Abstract
Alveolar macrophages (AMs) are critical for defense against airborne pathogens and AM dysfunction is thought to contribute to the increased burden of pulmonary infections observed in individuals living with HIV-1 (HIV). While HIV nucleic acids have been detected in AMs early in infection, circulating HIV during acute and chronic infection is usually CCR5 T cell-tropic (T-tropic) and enters macrophages inefficiently in vitro. The mechanism by which T-tropic viruses infect AMs remains unknown. We collected AMs by bronchoscopy performed in HIV-infected, antiretroviral therapy (ART)-naive and uninfected subjects. We found that viral constructs made with primary HIV envelope sequences isolated from both AMs and plasma were T-tropic and inefficiently infected macrophages. However, these isolates productively infected macrophages when co-cultured with HIV-infected CD4+ T cells. In addition, we provide evidence that T-tropic HIV is transmitted from infected CD4+ T cells to the AM cytosol. We conclude that AM-derived HIV isolates are T-tropic and can enter macrophages through contact with an infected CD4+ T cell, which results in productive infection of AMs. CD4+ T cell-dependent entry of HIV into AMs helps explain the presence of HIV in AMs despite inefficient cell-free infection, and may contribute to AM dysfunction in people living with HIV.
Similar content being viewed by others
Introduction
Individuals with untreated HIV-1 (HIV) are at increased risk of pulmonary infections with mycobacteria and other bacteria, viruses, and fungi1,2,3. The first line of defense against these infections in the lung are alveolar macrophages (AMs), which comprise the vast majority of immune cells in the alveolar space4,5. AMs from individuals with untreated HIV are dysfunctional, with impaired phagocytic activity6,7,8, proteolytic activity9, and bacterial killing10. The lung is an early site of HIV and simian immunodeficiency virus (SIV) replication in humans, humanized mice and non-human primates (NHPs)11,12,13,14,15, and HIV nucleic acids are detectable in AMs in both antiretroviral (ART)-naive and treated individuals9,16,17,18,19,20,21,22. Thus, HIV is found in the lung and in AMs early in the course of HIV-1 infection, but the mechanism of entry of HIV into AMs remains incompletely understood.
Cell tropism in HIV infection is determined by the coreceptor usage and affinity of the envelope (Env) surface protein for host CD423. These phenotypes are separate, with coreceptor usage determined by the Env V3 region and macrophage or T cell tropism (M- or T-tropism, respectively) determined by the Env CD4 binding site23,24. There are three common combinations of coreceptor usage and cellular tropism. The first is CCR5-utilizing T-tropic HIV, which accounts for transmitted/founder viruses and most circulating viruses24,25. The second, CXCR4-utilizing T-tropic HIV, arises late in infection in about 50% of people living with HIV26. The third, M-tropic HIV, can use CCR5, CXCR4 or both coreceptors, and arises in some individuals late in infection and in T cell-depleted environments such as in advanced HIV-1 disease, in the central nervous system, or in settings of experimental depletion of CD4+ T cells27,28,29. The main target cells in early infection are mucosal CD4+ T cells, which are activated, have high expression levels of both CD4 and CCR5, and are depleted during HIV infection15,30,31. Monocyte-derived macrophages (MDMs) have approximately 20-fold lower surface expression of CD4 than CD4+ T cells32,33,34, and T-tropic HIV strains enter33 and replicate35 inefficiently in MDMs. Importantly, M- or T-tropism can be recapitulated by cloning of specific env genes into a replication-competent backbone, demonstrating that Env is responsible for this tropism25. CD4 and CCR5 surface expression and HIV infection rates can vary widely in MDMs depending on the conditions used for their maturation34, and HIV receptor surface expression and infection rates may be different compared to tissue resident macrophages like AMs36,37. Therefore, it is important to understand M- and T-tropic HIV entry and infection rates of primary AMs. Prior reports have shown that AMs are susceptible to M-tropic HIV entry and replication in vitro38,39,40. However, no study has established the tropism of HIV isolated from AMs from HIV-infected individuals, nor has any study established a mechanism of AM infection by T-tropic HIV strains, which predominate during acute and chronic stages of infection.
Evidence from humanized mice and NHPs infected with T-tropic HIV or SIV shows that infection of tissue macrophages in the spleen, central nervous system and lymph nodes may be dependent on the presence of CD4+ T cells41,42,43. Humanized mice reconstituted with human macrophages and not with human lymphocytes can be infected with M-tropic but not T-tropic HIV strains41. In NHPs, T-tropic SIV DNA is only measurable in macrophages residing in lymphoid tissues that have high numbers of CD4+ T cells, including the spleen and mesenteric lymph nodes43. A possible explanation for these observations is that interactions between macrophages and infected CD4+ T cells are needed to achieve high levels of macrophage infection. Prior studies suggest that T-tropic HIV inefficiently infects MDMs in vitro25, and while M-tropic HIV can infect macrophages, this is more efficient through contact with infected CD4+ T cells44,45. HIV transmission to AMs through T cells has not been studied in human AMs.
Results
HIV can be detected in alveolar macrophages (AMs) from individuals with untreated HIV infection
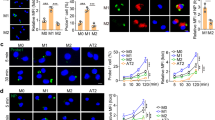
To determine levels of HIV nucleic acids in AMs from HIV-infected individuals, we performed bronchoscopies with bronchoalveolar lavage (BAL) in a cohort of HIV-infected ART naive and uninfected individuals living in Cape Town, South Africa (Table 1). All the participants in the cohort had evidence of immune sensitization to Mycobacterium tuberculosis but no indications of active disease. Cells from each bronchoscopy were adhered and non-adherent cells were removed; 99–100% of the remaining cells were AMs by microscopy (Supplementary Table 1). Using qPCR for HIV Gag, we detected Gag RNA in purified AMs from 4/11 (36.4%) participants and Gag DNA in AMs from 5/11 (45.5%) participants (Fig. 1A). While HIV RNA was not detected in all individuals, the BAL viral load was significantly higher in participants with detectable HIV RNA in the corresponding AM sample (Supplementary Fig. 1A). HIV RNA and DNA were both detected in samples from two participants, while two other participants had detectable HIV RNA only and three had detectable HIV DNA only. Overall, these findings were similar to prior reports using AMs from ART-naive participants in which HIV RNA and DNA was detectable in AMs from a median of 38.7% and 62.6% individuals, respectively, although the values varied widely across studies18,20,21,40,46,47,48,49,50. These data demonstrate that a significant number of HIV-infected individuals harbor AMs with detectable HIV RNA and DNA.
HIV RNA and DNA can be detected in AMs despite low T-tropic HIV entry and replication rates. (A) HIV Gag RNA and DNA are detectable in alveolar macrophages (AMs) from individuals with untreated HIV infection. HIV Gag RNA or DNA copies per cell by qRT-PCR in AMs from the Cape Town cohort. Dashed lines denote the limit of detection for the assay, and negative values are plotted at 50% of the limit of detection. (B, C) Viral entry was detected in AMs and BAL CD4+ T cells from HIV-uninfected subjects using gravity infection for 12 h with a BLaM-Vpr construct (HIV+) of JR-CSF (B) or 89.6 (C) or mock infection (HIV-) at a multiplicity of infection (MOI) of 1. HIV entry was detected by cleavage of a fluorescent BLaM substrate and measured by flow cytometry; n = 6. (D, E) CD4+ T cells, MDMs and AMs were infected with replication-competent JR-CSF (D) or 89.6 (E) at a MOI of 0.2 for 12 h. HIV p24 levels in the supernatant were measured by ELISA at the indicated time points and normalized to input at day 1; n = 11 for T cells, n = 12 for MDMs, and n = 6 for AMs. Statistics were done by Kruskal–Wallis tests with Dunn’s multiple test correction in B and C. *p < 0.05, **p < 0.01, ***p < 0.001.
