constructs were infiltrated into N. benthamiana leaves as described previously41. The leaves were detached 48 h post-infiltration (hpi) for fluorescence detection. Fluorescence signals were visualized under an inverted spectral confocal laser scanning microscope (LSM 710; Carl Zeiss AG, Oberkochen, Germany). The fluorescence of yellow (YFP) and red (RFP) fluorescent proteins was excited at 514 and 543 nm, respectively.
Western blotting analysis
Agro-infiltrated leaves were harvested for western blotting assay. Total protein was extracted from 0.2 g leavesusing the extraction buffer containing 20% glycerol, 20 mM Tris–HCl (pH 7.5), 1 mM EDTA, 150 mM NaCl, 1 mM PMSF, 1 × Protease inhibitor cocktail (Sigma, China). Total protein was separated in SDS–polyacrylamide gel electrophoresis, followed by transfer to nitrocellulose membranes. The membranes were probed using anti-PVX CP polyclonal antibodies followed by an HRP-conjugated secondary antibody. The detection signals were developed using an ECL reagent as instructed. PVX CP accumulation were photographed under a chemiluminescence apparatus (Amersham imager 680). CBB staining of the large subunit of RuBisCo served as a loading controls.
Quantitative RT-PCR
Total RNAs were extracted from harvested N. benthamiana protoplasts using Trizol reagent (invitrogen) and treated with RNase-free DNase I. First strand cDNA was synthesized using 1 μg total RNA(Promega). The reaction solution was prepared as follows: an oligo d(T) primer, random primer, and M-MLV reverse transcriptase as instructed. Ten-fold diluted cDNA products were used for qRT-PCR on an Eppendorf Real-Time PCR system using a SYBR Green master mix (Takara). The Nb-actin gene were used as internal reference. All the primers used for RT-PCR are listed in Table S7. The relative gene expression levels were calculated using the 2−△△CT method.
Statement
The study in the manuscript “Identification and functional analyses of host factors interacting with the 17-kDa protein of Barley yellow dwarf virus-GAV” is complied with local and national regulations.
References
Miller, W. A., Liu, S. & Beckett, R. Barley yellow dwarf virus: luteoviridae or tombusviridae?. Mol. Plant Pathol. 3, 177–183. https://doi.org/10.1046/j.1364-3703.2002.00112.x (2002).
Jin, Z. The complete nucleotide sequence and its organization of the genome of Barley yellow dwarf virus-GAV. Sci. China Ser. C 47, 175. https://doi.org/10.1360/03yc0076 (2004).
Liu, F. et al. A Chinese isolate of barley yellow dwarf virus-PAV represents a third distinct species within the PAV serotype. Arch. Virol. 152, 1365–1373. https://doi.org/10.1007/s00705-007-0947-8 (2007).
Wu, B. et al. Dynamics of molecular evolution and phylogeography of Barley yellow dwarf virus-PAV. PLoS ONE 6, e16896. https://doi.org/10.1371/journal.pone.0016896 (2011).
Zhang, W. et al. The complete nucleotide sequence of the barley yellow dwarf GPV isolate from China shows that it is a new member of the genus Polerovirus. Arch. Virol. 154, 1125–1128. https://doi.org/10.1007/s00705-009-0415-8 (2009).
Liu, Y., Sun, B., Wang, X., Zheng, C. & Zhou, G. Three digoxigenin-labeled cDNA probes for specific detection of the natural population of Barley yellow dwarf viruses in China by dot-blot hybridization. J. Virol. Methods 145, 22–29. https://doi.org/10.1016/j.jviromet.2007.05.006 (2007).
Ali, M., Hameed, S. & Tahir, M. Luteovirus: insights into pathogenicity. Arch. Virol. 159, 2853–2860. https://doi.org/10.1007/s00705-014-2172-6 (2014).
Fusaro, A. F. et al. The Luteovirus P4 Movement Protein Is a Suppressor of Systemic RNA Silencing. Viruses https://doi.org/10.3390/v9100294 (2017).
Gray, S. & Gildow, F. E. Luteovirus-aphid interactions. Annu. Rev. Phytopathol. 41, 539–566. https://doi.org/10.1146/annurev.phyto.41.012203.105815 (2003).
