Abstract
Bacterial cancer therapy was developed using probiotic Escherichia coli Nissle 1917 (EcN) for medical intervention of colorectal cancer. EcN was armed with HlyE, a small cytotoxic protein, under the control of the araBAD promoter (PBAD). The intrinsic limitation of PBAD for the gene expression is known to be negated by glucose and afflicted with all-or-nothing induction in host bacteria. This issue was addressed by metabolic engineering of EcN to uncouple the glucose-mediated control circuit and the L-arabinose transport-induction loop and to block L-arabinose catabolism. As a result, the reprogrammed strain (designated EcNe) enabled efficient expression of HlyE in a temporal control manner. The HlyE production was insensitive to glucose and reached a saturated level in response to L-arabinose at 30–50 μM. Moreover, the administrated EcNe exhibited tumor-specific colonization with the tumor-to-organ ratio of 106:1. Equipped with HlyE, EcNe significantly caused tumor regression in mice xenografted with human colorectal cancer cells. Overall, this study proposes a new strategy for the bacteria-mediated delivery of therapeutic proteins to tumors.
Similar content being viewed by others
Introduction
Tumor tissues are recalcitrant to the treatment of traditional chemotherapy and radiotherapy1. The emerging technology known as targeted therapy facilitates selective delivery of chemotherapeutic agents involving therapeutic genes and drugs to tumor sites, which enhances the therapeutic efficacy and reduces side effects2. However, this approach is usually applied for proliferating cells and the therapeutic cargos have a difficulty in diffusing deep into tumor tissues3. The occurrence of angiogenesis in tumors develops irregular and poorly-organized blood vessels. This in turn renders the supply of oxygen in tumor vasculature insufficient, consequently leading to formation of hypoxia or anoxia regions in malignant tumors. Hypoxic cells are generally non-proliferating and exhibit high resistance to radiotherapy4. Apparently, there is a pressing need to rationally design delivery systems for tumor therapy.
The microenvironment surrounding tumor tissues provides a favorable niche for bacteria to inhabit. Bacteria including Bifidobacterium5, Clostridium6, Salmonella7, and Escherichia8 have been illustrated to preferentially colonize in tumors after being administrated in mice. Following bloodstream clearance mediated by inflammation, bacteria are generally entrapped in the tumor vasculature. Obligate anaerobes such as Bifidobacterium and Clostridium survive in the anoxic region. In addition, the presence of available nutrients in necrotic tumor tissues attracts facultative anaerobes like Salmonella and Escherichia to the cancerous site via chemotaxis. Consequently, they thrived in the hypoxic/necrotic regions of tumors to evade clearance by the immune system. Bacterial therapy is not new9, and its implementation for tumor treatment has been recently acknowledged by the advent of synthetic biology. In general, the tumor-seeking bacteria are tailored to synthesize a variety of therapeutic agents12. By administration locally or systemically, the engineered bacteria target tumors where they reside, replicate, and continuously produce the payloads on site. It enables in situ delivery of the produced bioactive molecules to tumor site, which improves the therapeutic efficacy.
The tumor-targeting bacteria have been genetically instructed to deliver a variety of bioactive payloads, notably involving prodrugs-converted enzymes10, short hairpin RNA11, cytokines12, antigens13, antibodies14, and bacterial toxins15. These approaches generally show encouraging results. Nevertheless, they have intrinsic limitations that most of the produced payloads are restricted to proliferating cells or/and afflicted with tumor penetration. Hemolysin appears to be a promising protein payload. It is naturally produced in bacteria and displays a pore-forming activity that lyses mammalian erythrocytes16. As illustrated previously, Staphylococcus aureus α-hemolysin (SAH) was expressed in E. coli17. Recombinant SAH was shown to penetrate into tumor tissue and eradicate cancer cells. As a result, in situ delivery of SAH by E. coli reduced the volume of MCF7 tumor by 41%. Like SAH, hemolysin E (HlyE) is a pore-forming protein which naturally appears in E. coli18, S. enterica19, and Shigella flexneri20. HlyE is cytotoxic to cultured mammalian cells and macrophages21. It causes the formation of transmembrane pore on the host cell. The damaged cell membrane in turn induces cell apoptosis. The application of the HlyE-mediated cell lysis for cancer treatment was investigated in the later work. As first exemplified by 4T1 tumor, the administration of HlyE-expressing S. typhimurium significantly decreased the tumor volume15.
Colorectal cancer ranks the third most common malignant tumor and is marked with a low 5-year survival rate. The number of patients who newly contracted this disease accounts for almost 10% of new cancer cases worldwide22. It appears necessary to further explore a potential method for medical intervention of colorectal cancer. Probiotic bacteria have emerged as the most promising chassis for living therapeutics23. E. coli Nissle 1917 (EcN) is a probiotic strain free of enterotoxins and cytotoxins and is used for the conventional treatment of various gastrointestinal illnesses24. EcN that produces the therapeutic proteins has been illustrated for cancer therapy in the murine tumor model. Azurin is a cytotoxic protein which induces cancer cell apoptosis. The administration of azurin-producing EcN suppressed the growth of B16 melanoma and 4T1 breast cancer while prolonged the survival of tumor-bearing mice25. EcN with azurin also enabled to restrain pulmonary metastasis developed by 4T1 cancer cells. EcN has innate prodrug-converting enzymes. By intratumoural injection, EcN caused a significant reduction in tumor growth and an increase in survival of the CT26 colon cell-bearing mice after prodrugs were administrated26. In the human tumor model, tumor growth was suppressed by engineered EcN which produced cytotoxic compounds27. In this study, the issue was addressed by development of bacterial cancer therapy (BCT) based on HlyE-producing EcN. To approach the goal, HlyE was expressed under the control of the araBAD promoter (PBAD). The strategy of metabolic engineering was applied to EcN for the temporal and spatial control of the HlyE expression. As a result, the engineered EcN preferred colonization in tumor tissues and expressed HlyE that effectively caused tumor regression in mice xenografted with human colorectal cancer cells.