Cell free T-tropic HIV inefficiently enters and replicates in MDMs and AMs
Since most circulating HIV in acute and chronically infected individuals is T-tropic, we compared the entry and replication capacity of T-tropic and M-tropic HIV in AMs and MDMs from HIV-uninfected individuals. We first compared HIV receptor expression on AMs, MDMs, and BAL CD4+ T cells and observed that fewer AMs expressed CD4 and CCR5 compared to MDMs and BAL CD4+ T cells (Supplementary Fig. 1B and C). Using a beta-lactamase (BLaM)-Vpr reporter assay that measures viral entry, we found that the CCR5-using T-tropic HIV strain JR-CSF did not significantly enter AMs, but was able to enter BAL CD4+ T cells, suggesting that BAL CD4+ T cells are more permissive targets of T-tropic HIV entry in the lungs (Fig. 1B). This entry into BAL CD4+ T cells was blocked by the CCR5 inhibitor maraviroc (Supplementary Fig. 1D). Using the CCR5 and CXCR4-utilizing M-tropic HIV strain 89.6, in contrast, both AMs and BAL CD4+ T cells allowed viral entry (Fig. 1C). Next, we utilized replication competent T-tropic and M-tropic HIV and showed that T-tropic HIV JR-CSF replicated in blood CD4+ T cells but not in MDMs or AMs (Fig. 1D and Supplementary Fig. 1E). M-tropic HIV 89.6, on the other hand, replicated in blood CD4+ T cells, MDMs, and AMs (Fig. 1E and Supplementary Fig. 1F). These experiments indicate that relatively few AMs express HIV receptors and that HIV infection by a lab-adapted CCR5-using T-tropic HIV strain, which is similar to those that predominate in early stages of infection, inefficiently enters and replicates in AMs and MDMs.
AM- and plasma-derived HIV is T-tropic
In order to understand the entry properties of HIV derived from AMs, we isolated HIV env from lung and plasma samples. We sequenced HIV env RNA from AMs (27 isolates), BAL fluid (19 isolates), and plasma (21 isolates) from 3 HIV-infected ART-naive individuals (Supplementary Fig. 2). These sequences clustered by HIV-infected donor, but there was no compartmentalization when all AM-derived isolates were compared to all plasma-derived isolates. However, compartmentalization between AM and plasma derived sequences was observed in 2 of 3 donors (Supplementary Fig. 2). To test the ability of these AM- and plasma-derived Env proteins to allow infection of CD4+ T cells and MDMs, we cloned the env genes into an NL4-3 env deleted HIV backbone to generate replication-competent virus. We tested viruses containing Env from 8 AM and 9 plasma isolates selected to represent the sequence diversity found in the samples (arrows in Supplementary Fig. 2). Viruses containing all of the AM- and plasma-derived HIV Env isolates productively infected CD4+ T cells but did not infect MDMs (Fig. 2 and Supplementary Fig. 3). These data indicate that all AM- and plasma-derived HIV primary isolates found in our study were T-tropic.
AM- and plasma-derived HIV primary isolates are T-tropic. HIV env isolated from AMs or plasma from 3 HIV-infected donors was cloned into an HIV NL4-3 env deleted backbone plasmid. Replication-competent virus was added to activated CD4+ T cells or MDMs for 12 h and washed, and supernatant was collected 1 h after washing (Day 1) and after 7 or 14 days. Data is shown as fold change of HIV p24 in supernatant at the final timepoint (Day 7 for T cells, Day 14 for MDMs) compared to Day 1. Cells were infected with AM-derived viral isolates (A) from viral donor 1, 2 or 3 or plasma-derived viral isolates (B) from viral donor 1, 2 or 3. Data from all viral isolates from the same viral donor and anatomical site were pooled. CD4 + T cells and MDMs were isolated from six HIV-uninfected blood donors. In (A), n = 12 (6 cell donors infected with 2 viral isolates) for viral donor 1, n = 18 (6 cell donors infected with 3 viral isolates) for viral donor 2, and n = 18 (6 cell donors infected with 3 viral isolates) for viral donor 3; in (B), n = 12 (6 cell donors infected with 2 viral isolates) for viral donor 1, n = 12 (6 cell donors infected with 2 viral isolates) for viral donor 2, and n = 30 (6 cell donors infected with 5 viral isolates) for viral donor 3. Statistics by Kruskal–Wallis test. *p < 0.05, **p < 0.01, ***p < 0.001.
T-tropic HIV infection is more efficient through cell-to-cell than through cell-free transmission to MDMs and AMs. (A) MDMs were matured for 7 days or AMs were adhered for 1 h. CD4+ T cells were activated with PHA for 3 days, infected with HIV JR-CSF or 89.6 at a MOI of 0.2 overnight, washed, and cultured for 4 days, when approximately 10% of T cells were HIV Gag+ by flow cytometry. After the T cells were centrifuged, the supernatant from CD4+ T cells was used for the SN condition. The pellet of CD4+ T cells was resuspended and added directly (CTC) or across a transwell (TW) for 3 h and washed, using a ratio of 1:1 T cell:macrophage. (B–E) Supernatants from (B, D ) MDMs or (C, E) AMs were collected and HIV p24 was measured by ELISA. p value shows the value for cell-to-cell vs. supernatant conditions at the given timepoint using a two-way ANOVA with Tukey’s multiple comparisons test; n = 6 for MDMs and AMs. *p < 0.05, **p < 0.01, ****p < 0.0001.
T-tropic HIV efficiently infects AMs through contact with infected CD4+ T cells
Previous work has shown increased efficiency of M-tropic HIV entry into MDMs through uptake of or fusion with infected T cells44,45. However, whether this also occurs with T-tropic HIV, the primary virus during acute and chronic infection, and its relevance to primary AM infection remains unknown. To test T-tropic and M-tropic HIV entry into AMs via interaction with HIV-infected T cells, we infected PHA-activated CD4+ T cells with the CCR5-using T-tropic HIV strain JR-CSF. We then cultured HIV from these autologous infected CD4+ T cells with MDMs or AMs in the following conditions: (1) infected CD4+ T cells added directly to the macrophages (“cell-to-cell” or “CTC”); (2) infected CD4+ T cells separated from the macrophages by a transwell (“TW”); and (3) infected CD4+ T cell culture supernatant added to the macrophages (“SN”) (Fig. 3A). The cells or supernatant were added for 3 h before washing the macrophages. In MDMs and AMs, T-tropic HIV replication was significantly increased by contact with infected CD4+ T cells compared to the SN or TW conditions (Fig. 3B, C). Other reports have suggested that contact with T cells enhances M-tropic infection rates of MDMs44,45, but this has not been shown in AMs. To test this, we repeated the experiment with CD4+ T cells infected with the CCR5 and CXCR4-using M-tropic HIV strain 89.6. In MDMs, M-tropic infection was enhanced by contact with infected CD4+ T cells, similar to previous literature (Fig. 3D). However, in AMs, M-tropic HIV p24 levels were similar in the SN and CTC conditions by day 14 (Fig. 3E). In order to study whether productive HIV infection occurs in the T cell-macrophage co-cultures, we pre-treated the macrophages and T cells with the non-nucleoside reverse transcriptase inhibitor (NNRTI) efavirenz (EFV). We found that EFV pre-treatment inhibited T-tropic HIV replication by day 14 in the CTC condition in both MDMs and AMs (Supplementary Fig. 4A, B). These data demonstrate that T-tropic HIV infects MDM and AM cultures more efficiently through contact with an infected CD4+ T cell intermediate, and that co-culture of T-tropic HIV-infected CD4+ T cells with MDMs or AMs leads to productive infection.