Miller, W. A. & Rasochová, L. Barley yellow dwarf viruses. Annu. Rev. Phytopathol. 35, 167–190. https://doi.org/10.1146/annurev.phyto.35.1.167 (1997).
Smirnova, E. et al. Discovery of a small non-AUG-Initiated ORF in poleroviruses and luteoviruses that is required for long-distance movement. PLoS Pathog. 11, e1004868. https://doi.org/10.1371/journal.ppat.1004868 (2015).
Jin, H. et al. A distinct class of plant and animal viral proteins that disrupt mitosis by directly interrupting the mitotic entry switch Wee1-Cdc25-Cdk1. Sci Adv 6, eaba3418. https://doi.org/10.1126/sciadv.aba3418 (2020).
Chay, C. A., Gunasinge, U. B., Dinesh-Kumar, S. P., Miller, W. A. & Gray, S. M. Aphid transmission and systemic plant infection determinants of barley yellow dwarf luteovirus-PAV are contained in the coat protein readthrough domain and 17-kDa protein, respectively. Virology 219, 57–65. https://doi.org/10.1006/viro.1996.0222 (1996).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucl. Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Son, H. G. et al. Prefoldin 6 mediates longevity response from heat shock factor 1 to FOXO in C. elegans. Genes Dev. 32, 1562–1575. https://doi.org/10.1101/gad.317362.118 (2018).
Nakashima, K., Ito, Y. & Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 149, 88–95. https://doi.org/10.1104/pp.108.129791 (2009).
Liu, D., Shi, S., Hao, Z., Xiong, W. & Luo, M. OsbZIP81, a homologue of arabidopsis VIP1, may positively regulate JA levels by directly targetting the genes in JA signaling and metabolism pathway in rice. Int J Mol Sci https://doi.org/10.3390/ijms20092360 (2019).
Aeschbacher, R. A., Schrott, M., Potrykus, I. & Saul, M. W. Isolation and molecular characterization of PosF21, an Arabidopsis thaliana gene which shows characteristics of a b-Zip class transcription factor. Plant J. 1, 303–316 (1991).
Rong, W., Wang, X., Massart, S. & Zhang, Z. Molecular and ultrastructural mechanisms underlying yellow dwarf symptom formation in wheat after infection of barley yellow dwarf virus. Int. J. Mol. Sci. https://doi.org/10.3390/ijms19041187 (2018).
Yang, M. et al. Barley stripe Mosaic virus γb Interacts with glycolate oxidase and inhibits peroxisomal ROS production to facilitate virus infection. Mol. Plant 11, 338–341. https://doi.org/10.1016/j.molp.2017.10.007 (2018).
Yang, M. et al. Barley stripe mosaic virus γb protein subverts autophagy to promote viral infection by disrupting the ATG7-ATG8 interaction. Plant Cell 30, 1582–1595. https://doi.org/10.1105/tpc.18.00122 (2018).
Zhang, X. et al. Barley stripe mosaic virus infection requires PKA-mediated phosphorylation of γb for suppression of both RNA silencing and the host cell death response. New Phytol. 218, 1570–1585. https://doi.org/10.1111/nph.15065 (2018).
Kawarazaki, T. et al. A low temperature-inducible protein AtSRC2 enhances the ROS-producing activity of NADPH oxidase AtRbohF. Biochim. Biophys. Acta 1833, 2775–2780. https://doi.org/10.1016/j.bbamcr.2013.06.024 (2013).
Marcec, M. J., Gilroy, S., Poovaiah, B. W. & Tanaka, K. Mutual interplay of Ca(2+) and ROS signaling in plant immune response. Plant Sci. 283, 343–354. https://doi.org/10.1016/j.plantsci.2019.03.004 (2019).
Roose, J. L., Frankel, L. K. & Bricker, T. M. Developmental defects in mutants of the PsbP domain protein 5 in Arabidopsis thaliana. PLoS ONE 6, e28624. https://doi.org/10.1371/journal.pone.0028624 (2011).
Zhao, H. et al. The Arabidopsis thaliana nuclear factor Y transcription factors. Front. Plant Sci. 7, 2045. https://doi.org/10.3389/fpls.2016.02045 (2016).