Materials and methods
Bacterial strains and plasmids
The bacterial strain, EcNe (ΔaraBAD ΔaraFGH ΔptsG PEM7::araE lacZ::PBAD-T7 gene 1), was derived from EcN and applied for this study. It was constructed in several steps as follows. First, endogenous genes of EcN were knocked out according to the previous study28. In brief, the passenger DNA contained FRT-kan-FRT flanked by two homologous regions of the araBAD operon and was obtained by PCR with primers (5′-atggcgattgcaattggcctc and 5′-ttactgcccttaatatgccttc). By electroporation, the PCR DNA was introduced into EcN which carried helper plasmid pKD46 for expression of λ-Red recombinase. It resulted in the replacement of the araBAD operon with the FRT-kan-FRT via homologous recombination. The inserted kan marker was later removed by Flp using helper plasmid pCP2029. In a similar way, the araFGH operon was knocked out with the passenger DNA which was obtained by PCR with primers (5′-atgcacaaatttactaagc and 5′-tcagacagtgcgtttcg). P1 transduction was applied to remove ptsG, and transducing phages were prepared from JW1087-2 (△ptsG::FRT-kan-FRT) stain. Secondly, the araE expression was driven by the constitutive EM7 promoter (PEM7). This was carried out by the PCR-amplification of the FRT-kan-FRT-PEM7 fragment from plasmid pKD-EM7 using primers 5′-gtgaggaactaaaaatcgctcctggcaggaaaaaatggttac and 5′-ggtcgacggatccccggaatatagtg-aaaaaatacgtgaac). With the act of λ-Red recombinase, PEM7 was substituted for the native promoter of araE after electroporation of the FRT-kan-FRT-PEM7 fragment. Finally, the bacterial strain was equipped with a genomic copy of T7 gene 1 (encoding T7 RNA polymerase) under the control of PBAD.
Plasmid pBAD33-hlyE contained the hlyE gene of E. coli strain MG1655 under the control of PBAD and was constructed as follows. By PCR, the hlyE DNA was amplified with primers (5′-ttatttctagatgtaaaacaggagtttcattac and 5′-gcttgaagcttctcga-gaataatcgcttatc). The PCR DNA was treated with the XbaI-HindIII digestion and incorporated into plasmid pBAD33. In addition, EcNe which carried a variant of red fluorescent protein (DsRed) under the control of the T7 promoter (PT7) was constructed in the following. The PCR-amplification of a DNA containing DsRed fused to PT7 (PT7-DsRed) was carried out from plasmid pRed-Coh with primers (5′-actaggatcccgcgaaattaatacgac and 5′-ctcctgctagcaaaaaacccctcaagacc). By digestion with BamHI and NheI, the PCR DNA and integration plasmid pLam-LoxKm30 were spliced together to obtain plasmid pLam-Red. PT7-DsRed was integrated into EcNe genome at the prophage λ attachment site by plasmid pLam-Red.
Production of HlyE
To produce HlyE, EcNe was transformed with plasmid pBAD33-hlyE. A single colony was picked and inoculated into a shake flask containing LB medium31. The bacterial growth was conducted at 37 °C and measured turbidimetrically at OD550. In the next day, the overnight-grown bacteria were seeded into a fresh LB medium and induced for the protein production with 30 μM L-arabinose upon OD550 reaching 0.3. After induction for 4 h, bacteria were harvested by centrifugation and then resuspended in 10 mM sodium phosphate buffer (PBS) at pH 7.5. Bacteria were disrupted by sonication, followed by centrifugation to collect the supernatant part. Proteins were analyzed by sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie Blue staining and quantified using the Image Analyzer GA90000 (UVItech, England).
Real-time digital bio-imaging
Tumor cells were seeded into a 96-well plate (5 × 104 cells per well) and incubated with EcN, EcNe/HlyE, and HlyE upon reaching 80% confluence. The cell imaging was carried out by placing the plate in the multi-mode plate reader Cytation 5 (Biotek, Winooski, VT). The assay was performed at 37 °C in an atmosphere supplemented with 5% CO2. Images of each well were collected every 15 min over 1 h by using a 10X phase-contrast objective. Cell images were processed with GEN5 3.0 software (Biotek).
Analysis of nuclear fragmentation
Tumor cells were seeded into a 24-well plate with a 12 mm cover slide (1 × 105 cells per well) at 37 °C in an atmosphere supplemented with 5% CO2 overnight. After treatment with EcN or EcNe/HlyE, tumor cells were then washed twice with PBS and fixed with 2.5% paraformaldehyde for 20 min at room temperature. By washing with PBS, fixed cells were stained with 1 μg/ml diamidino-2-phenylindole (DAPI) at room temperature for 5 min. Slides were mounted with the mounting solution, followed by observing tumor cells with fluorescence microscope (Olympus IX71, Japan). Microphotographs were taken with Olympus camera.
Cell morphology
In a similar way, tumor cells in the plate were treated with EcN and EcNe/HlyE and stained with 2.5 mg/ml propidium iodide (PI). The analysis was conducted with the multi-mode plate reader Cytation 5 (Biotek, Winooski, VT). The assay was performed at 37 °C in an atmosphere supplemented with 5% CO2. Images of each well were collected every 15 min over 1 h by using a 10X phase-contrast objective. Cell images were processed with GEN5 3.0 software (Biotek).
In vitro assessment of cytotoxicity
Cell proliferation was measured using the methylthiazol tetrazolium (MTT) assay. Tumor cells in the plate were treated with EcN, EcNe/HlyE, or HlyE for 3 h and then washed twice with PBS. MTT (5 mg/ml) in PBS was added to tumor cells, followed by incubation for 4 h. In each well, the supernatant of cell culture (15 μl) was removed and replenished with the same volume of DMSO. The absorbance at 570 nm was measured with a microplate reader (BioTek EPOCH, Winooski, VT, USA).