Viral constructs made with patient-derived HIV env from AMs and plasma productively infect MDMs more efficiently through T cell contact. Experimental conditions were as in Fig. 3 with the HIV constructs described in Fig. 2. Cells were infected with AM-derived (A) or plasma-derived (B) viral isolates from viral Donors 1, 2 or 3. CD4+ T cells and MDMs were isolated from the same blood donors. In (A), n = 11 cell donors for viral donor 1, n = 6 cell donors for viral donor 2, and n = 10 cell donors for viral donor 3; in (B), n = 9 cell donors for viral donor 1, n = 9 cell donors for viral donor 2, and n = 7 cell donors for viral donor 3. p value from Kruskal–Wallis test. Statistics compare the fold change at the final timepoint to day 1. **p < 0.01, ***p < 0.001.
AM-derived HIV can productively infect MDMs more efficiently when there is contact with infected CD4+ T cells
Having established that infection of AMs and MDMs with a laboratory strain of T-tropic HIV can be enhanced by contact with an infected CD4+ T cell, we tested whether infection with primary patient-derived AM- or plasma-derived HIV isolates could also be enhanced in macrophages through this same route. Using the experimental setup described above, we demonstrate that both AM-derived and plasma-derived primary isolates did not productively infect MDMs in the SN condition (Fig. 4 and Supplementary Fig. 5), consistent with our observations of the T-tropic virus JR-CSF. On rare occasions, there was detectable p24 production in the cell-free virus (SN) condition. However, for all 3 AM-derived and 2 out of the 3 plasma-derived Envcontaining viruses, infection of MDMs was observed in the CTC condition (Fig. 4 and Supplementary Fig. 5). These data indicate that primary HIV isolates made with Env derived from AMs infect MDMs inefficiently on their own, but can infect MDMs more efficiently via contact with an infected CD4+ T cell intermediate.
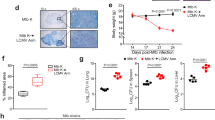
T-tropic HIV-infected T cell contact with AMs leads to HIV Gag staining in the AM cytosol. (A) AMs were co-cultured with HIV JR-CSF-infected autologous CD4+ T cells (cell-to-cell, CTC) or T cell supernatant (SN) for 3 h, washed, and cultured for an additional 14 days. Staining was performed with DAPI for cell nuclei, HIV Gag, CD3 for T cells, and CD68 for macrophages, and images were acquired by immunofluorescent confocal microscopy. White dotted outlines show the AM or T cell membrane. (B–D) % Gag+ cells out of all nucleated cells were quantified. (B) Gag + CD4+ T cells not associated with AMs (Gag+ T cells) were rare in both the SN and CTC conditions. (C) Cytosolic gag staining was increased in AMs associated with CD3+ T cells in the CTC condition compared to the SN condition. (D) Pie charts of all of the staining categories in SN and CTC conditions. Gag staining was rarely observed in solitary CD3+ T cells. Each category was compared between SN and CTC by the Friedman test with Dunn’s multiple comparison’s test. 100 cells per donor were quantified; n = 6 donors. *p < 0.05.
T cell-mediated infection of macrophages is mainly associated with CD4+ T cell contact
To further characterize the cellular location of HIV in infected macrophages, we imaged T cell-macrophage co-cultures using immunofluorescent confocal microscopy. As before, autologous CD4+ T cells were infected with T-tropic HIV, and T cell supernatant (SN) or HIV infected CD4+ T cells (CTC) were added to AMs. The samples were stained with anti-CD3 (T cells), anti-CD68 (macrophages) and anti-HIV Gag, and imaged after 14 days (Fig. 5A). Staining was quantified to localize HIV Gag. As expected, we detected Gag staining within AMs primarily in the CTC but not in the SN condition (Fig. 5A and Supplementary Fig. 6A). The CD3 staining was limited to a discrete intracellular area, suggestive of an endocytosed CD3+ T cell. Gag+ CD3+ T cells that were not associated with AMs were rare, with a median of 0% and 1.07% of all cells for JR-CSF SN and JR-CSF CTC, respectively (Fig. 5B), indicating that the increase in HIV release in co-culture experiments was likely not due to infection of T cells alone. Interestingly, the majority of the HIV-infected AMs were closely associated with T cells (CD68+ CD3+) and had HIV Gag staining within the macrophage cytosol (“CD68+ CD3+ with Gag in macrophage cytosol”; Fig. 5C and D). Some of these T cells were fully internalized into the macrophage, but other T cells were in close contact with the AM, suggesting that T cell internalization was not required for HIV Gag staining in the AM cytosol. HIV Gag was rarely observed only within the cytosol of T cells internalized by AMs without HIV Gag staining also in the macrophage cytosol (“CD68+ CD3+ Gag+ in T cell only”; Fig. 5D and Supplementary Fig. 6B). Both the SN and CTC conditions had a similar percentage of Gag+ AMs with no T cell staining (Fig. 5D, Supplementary Fig. 6C), which may be the result of archiving of HIV in endosomal compartments51. The percentage of Gag+ AMs with no T cell staining was the same on day 0 after 3 h of incubation and on day 14, supporting the conclusion that this staining in cell-free infection is due to phagocytosis or archiving (data not shown). These findings show that T-tropic HIV can be found in the AM cytosol after co-culture with infected T cells, suggesting that HIV is transmitted from an infected CD4+ T cell to the AM cytosol, and results in productive infection of AMs.
Discussion
While HIV nucleic acid has been detected in AMs during early stages of HIV infection, circulating HIV at this stage is primarily T-tropic and enters macrophages inefficiently in vitro. The tropism of HIV from AMs and the mechanism of HIV entry into AMs are both unknown, and may have a significant impact on the burden of pulmonary disease observed in those living with HIV. We studied AMs obtained by bronchoscopy in a cohort of ART-naive HIV-infected and uninfected individuals in Cape Town, South Africa and demonstrate that HIV RNA and DNA can be detected within AMs. We show that AM-derived HIV isolates are T-tropic and efficiently infect T cells but not macrophages; however, these isolates can productively infect macrophages through contact with infected CD4+ T cells. Our findings indicate that CD4+ T cell-dependent infection of AMs is a route of infection that may explain the presence of T-tropic HIV in AMs despite inefficient cell-free infection. This route of AM infection may be an important contributor to the burden of pulmonary disease observed in those living with HIV infection.
A few characteristics of the study population are notable, including sensitization to Mycobacterium tuberculosis and frequency of detection of HIV nucleic acids. The volunteers in the study were all sensitized to Mycobacterium tuberculosis, which could potentially increase the ability of AMs to be infected by HIV, including increased replication of HIV at sites of TB infection52,53,54,55. This reflects the high rates of latent tuberculosis infection seen in this population. However, the detection frequency of HIV Gag RNA and DNA were consistent with other studies, including those performed in non-TB sensitized populations. We observed HIV Gag RNA in AMs from 36.4% of participants and Gag DNA in AMs from 45.5% of participants, which is consistent with studies using qPCR to detect HIV nucleic acids in AMs18,20,21,40,46,47,48,49,50.