Myers, Z. A. et al. NUCLEAR FACTOR Y, subunit C (NF-YC) Transcription factors are positive regulators of photomorphogenesis in Arabidopsis thaliana. PLoS Genet. 12, e1006333. https://doi.org/10.1371/journal.pgen.1006333 (2016).
Robson, F. et al. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 28, 619–631. https://doi.org/10.1046/j.1365-313x.2001.01163.x (2001).
Min, J. H., Chung, J. S., Lee, K. H. & Kim, C. S. The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J. Integr. Plant Biol. 57, 313–324. https://doi.org/10.1111/jipb.12246 (2015).
Zobell, O., Coupland, G. & Reiss, B. The family of CONSTANS-like genes in Physcomitrella patens. Plant. Biol. (Stuttg) 7, 266–275. https://doi.org/10.1055/s-2005-865621 (2005).
Liu, H. et al. CONSTANS-like 9 (OsCOL9) interacts with receptor for activated C-Kinase 1(OsRACK1) to regulate blast resistance through salicylic acid and ethylene signaling pathways. PLoS ONE 11, e0166249. https://doi.org/10.1371/journal.pone.0166249 (2016).
Baena-González, E., Rolland, F., Thevelein, J. M. & Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942. https://doi.org/10.1038/nature06069 (2007).
Broeckx, T., Hulsmans, S. & Rolland, F. The plant energy sensor: evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J. Exp. Bot. 67, 6215–6252. https://doi.org/10.1093/jxb/erw416 (2016).
Hulsmans, S., Rodriguez, M., De Coninck, B. & Rolland, F. The SnRK1 energy sensor in plant biotic interactions. Trends Plant Sci. 21, 648–661. https://doi.org/10.1016/j.tplants.2016.04.008 (2016).
Polge, C. & Thomas, M. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control?. Trends Plant Sci. 12, 20–28. https://doi.org/10.1016/j.tplants.2006.11.005 (2007).
Shen, Q. et al. Tomato SlSnRK1 protein interacts with and phosphorylates βC1, a pathogenesis protein encoded by a geminivirus β-satellite. Plant Physiol. 157, 1394–1406. https://doi.org/10.1104/pp.111.184648 (2011).
Shen, W., Dallas, M. B., Goshe, M. B. & Hanley-Bowdoin, L. SnRK1 phosphorylation of AL2 delays Cabbage leaf curl virus infection in Arabidopsis. J. Virol. 88, 10598–10612. https://doi.org/10.1128/jvi.00761-14 (2014).
Zhong, X. et al. Mimic phosphorylation of a βC1 protein encoded by TYLCCNB impairs its functions as a viral suppressor of RNA silencing and a symptom determinant. J. Virol. https://doi.org/10.1128/jvi.00300-17 (2017).
Luo, H. et al. The Arabidopsis Botrytis Susceptible1 Interactor defines a subclass of RING E3 ligases that regulate pathogen and stress responses. Plant Physiol. 154, 1766–1782. https://doi.org/10.1104/pp.110.163915 (2010).
Cheng, Y. Q. et al. HC-Pro protein of sugar cane mosaic virus interacts specifically with maize ferredoxin-5 in vitro and in planta. J. Gen. Virol. 89, 2046–2054. https://doi.org/10.1099/vir.0.2008/001271-0 (2008).
Walter, M. et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. https://doi.org/10.1111/j.1365-313X.2004.02219.x (2004).
Acknowledgements
This work was funded by the Henan Science Fund for Excellent Young Scholars (Grant Number 202300410194) and the National Key Research and Development Program of China (2018YFD0200602).We’d like to thank Dr Chao Zhang for the critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
Y.S. designed the experiment. S.Y.C., X.Y.H., L.L.Y., Q.L.L., and Y.J.S. performed experiments. H.L.L. critically reviewed the manuscript, L.L.C., and B.J.S. contributed to the data discussion. Y.S. and X.Y. wrote the paper. All authors have read and approved to publish the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, S., Han, X., Yang, L. et al. Identification and functional analyses of host factors interacting with the 17-kDa protein of Barley yellow dwarf virus-GAV. Sci Rep 11, 8453 (2021). https://doi.org/10.1038/s41598-021-87836-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-87836-1
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.