In vivo antitumor activity
Animal experiments exclusively complied with the Guide stipulated by the Council of Agriculture Executive Yuan and ARRIVE guidelines for the Care and Use of Laboratory Animals and were approved by the Animal Ethics Committee of China Medical University (No. 104–117-N). Specific-pathogen-free (SPF) BALB/cAnN.Cg nude male mice (4 weeks old and the body weight of 20 g) were purchased from the National Laboratory Animal Center, Taiwan. Animals were fed with drinking water containing 100 mM L-arabinose and maintained under the SPF conditions in the Animal Center at China Medical University for at least 5 days before use. The xenograft mouse models were established by implanted with the subcutaneous injection of either HT29 or SW620 cancer cells (1 × 107/100 μl) on the right flank side. Bacteria were grown on LB medium for 6 h and harvested by centrifugation. Bacteria were washed by PBS, followed by centrifugation. Bacteria pellets were then resuspended in PBS for further use. EcN (106/100 μl) and EcNe/HlyE (106/100 μl) were administrated by two intraperitoneal or intratumoral injections on day 1 and day 5 per week or at the time when the tumor volume was > 150 mm3. Tumor-bearing mice were randomly assigned to 4 groups, each involving 5 mice. The body weight and tumor volume of mice were measured twice weekly. The tumor volume was calculated based on the following equation:
At the end of experiments, mice were sacrificed to have their tumors and organs weighed. All animal experiments were repeated three times.
Biodistribution of bacteria
Mice organs were weighed and homogenized aseptically in 3 ml of ice-cold and sterilized PBS. The homogenate was serially diluted and plated onto LB agar plates with or without chloramphenicol at 35 mg/ml. This allowed the examination of plasmid stability. Petri dishes were then incubated at 37℃ for 16 h. The colony forming unit (CFU) per gram tissue was determined by dividing bacterial by the weight of the organ.
Histopathological study
The method of hematoxylin and eosin (H&E) stains followed the previous report32. Mice were anesthetized by transcardial perfusion with 4% paraformaldehyde in PBS. Tumors were blocked in longitudinal sections and processed for paraffin embedding. H&E stains were applied to tumor sections with the thickness of 5 μm, and the images were photographed using a light microscopy (Olympus, Nagano, Japan). Tumor necrosis regions were analyzed using Image J (Media Cybernetics, Bethesda, MD).
Assessment of tumor necrosis
Tumor sections were dewaxed, rehydrated, and processed for H&E stains according to the standard histological method. Cell images were analyzed by the multi-mode plate reader Cytation 5 (Biotek, Winooski, VT) with GEN5 3.0 software (Biotek). After performing plasma disruption, organelle breakdown and cell lysis, Imag J software was applied to measure the total cross-sectional area of each tumor and the area of necrosis present in each tumor. The percentage of necrosis in each tumor section was calculated.
Serum cytokine analysis
The analysis was performed using female C57BL/6 mice (6–8 weeks old) from National Laboratory Animal Center, Taiwan. Mice were divided into four groups (n = 5 mice/group). Agents, including PBS, EcNe (106/100 μl), EcNe/HlyE (106/100 μl), and HlyE (3.4 μg/100 μl), were administered for two consecutive days through the tail vein. Blood was collected through the retro-orbital route at 3, 6, and 24 h post the injection. The serum cytokines, TNF-α, IL-6, and interferon (IFN)-γ were quantified using ELISA kits (R&D Systems). Duplicated readings of each sample at 450 nm were analyzed following the manufacturer’s instructions.
Statistical analysis
The statistical significance of experiments was determined by Student’s t-test. Results with P < 0.05 were considered significant.
Results
Design of the bacterial vector
The concern for HlyE safety calls for an expression system with tight regulation. PBAD is characterized with stringent regulation and exhibits a swift response to the induction of L-arabinose, which renders it useful for the expression of toxic proteins33. However, the expression of a PBAD-controlled gene displays the all-or-nothing induction pattern with a subsaturating level of L-arabinose34. This is ascribed to the “autocatalytic induction” mechanism triggered by L-arabinose which mediates the AraC-dependent activation of AraFGH for active transport of L-arabinose. Moreover, glucose interferes with the expression of PBAD via a regulatory circuit known as catabolite repression35. To address these issues, EcN was reshaped by tailoring its metabolism as shown in Fig. 1. The inducer transport-induction loop was first decoupled by inactivation of araFGH operon, and PEM7-driven araE was employed to serve for the uptake of L-arabinose. Next, ptsG was knocked down to dismantle the control circuit of catabolite repression36. Finally, the strain was deprived of araBAD operon involved in the catabolic pathway of L-arabinose. This ensures the persistent inducibility of L-arabinose which is not metabolized and maintained within the bacteria. Consequently, a genetically-modified strain was obtained and designated EcNe.
Schematic illustration of the strategy applied to rewire EcN metabolism. Shown were metabolic pathways involved in this study. Pathways of blockage and genes of deletion were marked with “X”. Through the PEP:carbohydrate phosphotransferase system (PTS), EIIAGlc receives the phosphoryl group from phosphoenolpyruvate (PEP) and becomes phosphorylated (P-EIIAGlc) associated with the production of pyruvate (PYR). During translocation via PTS, glucose (Glc) is phosphorylated to glucose 6-phosphate (G6P) by P-EIIAGlc. The araBAD operon which consists of araB (encoding L-ribulokinase), araA (encoding L-arabinose isomerase), and araD (encoding L-ribulose-5-phosphate 4-epimerase) mediates conversion of L-arabinose (L-Ara) to D-xylulose 5-phosphate (X5P). PBAD is activated (“+”) by regulator proteins involving AraC and CRP which complex with L-Ara and cAMP, respectively. Refer to text for more details. Notes: AraC, arabinose regulator; AraE, arabinose-proton symporter (encoded by araE); AraFGH, arabinose ABC transporter (encoded by araFGH operon); CRP, cAMP receptor protein; CyaA, adenylate cyclase; PtsG, glucose transporter (encoded by ptsG), PFGH, endogenous promoter of araFGH operon.