To better understand the phenotype of HIV derived from AMs, we generated replication-competent strains of HIV using env isolated from individuals with untreated HIV infection. We found that 8 separate AM-derived viruses and 9 plasma-derived viruses, selected to maximize sequence diversity, all had a T-tropic phenotype. This suggests that T-tropic HIV is infecting AMs in vivo despite inefficient infection of macrophages by these isolates in vitro. We used PCR-based methods to amplify virus, which could result in HIV recombination within the samples. However, the consistency of the entry phenotype across separate AM-derived Env constructs suggests that the tropism of macrophage-derived Env is more broadly applicable. In addition, the adherence method of isolating AMs from BAL, while resulting in 99–100% macrophage purity by RapidDiff stain, does not allow us to rule out a contribution of residual infected T cells to the HIV we detected and isolated. This method has been used in recent prior literature measuring HIV in AMs22,56 and led to minimal T cell inclusion in our co-culture studies, but could have resulted in leftover HIV-infected CD4+ T cells. A third caveat is that cell-free virus occasionally replicated over time in the cell-free conditions, although more rarely than with cell-to-cell contact. This phenomenon suggests that cell-free infection of MDMs by this T-tropic virus is possible but is less efficient than cell-to-cell infection. M-tropic HIV strains have 30-fold more efficient entry into macrophages than T-tropic strains do23, which is consistent with the idea that T-tropic strains occasionally enter MDMs.
Our data suggests that the p24 increase in AM-T cell co-cultures is due to productive infection of AMs. We cannot rule out new infection of CD4+ T cells that remained in the co-cultures after three rounds of washing. Treatment of these co-cultures with the reverse transcriptase inhibitor efavirenz inhibited viral replication. This shows that new viral production originated from cells infected after co-culture of CD4+ T cells and macrophages, and did not come from viral release from residual infected cells. It is possible that the newly infected cells are CD4+ T cells. However, in our microscopy staining, CD4+ T cells were at low prevalence (Fig. 5B), and most of the HIV Gag staining was localized to macrophages (Fig. 5D). Our work indicates that cell-to-cell infection of macrophages may be an important mechanism of T-tropic viral entry into AMs in the acute and chronic stages of infection.
While dendritic cells, alveolar epithelial cells, CD8+ T cells, NK cells and neutrophils may impact HIV infection dynamics in the alveolar space, either directly by acting as targets for infection or indirectly through cytokine secretion57, the main cells infected with HIV in the lung in vivo are CD4+ T cells and alveolar macrophages58,59.
A number of possibilities may explain the mechanism of the observed enhancement. The formation of an immunological and/or virological synapse, which involves the clustering of immune receptors and HIV virions at the interface between an AM and CD4+ T cell, may help overcome the low density of HIV entry receptors on AMs44,60,61. Fusion of the infected CD4+ T cell with the macrophage is another proposed mechanism of HIV infection of macrophages45, although we did not observe multinucleated giant cells or macrophages with surface CD3 expression, suggesting that this may not be occurring in our system with high frequency. It is also possible that autologous CD4+ T cells may activate AMs and increase HIV transcription62, as has been shown in dendritic cells63. In the CTC condition, we observed AMs with cytosolic Gag staining with T cells closely apposed to the membrane or internalized. This implies that AM phagocytosis of T cells occurs but may not be required for transmission of HIV Gag to the AM. Future work is needed to clarify this mechanism and understand its implications for HIV compartmentalization within AMs.
Infected AMs have been observed both in ART-naïve and ART-treated individuals with HIV9,16,17,18,19,20,21,22,64,65. ART may be able to inhibit HIV infection of new AMs in vivo as well as in vitro, as suggested by the ability of efavirenz to inhibit productive MDM infection (Supplementary Fig. 3A). However, HIV-infected macrophages are known to persist for long periods because they are more resistant to the cytopathic effects of HIV35, the cytotoxic activity of CD8+ T cells66, and apoptosis67,68,69. Infected AMs may persist during ART treatment and may contribute to the dysfunction22,69 either directly or through indirect effects of HIV components or other factors on bystander AMs70. Persistent AM dysfunction may contribute to the increased rate of lung inflammation and infectious disease seen in people with HIV on ART71,72,73,74. An understanding of the mechanism behind AM infection may help efforts to address persistent sources of inflammation in the lung in people with HIV on ART.
ART reverses some but not all of the functional phagocytic defects in alveolar macrophages, which may be due to a differential impact of ART on infected and bystander AMs75. Direct infection of macrophages can activate immune pathways including type I interferons that affect bystander function76,77 and chemokines which recruit other immune cells66,78. A number of papers have explored mechanisms of macrophage functional impairment in people with HIV on ART that are not dependent on direct infection, which include gp120-induced inhibition of apoptosis69, nef-induced inhibition of phagocytosis8,79 and downregulation of CD3680, as well as post-translational modification of Fc receptors81 and changes in reactive oxygen species generation82. Notably, HIV proteins including gp120 and nef persist in bronchoalveolar lavage fluid in people with HIV on ART69,83. Finally, data from ART-naïve macaques shows that only about 1 in 20,000–100,000 AMs is productively infected with SIV84, so HIV-infected AMs likely only make up a small minority of AMs in the lung in ART-naïve or treated individuals. The mechanisms of HIV-mediated AM impairment, even in the absence of ART, are most likely attributable to an indirect effect of either HIV or soluble factors produced by infected AMs on uninfected AMs.
The finding that HIV isolated from AMs is T-tropic has important implications for our understanding of the HIV reservoir. The vast majority of HIV strains, including transmitted/founder strains, are T-tropic25,33. If T-tropic HIV is able to infect AMs, the majority of people with HIV have the potential to have infected AMs, as well as infected macrophages in other tissues. This has implications for macrophages as a reservoir for HIV. It also suggests that tissue macrophages may need to be targeted in cure strategies.
The potential for AMs to be productively infected by contact with an infected CD4+ T cell intermediate has several important implications for our understanding of HIV pathogenesis. Productive HIV infection has been shown to have many effects on macrophage function, including the generation of defects in phagocytosis, proteolysis, and cytokine production9. Additionally, infected macrophages may efficiently transmit HIV to other CD4+ T cells and macrophages, as has been shown for M-tropic HIV, through efferocytosis of infected macrophages or cell-to-cell contact61,85. HIV-infected macrophages are resistant to cell death and in non-human primates can harbor replication-competent SIV after ART65, leading to a cellular reservoir that may play a role in the failure of current attempts at HIV cure. Finally, HIV infection in macrophages via contact with T cells may play a role in the chronic immune activation that is seen with HIV infection. Multiple studies have associated macrophage-derived products with increased morbidity and mortality during HIV infection, including those on ART. These include IL-6, soluble CD14 (sCD14), soluble tumor necrosis factor receptor 1 (sTNFR1), sTNFR2, and indoleamine 2,3-dioxygenase (IDO) activity86,87,88,89. Therefore, AMs infected with T-tropic HIV via T cell contact may lead to multiple immune defects which may contribute to an increased incidence of lung disease and promotion of HIV disease progression in people living with HIV.
Methods
Human participants
Bronchoscopies were performed in a cohort of participants with HIV infection and uninfected control individuals residing in Cape Town, South Africa, as previously described in90. All participants in the cohort were sensitized to Mycobacterium tuberculosis as defined by positive results of an interferon γ release assay (IGRA; Quantiferon, Cellestis), without active TB as defined by absence of signs or symptoms of active TB, lack of clinical findings by chest X-ray, and a negative BAL TB culture. All volunteers who had previously been diagnosed with or treated for TB were excluded. The HIV-infected participants in the Cape Town cohort were asymptomatic and not using antiretroviral therapy. All HIV + IGRA + volunteers were eligible for INH prophylaxis according to South African National guidelines, and were offered isoniazid prophylactic treatment for 6 months, which was the indication at the time. All samples were taken prior to INH treatment.