EcNe which harbored plasmid pBAD33-hlyE (i.e., EcNe/HlyE) was investigated for the HlyE production. As shown in Fig. 2a,b, EcNe/HlyE produced HlyE upon induction with L-arabinose and the protein production remained unaffected in the presence of glucose. The HlyE production reached a saturated level with L-arabinose at 30–50 μM. In contrast, the HlyE production in the original strain EcN bearing pBAD33-hlyE (designated EcN/HlyE) was negated by glucose (Fig. 2c). In addition, the protein production increased with increasing L-arabinose and was saturated with L-arabinose exceeding 1 mM (Fig. 2a). In agreement with the previous report37, HlyE mostly accumulated within the bacterial cells. The functionality of intracellular and extracellular HlyE was further investigated by the haemolysis assay. The result showed that two forms of HlyE displayed a hemolytic zone on sheep blood agar plates (SI Fig. S1a). Furthermore, RBCs were treated with various concentrations of HlyE. The haemolysis of RBCs positively correlated to the HlyE dose (SI Fig. S1b).
HlyE production and functional activity. (a)The SDS-PAGE analysis of HlyE. The production of HlyE in either EcNe/HlyE (top) or EcN/HlyE (bottom) was conducted with LB medium. The arrow indicates the location of HlyE in SDS-PAGE. Lane 1, without induction (−); lane 2, induction (+)/10 μM L-arabinose; lane 3, (+)/30 μM L-arabinose; lane 4, (+)/50 μM L-arabinose; lane 5, (+)/100 μM L-arabinose; lane 6, (+)/1 mM L-arabinose; lane 7, (+)/10 mM L-arabinose. (b) The effect of glucose on the HlyE production in EcNe/HlyE. EcNe/HlyE was grown on LB medium plus various amounts of glucose and induced for the HlyE production by adding 50 μM L-arabinose. Lane 1, (−)/nil glucose; lane 2, (+)/nil glucose; lane 3, (+)/glucose of 4 g/100 ml; lane 4, (+)/glucose of 10 g/100 ml. (c) The effect of glucose on the HlyE production in EcN/HlyE. EcN/HlyE was grown on LB medium plus various amounts of glucose and induced for the HlyE production by adding 1 mM L-arabinose. Lane 1, (−)/nil glucose; lane 2, (+)/nil glucose; lane 3, (+)/glucose of 4 g/100 ml; lane 4, (+)/glucose of 10 g/100 ml; lane 5, (+)/nil glucose; lane 6, (+)/glucose of 0.1 g/100 ml; lane 7, (+)/glucose of 0.2 g/100 ml.
HlyE cytotoxicity
The trypan blue exclusion assay provides a simple and quick examination of the HlyE-mediated effect on colorectal cancer cells. Most of SW620 and HT29 cancer cells showed a tryphan blue-stained cytoplasm after the HlyE treatment (SI Fig. S2). The cytoplasm of untreated cancer cells remained clear. The result clearly indicates that functional HlyE reduces the viability of tumor cells. The cytotoxic effect of HlyE on cell viability was further investigated by the MTT assay. Cancer cells were exposed to various concentrations of HlyE for 3 h. In general, cell viability reduced with the increasing amount of HlyE (Fig. 3a). The IC50 concentration of HlyE was estimated to be 0.84 and 2.36 μg/ml for SW620 and HT29 cells, respectively.
In vitro toxicity of HlyE. (a) Examination of cell viability by trypan blue exclusion assay. After the treatment for 30 min, cell suspension was mixed with an equal amount of 0.4% trypan blue dye. The mixture was incubated 2 min at room temperature. 1, Mock; 2, LB medium; 3, extracellular HlyE from induced EcNe/HlyE; 4, intracellular HlyE from induced EcNe/HlyE. (b) Examination of cell viability by MTT assay. Tumor cells were treated with HlyE from induced EcNe/HlyE for 3 h. SW620 or HT29 cells were administrated with the various levels of HlyE as indicated. (c) Time-lapse images of cell morphology. Images of SW620 (upper) and HT29 cells (bottom) were taken over time as indicated on the left upon the HlyE treatment. The administration dose of HlyE was shown on the top of each panel. Arrows indicate internal rearrangement of organelles and loss of membrane integrity. Cell images were processed with GEN5 3.0 software (Biotek).
The change in cell morphology was followed along the time course by using an optical microscope. As indicated in Fig. 3b,c, SW620 and HT29 cells underwent oncosis after the HlyE treatment for 15–45 min. Cell swelling occurred in the HlyE-treated cells, followed by condensation of cellular materials at one side of cells. In general, it took 15 min for cell membranes to completely destruct after the onset of cell death.
In vitro cytotoxicity of HlyE-producing bacteria
It was informative to learn the interaction of EcNe with tumor cells. EcNe was equipped with a genomic copy of PT7-DsRed, resulting in EcNe/DxRed. Subsequently, either SW620 or HT29 cells were incubated with EcNe/DxRed for 1 h. As revealed by a fluorescence microscope, red signals emitted by EcNe/DxRed scattered around the cell’s periphery (Fig. 4a). It suggests that EcNe is not invasive and adheres to these colorectal cancer cells. The cytotoxic effect of EcNe/HlyE on cancer cells was further investigated. Cells were incubated with EcNe/HlyE for 3 h. EcNe free of HlyE was used for comparison. The viability of tumor cells remained unaffected by EcNe (Fig. 4b). In contrast, EcNe/HlyE displayed a strong cytotoxic activity toward tumor cells. SW620 cells were highly susceptible to EcNe/HlyE with multiplicity of infection (MOI) of 25 while the viability of HT29 cells dropped to 45% with MOI of 50.
In vitro toxicity of EcNe/HlyE. (a) The specific attachment of EcNe to tumor cells. (b) Examination of cell viability by MTT assay. SW620 or HT29 cells were incubated with either EcNe or induced EcNe/HlyE at various MOI as indicated for 3 h. (c) Time-lapse images of cell nuclear morphology by DAPI staining. Images of SW620 and HT29 cells were taken over time as indicated on the top upon administration with either EcNe or induced EcNe/HlyE at MOI of 100. Bacteria were identified in red. (d) Time-lapse images of cell nuclear morphology by PI staining. Arrows indicate internal rearrangement of organelles and loss of membrane integrity. Cell images were processed with GEN5 3.0 software (Biotek).
As revealed by the PI or DAPI staining, EcNe/HlyE-treated cells exhibited the characteristic morphology of oncosis (Fig. 4c,d). The variants of oncotic cells were identified in the following. The time-lapse images showed that small membrane blebs and higher mass of cells occurred for SW620 and HT29 cells at 5 and 15 min post treatment, respectively. After the treatment for 0.5–1 h, SW620 and HT29 cells were more frequent to show larger plasmatic blebs, gradual loss of cell shape, and clumping of chromatin. In cells with the treatment for 2–3 h, it was more frequent to observe larger plasmatic blebs, lower mass of cells, significant swelling of nucleus where nuclear membrane and chromatin condensation still remained distinguishable. The homogenization of nuclear structure, cell membrane disruption with a partial extrusion of cell contents, and the morphological characteristics of necrosis were more prevalent in SW620 and HT29 cells that received the bacterial treatment for 3 and 6 h, respectively. The intensity of PI staining increased in the treated cells, indicating permeabilized cell membranes and cell death (Fig. 4d).
Tumor regression by HlyE-producing bacteria
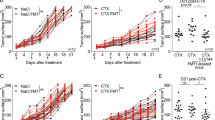
The antitumor activity of EcNe/HlyE was investigated in tumor-bearing mice. The mice xenografted with either SW620 or HT29 cells were randomly divided into four groups (n = 5). Each group was administrated with EcNe or EcNe/HlyE while PBS was used as a control. As a result, there was no significant difference in body weights of mice that received any treatments (Fig. 5a). The administration of EcNe/HlyE readily caused tumor regression (Fig. 5b). The final tumor volume in mice bearing either SW620 or HT29 cells was reduced to 11 ± 4 and 369 ± 65 mm3, respectively. In contrast, tumors in control mice grew continuously. The volume of SW620 and HT29 tumors in the PBS-treated (control) group increased to 859 ± 70 and 1508 ± 56 mm3, respectively. As compared to the control, the administration of EcNe/HlyE reduced the tumor volume of SW620 cells and of HT29 cells by approximately 98% and 76%, respectively. Interestingly, EcNe without HlyE enabled to retard the growth of HT29 tumor. It suggests that SW620 tumor is more susceptible to HlyE than HT29 tumor. Moreover, the administration of EcNe/HlyE by intratumoral injection also reduced tumor volumes (SI Fig. S3). The result indicates that BCT based on EcNe/HlyE is not affected by the injection route.
Tumor regression by EcNe/HlyE. (a) The change in the body weight of mice receiving intraperitoneal injection of bacteria. (b) Therapeutic effect of EcNe/HlyE on mice (n = 5) bearing SW620 or HT29 tumor cells. Tumor volumes (mm3) were estimated using external calipers, and values were expressed as means ± standard deviations (SD). The statistical significance was analyzed by Student's t-test (*p < 0.05; **p < 0.01; and ***p < 0.001 vs. the PBS group). (c) Distribution of bacteria in vivo.
The bacterial CFU count was implemented to determine the distribution of either EcNe or EcNe/HlyE in mice bearing SW620 or HT29 tumors. On the basis of per gram tissue, 108–109 CFU were found in the tumor and 102–103 CFU in the organ involving heart, liver, spleen, lung, and kidney (Fig. 5c). It clearly indicates preferential accumulation of EcNe and EcNe/HlyE in tumors.
Histological morphology of tumor tissues
The morphological change in tumor tissues of mice was observed by the H&E staining. SW620 tumor tissues assumed a fairly complete structure and regular shapes. The administration with EcNe/HlyE increased the extent of necrosis, consequently leading to the reduction of tumor (region N in Fig. 6a,b). Necrosis was surrounded by viable tissue (region V). In contrast, the necrotic region was present in HT29 tumor tissues irrespective of the treatment (Fig. 6c,d). In general, the necrotic cell type of pkynosis was more common in the EcNe/HlyE-caused tumor necrosis. Cells with karyorrhexis and karyolysis also appeared in the necrotic region. The necrotic region was bounded by cells with karyorrhexis. Overall, the fraction of viable tissue was largely reduced after the treatment of EcNe/HlyE.
H&E staining of tumor tissues and organs. H&E staining was applied to analyze nuclear chromatin (purple-blue) and cytoplasm (red). (a) Tumor tissues of SW620 cells after receiving various treatments. The necrosis (N) and viable (V) regions of tissues were marked (bar = 3 mm). Imag J software was applied to measure the total cross-sectional area of each tumor and the area of necrosis present in each tumor. (b) High-resolution micrographs for necrosis regions of SW620 tissues (bar = 200 μm). Nuclear condensation (pyknosis), nuclear fragmentation (karyorrhexis), and nuclear dissolution (karyolysis) were observed as indicated by white, yellow, and green arrows, respectively. (c) Tumor tissues of HT29 cells after receiving various treatments. (d) High-resolution micrographs for necrosis regions of HT29 tissues. (e) Histopathological morphology of liver. (f) Histopathological morphology of spleen.
The histopathological study showed that the morphology of organs remained unaffected for all types of treatment (Fig. 6e,f). Liver and spleen were all free from damage.
Investigation of immune responses
Finally, the safety issue of the HlyE-producing bacteria was further addressed by investigation of the immune response in mice. This was conducted with C57BL/6 mice that received two intravenous injections of HlyE and either EcNe or EcNe/HlyE. As a result, serum TNF-α, TNF-β and IL6 levels slightly increased in the first 6 h after the final injection. Their levels returned to nearly the basal level at 24 h (Fig. 7). The result indicates that the severe host immune response and the prolonged inflammatory cytokine response are not induced in EcNe/HlyE-treated animals.
Investigation of immune responses in mice. C57BL/6 mice were treated with systemic administration of PBS, EcNe or EcNe/HlyE for four consecutive days. Since then, their serum levels of TNF-α, TNF-γ and IL6 in C57BL/6 mice were quantified at the indicated time three time points. Values were expressed as means ± SD (*p < 0.05; **p < 0.01; and ***p < 0.001 vs. the PBS group).