For in vitro experiments, BAL was also collected from a cohort of HIV-uninfected individuals, recruited from outpatient clinics at local Boston hospitals, following institutional review board approval (IRB protocol # 2013P000063) and written informed consent. For HIV infection experiments involving monocyte derived macrophages (MDMs), cells were isolated from buffy coats of anonymous healthy blood donors obtained from the Massachusetts General Hospital (MGH) blood donor center (Boston, MA) under protocol # 2005P001218.
Bronchoalveolar lavage (BAL)
Bronchoscopies were performed under conscious sedation via standard technique15,90. For each BAL in the Cape Town cohort, samples were collected from the right middle lobe by washing with 160 ml of normal saline. After adherence, approximately 10,000 cells were centrifuged onto a coated microscope slide using a Cytospin Centrifuge (Shandon 3.0). The slide was then allowed to dry, dipped repeatedly in fixative and differentially stained by dipping the slide consecutively in two contrasting dyes (RapidDiff Kit, Clinical Sciences Diagnostics). The differential stain allows characterization of lymphocytes, macrophages and neutrophils, based on morphology. The slide was then viewed under immersion oil magnification using a light microscope and a total of 200 cells were counted. The BAL samples consisted of a median of 96% and 93% macrophages prior to adherence, in the HIV+ and HIV- BAL samples, respectively. This was enriched further by adherence for 20 min in order to eliminate the non-adherent cell fraction.
For the Boston cohort, samples were collected from the lingula and the right middle lobe by washing 120 ml of normal saline in each segment. Alveolar macrophages (AMs) from bronchoscopy participants were isolated by 20–60 min of adherence. AMs for in vitro infection were washed 3 times with PBS and cultured in RPMI with 10% (v/v) FCS. BAL CD4+ T cells were obtained at bronchoscopy of ART-naive uninfected participants using a Human CD4+ T cell Enrichment kit (StemCell EasySep) on non-adherent BAL cells.
Cell isolation from blood
Peripheral blood from the bronchoscopy study participants recruited in Boston was obtained at least 7 days before the bronchoscopy and PBMCs were processed and cryopreserved. Ficoll gradients were used to isolate peripheral blood mononuclear cells (PBMCs). Monocytes and CD4+ T cells were then isolated by CD14+ positive selection (Miltenyi) and CD4+ T cell negative selection (StemCell EasySep), respectively. MDMs were obtained by maturing monocytes in RPMI with 10% GemCell US Origin Human Serum AB (GemBio) for 7 days.
Measurement of HIV nucleic acids
For measurement of HIV RNA and DNA, AMs were lysed in RLT Plus lysis buffer (QIAGEN) with 1% β-mercaptoethanol (Sigma). The lysate was run through QIAShredder columns (QIAGEN), and isolated with AllPrep Micro DNA/RNA kits (QIAGEN). HIV Gag primers were used to obtain the number of copies of HIV RNA and DNA, and CCR5 primers were used to determine the number of cells per sample using TaqMan qRT-PCR (Applied Biosystems)91. Standard curves were prepared using preamplification of a purified restriction digested HxB2 plasmid for HIV gag, kindly provided by Todd Allen and Karen Power, and by preamplification of a CCR5 plasmid91.
HIV production
Cells were infected with CCR5-utilizing T-tropic (JR-CSF) or CCR5/CXCR4-utilizing M-tropic (89.6) HIV constructs. JR-CSF was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HIV JR-CSF Infectious Molecular Clone (pYK-JRCSF) (Cat# 2708) from Dr. Irvin SY Chen and Dr. Yoshio Koyanagi92,93,94; 89.6 was obtained as above as the HIV 89.6 Infectious Molecular Clone (p89.6) from Ronald G. Collman, MD (cat# 3552)95,96,97. For quantification of entry, the JR-CSF and 89.6 viruses were produced with the BLaM-Vpr reporter plasmid98, kindly provided by Blandine Monel. Production of HIV was performed by transfecting viral plasmids with Hilymax (Dojindo) into HEK293T cells (ATCC), changing media after 12 h, harvesting cell culture supernatant after 48 h, and concentrating 50 times with PEG-it (System Biosciences). Supernatants were aliquoted and stored at − 80 °C until further usage. Titers of HIV infectious particles/ml were determined by infection of CD4, CCR5 and CXCR4-expressing GHOST cells (NIH AIDS Research and Reference Reagent Program)99.
HIV entry assay
The HIV entry assay was performed by infecting 100,000 BAL CD4+ T cells, AMs or MDMs with 105 infectious units of BLaM-Vpr HIV in a polypropylene FACS tube in a total volume of 100 μl after 1 h of pretreatment with DMSO or 40 μM maraviroc (MVC, Sigma). The cells were incubated at 37 °C for 12 h, then washed three times. Staining was performed by adding CCF2-AM along with Fixable Viability Dye eFluor 780 (eBioscience) to the cells and assaying for cleavage by flow cytometry98.
HIV infection of CD4+ T cells and macrophages
For cell-free infection experiments, 2 × 104 infectious units of HIV, or for mock infection, media, were added to 105 cells (MDMs, T cells or AMs) in 100 μl total volume for 12 h at 37 °C, then washed three times with PBS and replaced with 200 μl media. Supernatant was collected for p24 measurement at days 1 (1 h after changing media), 2, 7, and 14. CD4 + T cell cultures had low viability at day 14, consistent with prior literature100, so samples were not collected at this point, and day 7 was the final timepoint. HIV p24 levels were measured by p24 ELISA (PerkinElmer).
Co-culture of HIV infected CD4+ T cells and macrophages
For co-culture experiments with HIV-infected T cells, CD4+ T cells from matched donors were activated for 3 days with 2 ng/ml phytohemagglutinin (PHA-P) (ThermoFisher) and 10 ng/ml IL-2 (kindly provided by Alicja Trocha and the Ragon Institute Protein Core). 2 × 104 infectious units of HIV were added to 105 cells in each well and spin-infected by centrifugation at 800 ×g for 90 min at 4 °C; the cells were cultured for 12 h, washed and the media was replaced15. On day 4 after infection, autologous HIV- or mock-infected T cells were spun at 500 ×g for 3 min, the supernatant was collected, and the T cells were resuspended in fresh media. MDMs or AMs were cultured with one of three preparations for a 1:1 ratio of T cells to macrophages: supernatant collected from the CD4+ T cells (supernatant or “SN”), washed CD4+ T cells (cell-to-cell or “CTC”), or washed CD4+ T cells in the upper chamber of a 0.4 μm transwell (Transwell Costar) (transwell or “TW”). After incubation for 3 h at 37 °C, the cells were washed three times for 60 s with ice cold PBS with 10 mM EDTA, and media was replaced. Supernatant was collected for p24 measurement at days 0 (1 h after changing media), 2, 7 and 14 from the same wells with collection and replacement of 25% of the media volume. p24 levels were measured by ELISA (PerkinElmer). For experiments with AMs, autologous PBMCs were thawed 7 days before the bronchoscopy, and CD4+ T cells were isolated and activated for three days as above. In Fig. 4 and Supplementary Fig. 5, samples were only included in the final dataset if there was detectable HIV infection in the CD4+ T cells before they were added to the macrophages.
HIV primary isolate sequencing and production of primary isolates
Viral RNA was extracted from 140 μl of plasma and BAL fluid using a QIAamp viral RNA minikit (QIAGEN), and cDNA copies of the vRNA were generated by reverse transcription using the SuperScript III protocol (Invitrogen). Nested PCR was performed for the env gene as described in101, and amplicons were cloned into a pHDM vector using the In-Fusion HD Cloning kit (Clontech) and sequenced on an Illumina MiSeq.
Based upon analysis of env sequence diversity, 22 isolates were selected to represent the diversity of sequences from the 3 donors, with at least one sequence from each clade (arrows in Supplementary Fig. 2). pNL4-3 ΔEnv GFP was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HIV NL4-3 Δenv EGFP Reporter Vector from Drs. Haili Zhang, Yan Zhou, and Robert Siliciano (cat# 11100)102. The amplicons were PCR amplified from the pHDM vectors and cloned into the pNL4-3 vector. HIV stocks were produced, frozen and titered on GHOST cells as above. Experiments in Fig. 2 and Supplementary Fig. 3 used 2 AM-derived and 2 plasma-derived viral isolates from donor 1, 3 AM derived and 2 plasma-derived viral isolates from donor 2, and 3 AM derived and 5 plasma-derived viral isolates from donor 3, which are indicated by arrows with and without a black border in Supplementary Fig. 2. Experiments in Figs. 4 and 5 used one viral isolate from each viral donor and compartment, which are indicated by arrows without a black border.
Microscopy
Poly-L-lysine coated coverslips were used to culture MDMs and AMs with infected CD4+ T cells or supernatant from infected CD4+ T cells under SN or CTC conditions as described above. Cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% saponin, blocked with 1% BSA, and stained with antibodies against HIV Gag (conjugated to FITC, Beckman Coulter), CD3 (rabbit polyclonal, Dako), and CD68 (KP1, BioCare Medical), then Alexa Fluor 568 Goat anti-Rabbit and Alexa Fluor 647 Goat anti-Mouse (Invitrogen), then DAPI and mounted with ProLong Diamond mounting media (Thermo Fisher). Slides were scanned with a TissueFAXS SL Q spinning disc Confocal microscope (TissueGnostics USA) using a Zeiss Plan-apochromatic 63 × 1.4NA oil immersion objective. Image analysis involved counting 100 cells from each slide on a single Z-stack image and classifying each cell as CD68± , CD3± , and Gag±, and if Gag+ , whether it was present in CD68+ and/or CD3+ areas of the cell.
Quantification and statistical analysis
Statistical details of experiments can be found in each figure legend. Nonparametric tests were used to compare medians between groups unless noted otherwise. The Mann–Whitney test was used for 2 groups and the Kruskal–Wallis test followed by Dunn’s multiple comparison post-test was used for > 2 groups. Wilcoxon signed rank was used to compare continuous data between two time points. Spearman’s correlation coefficients were used to examine associations between variables. Differences were considered significant at p < 0.05. For figures marked “fold increase”, the value at the final timepoint was compared to the initial timepoint; rare values below 1 were normalized to the value of 1. Prism 8 (Graphpad) was used for all analyses15.
Ethical statement
All participants provided prior written informed consent. The study was conducted in accordance with the Declaration of Helsinki. The work involving bronchoalveolar lavage samples from Cape Town, South Africa was approved by the Research Ethics Committees of the University of Cape Town (REF158/2010) and Stellenbosch University (N10/08/275). All work involving material from human subjects performed in Boston, MA, USA was approved by the Institutional Review Board (IRB) at Massachusetts General Hospital (MGH). The work involving bronchoalveolar lavage samples from Boston, USA was approved by the Partners Human Research Committee (2013P002436), and the collection of blood samples for PBMC isolation was approved by the Partners Human Research Committee (2005P001218).
References
Afessa, B., Green, W., Chiao, J. & Frederick, W. Pulmonary complications of HIV infection: autopsy findings. Chest 113, 1225–1229 (1998).
Hirschtick, R. E. et al. Bacterial pneumonia in persons infected with the human immunodeficiency virus. Pulmonary complications of HIV infection study group. N. Engl. J. Med. 333, 845–851. https://doi.org/10.1056/NEJM199509283331305 (1995).
Sonnenberg, P. et al. How soon after infection with HIV does the risk of tuberculosis start to increase? A retrospective cohort study in South African gold miners. J. Infect. Dis. 191, 150–158. https://doi.org/10.1086/426827 (2005).
Murphy, J., Summer, R., Wilson, A. A., Kotton, D. N. & Fine, A. The prolonged life-span of alveolar macrophages. Am. J. Respir. Cell Mol. Biol. 38, 380–385. https://doi.org/10.1165/rcmb.2007-0224RC (2008).
Nayak, D. K. et al. Long-term persistence of donor alveolar macrophages in human lung transplant recipients that influences donor-specific immune responses. Am. J. Transplant. 16, 2300–2311. https://doi.org/10.1111/ajt.13819 (2016).
Kedzierska, K. et al. Defective phagocytosis by human monocyte/macrophages following HIV-1 infection: underlying mechanisms and modulation by adjunctive cytokine therapy. J. Clin. Virol. 26, 247–263 (2003).
Koziel, H. et al. Reduced binding and phagocytosis of Pneumocystis carinii by alveolar macrophages from persons infected with HIV-1 correlates with mannose receptor downregulation. J. Clin. Invest. 102, 1332–1344. https://doi.org/10.1172/JCI560 (1998).
Mazzolini, J. et al. Inhibition of phagocytosis in HIV-1-infected macrophages relies on Nef-dependent alteration of focal delivery of recycling compartments. Blood 115, 4226–4236. https://doi.org/10.1182/blood-2009-12-259473 (2010).
Jambo, K. C. et al. Small alveolar macrophages are infected preferentially by HIV and exhibit impaired phagocytic function. Mucosal Immunol. 7, 1116–1126. https://doi.org/10.1038/mi.2013.127 (2014).
Reardon, C. C., Kim, S. J., Wagner, R. P., Koziel, H. & Kornfeld, H. Phagocytosis and growth inhibition of Cryptococcus neoformans by human alveolar macrophages: effects of HIV-1 infection. AIDS 10, 613–618 (1996).
Kalsdorf, B. et al. Relationship between chemokine receptor expression, chemokine levels and HIV-1 replication in the lungs of persons exposed to Mycobacterium tuberculosis. Eur. J. Immunol. 43, 540–549. https://doi.org/10.1002/eji.201242804 (2013).
Wood, K. L. et al. Measurements of HIV viral loads from different levels of the respiratory tract. Chest 124, 536–542 (2003).
Barber, S. A. et al. Longitudinal analysis of simian immunodeficiency virus (SIV) replication in the lungs: compartmentalized regulation of SIV. J. Infect. Dis. 194, 931–938. https://doi.org/10.1086/507429 (2006).
Santangelo, P. J. et al. Whole-body immunoPET reveals active SIV dynamics in viremic and antiretroviral therapy-treated macaques. Nat. Methods 12, 427–432. https://doi.org/10.1038/nmeth.3320 (2015).
Corleis, B. et al. HIV-1 and SIV infection are associated with early loss of lung interstitial CD4+ T cells and dissemination of pulmonary tuberculosis. Cell Rep. 26, 1409–1418. https://doi.org/10.1016/j.celrep.2019.01.021 (2019).
Clarke, J. R. et al. HIV-1 proviral DNA copy number in peripheral blood leucocytes and bronchoalveolar lavage cells of AIDS patients. Clin Exp. Immunol. 96, 182–186 (1994).
Clarke, J. R., Krishnan, V., Bennett, J., Mitchell, D. M. & Jeffries, D. J. Detection of HIV-1 in human lung macrophages using the polymerase chain reaction. AIDS 4, 1133–1136 (1990).
Lebargy, F., Branellec, A., Deforges, L., Bignon, J. & Bernaudin, J. HIV-1 in human alveolar macrophages from infected patients is latent in vivo but replicates after in vitro stimulation. Am. J. Respir. Cell Mol. Biol. 10, 72–78 (1994).