Discussion
The platform for BCT based on therapeutic proteins basically consists of a bacterial vector, a protein payload, and a protein-expressing system. The bacterial vector is mainly responsible for the synthesis and in situ delivery of the protein payload. In this context, the bacterial ability of seeking tumor turns out to be a perquisite. The potential of probiotic EcN for cancer therapy was first acknowledged by recognizing its high efficiency of 4T1 breast tumor-specific colonization and replication38. In particular, the result of tumor colonization was not affected by the administration routes involving intraperitoneal or intratumoral injection of EcN. In this study, EcN was developed by metabolic engineering for medical intervention of colorectal cancer. The engineered strain (i.e., EcNe) specifically adhered to SW620 and HT29 cancer cells in vitro (Fig. 4a) whereas cancer cells were free from detriment (Fig. 4b). EcN has an extracellular structure composed of K5 capsule39 and possesses F1C fimbriae for formation biofilm40, which confers on EcN the intestinal colonization ability. These characteristic structures of EcNe likely interact with these colorectal cancer cells. EcNe preferentially colonized in SW620 and HT29 tumors (Fig. 5c) while retarded the growth of HT29 tumor (Fig. 5b). It was reported that EcN displayed high efficiency of tumor-selective colonization and amplification in both immunocompetent and immunocompromised mice38. Therefore, the reduced growth rate of HT29 tumor by EcNe is likely attributed to a high density of colonized bacteria that competes for available nutrients.
A potential protein payload needs to be secreted from the bacterial vector, highly cytotoxic against malignant cancer cells, and capable of diffusing deep into tumor tissue. Recognized as a small hemolytic protein, HlyE is exported from E. coli via the outer-membrane vesicles (OMVs)-mediated transport route41. OMVs-released HlyE displayed a high cytotoxic activity toward mammalian cells. Accordingly, HlyE has been applied for cancer therapy. The administration of S. typhimurium which expressed HlyE with a hypoxia-inducible promoter significantly increased necrosis of 4T1 breast tumor15. In another study, the growth of 4T1 breast and CT26 murine colon tumors were marginally affected by E. coli strain K-12 with HlyE under the control of a constitutive promoter42. In this study, E. coli HlyE was produced by a PBAD–based expression system in EcNe. PBAD is known to be stringently regulated and promptly responding to the induction of L-arabinose33. In addition, L-arabinose enables passive diffusion into tumor tissue and administration via the oral route. EcN was reprogrammed to improve the performance of the PBAD-based expression system, resulting in EcNe. EcN displayed a homogenous and glucose-insensitive induction profile and achieved a saturated production at a very low level of inducer. As shown in Fig. 2, EcNe/HlyE was far superior to EcN/HlyE in terms of the HlyE production. The production of PBAD-regulated HlyE in EcNe/HlyE is insensitive to glucose and saturated at 30–50 μM L-arabinose (indicative of bacterial population homogeneity). Consequently, EcNe/HlyE exhibited the tumor-specific colonization with the tumor-to-organ ratio of 106:1 and significantly reduced the volume of SW620 tumor and HT29 tumor by 98% and 76% (Fig. 5b,c), respectively.
E. coli HlyE and S. aureus SAH share a similar function of pore-forming. The previous study reported that most of SAH regulated by PBAD was released outside from E. coli strain K17. Mice were injected with L-arabinose (2.67 M), and the treatment with SAH-producing E. coli significantly reduced the volume of MCF7 breast tumor and increased the extent of cell necrosis. In contrast to the injection route, the oral administration of L-arabinose at 100 mM in this study was effective to induce the protein payload in mice. Note that S. typhimurium were cleared from the blood at 6 h and the liver and spleen at 48 h post injection7. Similarly, EcN were mostly found in tumors at 24 h after injection38. This is advantageous for a temporal control of the protein expression by PBAD, which enables constraint of the expressed payload to tumor. In their SAH study, a mathematical model was proposed and predicted the positive linkage of bacterial colony size and the protein production rate to SAH efficacy17. However, the colony size was found to increase with decreasing the production rate. The bacterial growth is usually afflicted by the protein overproduction, known as “metabolic burden”43. This physiological stress would exaggerate the characteristic heterogeneity of PBAD as described earlier. Therefore, a large colony size mainly consists of uninduced bacteria due to their fitness for growth. The interference of the blood sugar level with PBAD also negatively affects the protein production rate. Colonized bacteria survive in the transition zone between the proliferative and necrotic regions of tumors where the microenvironment constantly varies44. This further renders the spatial control of the protein expression by PBAD complicate. Reprogrammed EcNe appears feasible to address the issue (see discussion earlier). As analyzed by time-lapse images, tumor cells readily displayed a characteristic morphological change of oncosis upon treatment with EcNe/HlyE (Fig. 4c,d). In tumor-bearing mice, colonized EcNe/HlyE maintained L-arabinose inside and continuously expressed HlyE with a sufficient level to kill tumor cells. The necrotic regions of tumors thus enlarged (Fig. 6a–d), and EcNe/HlyE which lived in the border regions between viable and necrotic tissues moved toward to the tumor peripheral. The tumor microenvironment changed with the dynamics of the bacteria-tumor cells interaction. Consequently, the tumor growth of HT29 tumor was arrested while SW620 tumor was nearly removed (Fig. 5b).
In contrast to previous studies conducted with murine tumor models25,26, this study investigated the cancer therapeutics of HlyE-producing EcN (i.e., EcNe/HlyE) in human tumor models. A study also reported that a fusion protein-expressing EcN exhibited efficient inhibition of the growth of human hepatoma SMMC-7721 tumor45. This hybrid protein consisted of the tumor suppressor protein p53 and the anti-angiogenic peptide Tum-5 under the control of the oxygen-dependent promoter from Vitreoscilla. Overall, the encouraging results from these various studies suggest the potential application of EcN for cancer therapy. As shown in Fig. 7, the induction of acute infection was absent from injection of EcNe/HlyE. By regulation of newly-recruited T cells, EcN was able to reduce intestinal inflammation46. Therefore, probiotic EcN manifests itself as an alternative to Salmonella that has been intensively investigated for conveyance of antitumor drugs47.