Landay, A. L. et al. Detection of HIV-1 provirus in bronchoalveolar lavage cells by polymerase chain reaction. J. Acquir. Immune Defic. Syndr. 6, 171–175 (1993).
Nakata, K., Weiden, M., Harkin, T., Ho, D. D. & Rom, W. N. Low copy number and limited variability of proviral DNA in alveolar macrophages from HIV-q-infected patients: evidence for genetic differences in HIV-1 between lung and blood macrophage populations. Mol. Med. 1, 744–757 (1995).
Sierra-Madero, J. G. et al. Relationship between load of virus in alveolar macrophages from human immunodeficiency virus type 1-infected persons, production of cytokines, and clinical status. J. Infect. Dis. 169, 18–27 (1994).
Cribbs, S. K., Lennox, J., Caliendo, A. M., Brown, L. A. & Guidot, D. M. Healthy HIV-1-infected individuals on highly active antiretroviral therapy harbor HIV-1 in their alveolar macrophages. AIDS Res. Hum. Retrovir. 31, 64–70. https://doi.org/10.1089/AID.2014.0133 (2015).
Joseph, S. B. & Swanstrom, R. The evolution of HIV-1 entry phenotypes as a guide to changing target cells. J. Leukoc. Biol. 103, 421–431. https://doi.org/10.1002/JLB.2RI0517-200R (2018).
Arrildt, K. T. et al. Phenotypic correlates of HIV-1 macrophage tropism. J. Virol. 89, 11294–11311. https://doi.org/10.1128/JVI.00946-15 (2015).
Ochsenbauer, C. et al. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J. Virol. 86, 2715–2728. https://doi.org/10.1128/JVI.06157-11 (2012).
Brumme, Z. L. et al. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naive individuals. J. Infect. Dis. 192, 466–474 (2005).
Micci, L. et al. CD4 depletion in SIV-infected macaques results in macrophage and microglia infection with rapid turnover of infected cells. PLoS Pathog. 10, e1004467. https://doi.org/10.1371/journal.ppat.1004467 (2014).
DiNapoli, S. R., Hirsch, V. M. & Brenchley, J. M. Macrophages in progressive human immunodeficiency virus/simian immunodeficiency virus infections. J. Virol. 90, 7596–7606. https://doi.org/10.1128/JVI.00672-16 (2016).
Schnell, G., Joseph, S., Spudich, S., Price, R. W. & Swanstrom, R. HIV-1 replication in the central nervous system occurs in two distinct cell types. PLoS Pathog. 7, e1002286. https://doi.org/10.1371/journal.ppat.1002286 (2011).
Mattapallil, J. J. et al. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature 434, 1093–1097. https://doi.org/10.1038/nature03501 (2005).
Brenchley, J. M. et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200, 749–759. https://doi.org/10.1084/jem.20040874 (2004).
Campbell, J. H., Hearps, A. C., Martin, G. E., Williams, K. C. & Crowe, S. M. The importance of monocytes and macrophages in HIV pathogenesis, treatment, and cure. AIDS 28, 2175–2187. https://doi.org/10.1097/QAD.0000000000000408 (2014).
Joseph, S. B. et al. Quantification of entry phenotypes of macrophage-tropic HIV-1 across a wide range of CD4 densities. J. Virol. 88, 1858–1869. https://doi.org/10.1128/JVI.02477-13 (2014).
Lee, B., Sharron, M., Montaner, L. J., Weissman, D. & Doms, R. W. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. USA 96, 5215–5220 (1999).
Gartner, S. et al. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233, 215–219 (1986).
Xue, J. et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40, 274–288. https://doi.org/10.1016/j.immuni.2014.01.006 (2014).
Roberts, A. W. et al. Tissue-resident macrophages are locally programmed for silent clearance of apoptotic cells. Immunity 47, 913–927. https://doi.org/10.1016/j.immuni.2017.10.006 (2017).
Park, I. et al. CD4 receptor-dependent entry of human immunodeficiency virus type-1 env-Pseudotypes into CCR5-, CCR3, and CXCR4-expressing human alveolar macrophages is preferentially mediated by the CCR5 coreceptor. Am. J. Respir. Cell Mol. Biol. 20, 864–871 (1999).
Valentin, A., Trivedi, H., Lu, W., Kostrikis, L. G. & Pavlakis, G. N. CXCR4 mediates entry and productive infection of syncytia-inducing (X4) HIV-1 strains in primary macrophages. Virology 269, 294–304. https://doi.org/10.1006/viro.1999.0136 (2000).
Salahuddin, S. Z., Rose, R. M., Groopman, J. E., Markham, P. D. & Gallo, R. C. Human T lymphotropic virus type III infection of human alveolar macrophages. Blood 68, 281–284 (1986).
Honeycutt, J. B. et al. Macrophages sustain HIV replication in vivo independently of T cells. J. Clin. Invest. 126, 1353–1366. https://doi.org/10.1172/JCI84456 (2016).
Honeycutt, J. B. et al. HIV persistence in tissue macrophages of humanized myeloid-only mice during antiretroviral therapy. Nat. Med. 23, 638–643. https://doi.org/10.1038/nm.4319 (2017).
Calantone, N. et al. Tissue myeloid cells in SIV-infected primates acquire viral DNA through phagocytosis of infected T cells. Immunity 41, 493–502. https://doi.org/10.1016/j.immuni.2014.08.014 (2014).
Baxter, A. E. et al. Macrophage infection via selective capture of HIV-1-infected CD4+ T cells. Cell Host Microbe. 16, 711–721. https://doi.org/10.1016/j.chom.2014.10.010 (2014).
Bracq, L. et al. T cell-macrophage fusion triggers multinucleated giant cell formation for HIV-1 spreading. J. Virol. https://doi.org/10.1128/JVI.01237-17 (2017).
Jeffrey, A. A., Israel-Biet, D., Andrieu, J. M., Even, P. & Venet, A. HIV isolation from pulmonary cells derived from bronchoalveolar lavage. Clin. Exp. Immunol. 84, 488–492 (1991).
Koziel, H. et al. Enhanced in vivo human immunodeficiency virus-1 replication in the lungs of human immunodeficiency virus-infected persons with Pneumocystis carinii pneumonia. Am. J. Respir. Crit. Care Med. 160, 2048–2055. https://doi.org/10.1164/ajrccm.160.6.9902099 (1999).
Clarke, J. R. et al. HIV-1 proviral DNA copy number in peripheral blood leucocytes and bronchoalveolar lavage cells of AIDS patients. Clin. Exp. Immunol. 96, 182–186 (1994).
Chayt, K. J. et al. Detection of HTLV-III RNA in lungs of patients with AIDS and pulmonary involvement. JAMA 256, 2356–2359 (1986).
Lewin, S. R. et al. HIV-1 DNA and mRNA concentrations are similar in peripheral blood monocytes and alveolar macrophages in HIV-1-infected individuals. AIDS 12, 719–727 (1998).
Sharova, N., Swingler, C., Sharkey, M. & Stevenson, M. Macrophages archive HIV-1 virions for dissemination in trans. EMBO J. 24, 2481–2489. https://doi.org/10.1038/sj.emboj.7600707 (2005).
Toossi, Z., Nicolacakis, K., Xia, L., Ferrari, N. A. & Rich, E. A. Activation of latent HIV-1 by Mycobacterium tuberculosis and its purified protein derivative in alveolar macrophages from HIV-infected individuals in vitro. J. Acquir. Immune Defic. Syndr. Human Retrovirol. 15, 325–331. https://doi.org/10.1097/00042560-199708150-00001 (1997).