In summary, colorectal cancer is a life-threatening disease due to the failure of conventional therapies in many cases. In this study, a new method of bacteria-mediated tumor therapy was successfully developed by exploiting EcN for selective delivery of a small cytotoxic protein (HlyE). To approach the goal, EcN was finely tailored by metabolic engineering. The resulting EcNe enabled the temporal and spatial control of the PBAD-regulated HlyE expression to improve its therapeutic efficacy. Production of recombinant proteins has been most commonly practiced in E. coli. Likewise, a tune-up of EcN is feasible to achieve efficient production of the therapeutic protein. Metabolic engineering provides a powerful method for rational design of a bacterial strain with the desired trait. We have applied this strategy to improve the protein production and alleviate the stress of metabolic load in E. coli48. With availability of the synthetic biology toolbox49, these technology advances are expected to design a robust system based on EcN for medical intervention of various tumors.
References
Tredan, O., Galmarini, C. M., Patel, K. & Tannock, I. F. Drug resistance and the solid tumor microenvironment. J. Natl. Cancer Inst. 99, 1441–1454 (2007).
Baudino, T. A. Targeted cancer therapy: The next generation of cancer treatment. Curr. Drug Discov. Technol. 12, 3–20 (2015).
Tannock, I. F., Lee, C. M., Tunggal, J. K., Cowan, D. S. & Egorin, M. J. Limited penetration of anticancer drugs through tumor tissue: A potential cause of resistance of solid tumors to chemotherapy. Clin. Cancer Res. 8, 878–884 (2002).
Wachsberger, P., Burd, R. & Dicker, A. P. Tumor response to ionizing radiation combined with antiangiogenesis or vascular targeting agents: exploring mechanisms of interaction. Clin Cancer Res 9, 1957–1971 (2003).
Yazawa, K. et al. Bifidobacterium longum as a delivery system for gene therapy of chemically induced ratmammary tumors. Breast Cancer Res Treat 66, 165–170 (2001).
Malmgren, R. A. & Flanigan, C. C. Localization of the vegetative form of Clostridium tetani in mouse tumors following intravenous spore administration. Cancer Res 15, 473–478 (1955).
Forbes, N. S., Munn, L. L., Fukumura, D. & Jain, R. K. Sparse initial entrapment of systemically injected Salmonella typhimurium leads to heterogeneous accumulation within tumors. Cancer Res 63, 5188–5193 (2003).
Min, J. J., Nguyen, V. H., Kim, H. J., Hong, Y. & Choy, H. E. Quantitative bioluminescence imaging of tumor-targeting bacteria in living animals. Nat. Protoc. 3, 629–636 (2008).
Nauts, H. C., Swift, W. E. & Coley, B. L. The treatment of malignant tumors by bacterial toxins as developed by the late William B. Coley, MD, reviewed in the light of modern research. Cancer Res. 6, 205–216 (1946).
Lehouritis, P., Springer, C. & Tangney, M. Bacterial-directed enzyme prodrug therapy. J. Control Release 170, 120–131 (2013).
Xiang, S., Fruehauf, J. & Li, C. J. Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nat. Biotechnol. 24, 697–702 (2006).
Loeffler, M., Le’Negrate, G., Krajewska, M. & Reed, J. C. Attenuated Salmonella engineered to produce human cytokine LIGHT inhibit tumor growth. Proc. Natl. Acad. Sci. USA 104, 12879–12883 (2007).
Fensterle, J. et al. Cancer immunotherapy based on recombinant Salmonella enterica serovar Typhimurium aroA strains secreting prostate-specific antigen and cholera toxin subunit B. Cancer Gene Ther. 15, 85–93 (2008).
Groot, A. J. et al. Functional antibodies produced by oncolytic clostridia. Biochem. Biophys. Res. Commun. 364, 985–989 (2007).
Ryan, R. M. et al. Bacterial delivery of a novel cytolysin to hypoxic areas of solid tumors. Gene Ther. 16, 329–339 (2009).
Provoda, C. J. & Lee, K. D. Bacterial pore-forming hemolysins and their use in the cytosolic delivery of macromolecules. Adv. Drug Deliv. Rev. 4, 209–221 (2000).
St. Jean, A. T., Swofford, C. A., Panteli, J. T., Brentzel, Z. J. & Forbes, N. S. Bacterial delivery of Staphylococcus aureus α-hemolysin causes regression and necrosis in murine tumors. Mol. Ther. 22, 1266–1274 (2014).
del Castillo, F. J., Leal, S. C., Moreno, F. & del Castillo, I. The Escherichia coli K-12 sheA gene encodes a 34-kDa secreted haemolysin. Mol. Microbiol. 25, 107–115 (1997).
Oscarsson, J. et al. Characterization of a pore-forming cytotoxin expressed by Salmonella enterica serovars Typhi and Paratyphi A. Infect. Immun. 70, 5759–5769 (2002).
del Castillo, F. J., Moreno, F. & del Castillo, I. Characterization of the genes encoding the sheA haemolysin in Escherichia coli O157:H7 and Shigella flexneri 2a. Res. Microbiol. 151, 229–230 (2000).
Lai, X. H. et al. Cytocidal and apoptotic effects of the ClyA protein from Escherichia coli on primary and cultured monocytes and macrophages. Infect. Immun, 68, 4363–4367. https://doi.org/10.1128/iai.68.7.4363-4367.2000 (2000).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics. CA Cancer J. Clin. 69, 7–34 (2019).
Claesen, J. & Fischbach, M. A. Synthetic microbes as drug delivery systems. ACS Synth. Biol. 4, 358–364 (2015).
Beimfohr, C. A review of research conducted with probiotic E. coli marketed as symbioflor. Int. J. Bacteriol. 2016, 3535621 (2016).
Zhang, Y. et al. Escherichia coli Nissle 1917 targets and restrains mouse B16 melanoma and 4 T1 breast tumors through expression of azurin protein. Appl. Environ. Microbiol. 78, 7603–7610 (2012).
Lehouritis, P., Stanton, M., McCarthy, F. O., Jeavons, M. & Tangney, M. Activation of multiple chemotherapeutic prodrugs by the natural enzymolome of tumour-localised probiotic bacteria. J. Control. Release 222, 9–17 (2016).
Li, R. et al. Expressing cytotoxic compounds in Escherichia coli Nissle 1917 for tumor-targeting therapy. Res. Microbiol. 170, 74–79 (2019).
Wang, Z. W., Lai, C. B., Chang, C. H., Chiang, C. J. & Chao, Y. P. A glucose-insensitive T7 expression system for fully-induced expression of proteins at a subsaturating level of L-arabinose. J. Agric. Food Chem 59, 6534–6542 (2011).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645 (2000).
Chiang, C. J., Lin, L. J., Wang, Z. W., Lee, T. T. & Chao, Y. P. Design of a noncovalently linked bifunctional enzyme for whole-cell biotransformation. Proc. Biochem. 49, 1122–1128 (2014).
Miller, J. H. Experiments in Molecular Genetics (Cold Spring Harbor Laboratory, 1972).
Chiang, C. J., Lin, L. J., Wu, C. P., Chen, C. J. & Chao, Y. P. Development of nanoscale oil bodies for targeted treatment of lung cancer. J. Agric. Food Chem. 66, 9438–9445 (2018).
Guzman, L. M., Belin, D., Carson, M. J. & Bechwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 (1995).
Morgan-Kiss, R. M., Wadler, C. & Cronan, J. E. Long-term and homogeneous regulation of the Escherichia coli araBAD promoter by use of a lactose transporter of relaxed specificity. Proc. Natl. Acad. Sci. USA 99, 7373–7377 (2002).
Schleif, R. Regulation of the L-arabinose operon of Escherichia coli. Trends Genet. 16, 559–565 (2000).
Picon, A., Teixeira de Mattos, M. J. & Postma, P. W. Reducing the glucose uptake rate in Escherichia coli affects growth rate but not protein production. Biotechnol. Bioeng. 90, 191–200 (2005).
Ludwig, A., Bauer, S., Benz, R., Bergmann, B. & Goebel, W. Analysis of the SlyA-controlled expression, subcellular localization and pore-forming activity of a 34 kDa haemolysin (ClyA) from Escherichia coli K-12. Mol. Microbiol. 31, 557–567 (1999).
Stritzker, J. et al. Tumor-specific colonization, tissue distribution, and gene induction by probiotic Escherichia coli Nissle 1917 in live mice. Int. J. Med. Microbiol. 297, 151–162 (2007).
Burns, S. M. & Hull, S. I. Comparison of loss of serum resistance by defined lipopolysaccharide mutants and an acapsular mutant of uropathogenic Escherichia coli O75:K5. Infect. Immun. 66, 4244–4253 (1988).
Lasaro, M. A. et al. F1C fimbriae play an importantrole in biofilm formation and intestinal colonization by the Escherichia coli commensal strain Nissle 1917. Appl. Environ. Microbiol. 75, 246–251 (2009).
Wai, S. N. et al. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115, 25–35 (2003).
Jiang, S. et al. Inhibition of tumor growth and metastasis by a combination of Escherichia coli-mediated cytolytic therapy and radiotherapy. Mol. Ther. 18, 635–642 (2010).
Hoffmann, F. & Rinas, U. On-line estimation of the metabolic burden resulting from the synthesis of plasmid-encoded and heat-shock proteins by monitoring respiratory energy generation. Biotechnol. Bioeng. 76, 333–340 (2001).
Weibel, S., Stritzker, J., Eck, M., Goebel, W. & Szalay, A. A. Colonization of experimental murine breast tumours by Escherichia coli K-12 significantly alters the tumour microenvironment. Cell Microbiol. 10, 1235–1248 (2008).
He, L. et al. Intestinal probiotics E. coli Nissle 1917 as a targeted vehicle for delivery of p53 and Tum-5 to solid tumors for cancer therapy. J. Biol. Eng. 13, 58 (2019).
Sturm, A. et al. Escherichia coli Nissle 1917 distinctively modulates T-cell cycling and expansion via Toll-like receptor 2 signaling. Infect. Immun. 73, 1452–1465 (2005).
Zhou, S., Gravekamp, C., Bermudes, D. & Liu, K. Tumour-targeting bacteria engineered to fight cancer. Nat. Rev. Cancer 18, 727–743 (2018).
Chiang, C. J., Hu, M. C. & Chao, Y. P. A strategy to improve production of recombinant proteins in Escherichia coli based on the glucose-glycerol mixture and glutamate. J. Agric. Food Chem. 68, 8883–8889 (2020).
Pedrolli, D. B., Ribeiro, N. V., Squizato, P. N., de Jesus, V. N. & Cozetto, D. A. Engineering microbial living therapeutics: The synthetic biology toolbox. Trends Biotechnol. 37, 674–681 (2019).
Acknowledgements
We acknowledge Mu-Chen Hu for his technical assistance. This work was supported by China Medical University (CMU-105-S-50) and Ministry of Science and Technology (MOST 108-2221-E-035-052-MY3), Taiwan.
Author information
Authors and Affiliations
Contributions
Conceptualization: C.J.C. Data curation: P.H.H. Formal analysis: P.H.H. Funding acquisition: C.J.C. In silico investigation: P.H.H. Methodology: P.H.H., C.J.C. Project administration: C.J.C. Supervision: C.J.C. Writing original draft: P.H.H. Writing review and editing: P.H.H., C.J.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chiang, CJ., Huang, PH. Metabolic engineering of probiotic Escherichia coli for cytolytic therapy of tumors. Sci Rep 11, 5853 (2021). https://doi.org/10.1038/s41598-021-85372-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-85372-6
This article is cited by
-
Engineered Cancer Nanovaccines: A New Frontier in Cancer Therapy
Nano-Micro Letters (2025)
-
Bacteria extracellular vesicle as nanopharmaceuticals for versatile biomedical potential
Nano Convergence (2024)
-
Enhancing tumor-specific recognition of programmable synthetic bacterial consortium for precision therapy of colorectal cancer
npj Biofilms and Microbiomes (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.