Toossi, Z. et al. Increased replication of HIV-1 at sites of Mycobacterium tuberculosis infection: potential mechanisms of viral activation. J. Acquir. Immune Defic. Syndr. 28, 1–8. https://doi.org/10.1097/00042560-200109010-00001 (2001).
Sun, J. et al. Protein phosphatase, Mg2+/Mn2+-dependent 1A controls the innate antiviral and antibacterial response of macrophages during HIV-1 and Mycobacterium tuberculosis infection. Oncotarget 7, 15394–15409. https://doi.org/10.18632/oncotarget.8190 (2016).
Benjamin, R. et al. Mycobacterial and HIV infections up-regulated human zinc finger protein 134, a novel positive regulator of HIV-1 LTR activity and viral propagation. PLoS ONE 9, e104908. https://doi.org/10.1371/journal.pone.0104908 (2014).
Hansen, E. C. et al. Diverse fates of uracilated HIV-1 DNA during infection of myeloid lineage cells. eLife https://doi.org/10.7554/eLife.18447 (2016).
Cribbs, S. K., Crothers, K. & Morris, A. Pathogenesis of HIV-related lung disease: immunity, infection, and inflammation. Physiol. Rev. 100, 603–632. https://doi.org/10.1152/physrev.00039.2018 (2020).
Charles, T. P. & Shellito, J. E. Human immunodeficiency virus infection and host defense in the lungs. Semin. Respir. Crit. Care Med. 37, 147–156. https://doi.org/10.1055/s-0036-1572553 (2016).
Iordanskiy, S., Santos, S. & Bukrinsky, M. Nature, nurture and HIV: the effect of producer cell on viral physiology. Virology 443, 208–213. https://doi.org/10.1016/j.virol.2013.05.023 (2013).
Kulpa, D. A. et al. The immunological synapse: the gateway to the HIV reservoir. Immunol. Rev. 254, 305–325. https://doi.org/10.1111/imr.12080 (2013).
Groot, F., Welsch, S. & Sattentau, Q. J. Efficient HIV-1 transmission from macrophages to T cells across transient virological synapses. Blood 111, 4660–4663. https://doi.org/10.1182/blood-2007-12-130070 (2008).
Hoshino, Y. et al. Maximal HIV-1 replication in alveolar macrophages during tuberculosis requires both lymphocyte contact and cytokines. J. Exp. Med. 195, 495–505 (2002).
Su, B. et al. Dendritic cell-lymphocyte cross talk downregulates host restriction factor SAMHD1 and stimulates HIV-1 replication in dendritic cells. J. Virol. 88, 5109–5121. https://doi.org/10.1128/JVI.03057-13 (2014).
DiNapoli, S. R. et al. Tissue-resident macrophages can contain replication-competent virus in antiretroviral-naïve, SIV-infected Asian macaques. JCI Insight 2, e91214. https://doi.org/10.1172/jci.insight.91214 (2017).
Abreu, C. M. et al. Infectious virus persists in CD4(+) T cells and macrophages in antiretroviral therapy-suppressed simian immunodeficiency virus-infected macaques. J. Virol. https://doi.org/10.1128/JVI.00065-19 (2019).
Clayton, K. L. et al. Resistance of HIV-infected macrophages to CD8(+) T lymphocyte-mediated killing drives activation of the immune system. Nat. Immunol. 19, 475–486. https://doi.org/10.1038/s41590-018-0085-3 (2018).
Busca, A., Saxena, M. & Kumar, A. Critical role for antiapoptotic Bcl-xL and Mcl-1 in human macrophage survival and cellular IAP1/2 (cIAP1/2) in resistance to HIV-Vpr-induced apoptosis. J. Biol. Chem. 287, 15118–15133. https://doi.org/10.1074/jbc.M111.312660 (2012).
Yuan, Z. et al. HIV-related proteins prolong macrophage survival through induction of Triggering receptor expressed on myeloid cells-1. Sci. Rep. 7, 42028. https://doi.org/10.1038/srep42028 (2017).
Collini, P. J. et al. HIV gp120 in the lungs of antiretroviral therapy-treated individuals impairs alveolar macrophage responses to Pneumococci. Am. J. Respir. Crit. Care Med. 197, 1604–1615. https://doi.org/10.1164/rccm.201708-1755OC (2018).
Le-Bury, G. & Niedergang, F. Defective phagocytic properties of hiv-infected macrophages: how might they be implicated in the development of invasive salmonella typhimurium?. Front. Immunol. 9, 531. https://doi.org/10.3389/fimmu.2018.00531 (2018).
Crothers, K. et al. HIV-associated lung infections and complications in the era of combination antiretroviral therapy. Proc. Am. Thorac. Soc. 8, 275–281. https://doi.org/10.1513/pats.201009-059WR (2011).
Fitzpatrick, M., Brooks, J. T. & Kaplan, J. E. Epidemiology of HIV-associated lung disease in the United States. Semin. Respir. Crit. Care Med. 37, 181–198. https://doi.org/10.1055/s-0036-1572556 (2016).
Stover, D. E. Approach to the HIV-infected patient with pulmonary symptoms. UpToDate (eds., J.G. Bartlett, T.E. King). https://www.uptodate.com/contents/approach-to-the-hiv-infected-patient-with-pulmonary-symptoms (2019).
Kruize, Z. & Kootstra, N. A. The role of macrophages in HIV-1 persistence and pathogenesis. Front. Microbiol. 10, 2828. https://doi.org/10.3389/fmicb.2019.02828 (2019).
Pugliese, A., Vidotto, V., Beltramo, T. & Torre, D. Phagocytic activity in human immunodeficiency virus type 1 infection. Clin. Diagn. Lab. Immunol. 12, 889–895. https://doi.org/10.1128/CDLI.12.8.889-895.2005 (2005).
Deshiere, A. et al. Global mapping of the macrophage-HIV-1 transcriptome reveals that productive infection induces remodeling of host cell DNA and chromatin. Sci. Rep. 7, 5238. https://doi.org/10.1038/s41598-017-05566-9 (2017).
Decalf, J. et al. Sensing of HIV-1 entry triggers a type I interferon response in human primary macrophages. J. Virol. https://doi.org/10.1128/JVI.00147-17 (2017).
Foley, J. F. et al. Roles for CXC chemokine ligands 10 and 11 in recruiting CD4+ T cells to HIV-1-infected monocyte-derived macrophages, dendritic cells, and lymph nodes. J. Immunol. 174, 4892–4900. https://doi.org/10.4049/jimmunol.174.8.4892 (2005).
Kedzierska, K. et al. nef-deleted HIV-1 inhibits phagocytosis by monocyte-derived macrophages in vitro but not by peripheral blood monocytes in vivo. AIDS 15, 945–955 (2001).
Olivetta, E. et al. HIV-1 Nef impairs key functional activities in human macrophages through CD36 downregulation. PLoS ONE 9, e93699. https://doi.org/10.1371/journal.pone.0093699 (2014).
Leeansyah, E., Wines, B. D., Crowe, S. M. & Jaworowski, A. The mechanism underlying defective Fcgamma receptor-mediated phagocytosis by HIV-1-infected human monocyte-derived macrophages. J. Immunol. 178, 1096–1104. https://doi.org/10.4049/jimmunol.178.2.1096 (2007).
Yeligar, S. M. et al. Dysregulation of alveolar macrophage PPARgamma, NADPH oxidases, and TGFbeta1 in otherwise healthy HIV-infected individuals. AIDS Res. Hum. Retrovir. 33, 1018–1026. https://doi.org/10.1089/AID.2016.0030 (2017).
Chelvanambi, S. et al. HIV-nef protein persists in the lungs of aviremic patients with HIV and induces endothelial cell death. Am. J. Respir. Cell Mol. Biol. 60, 357–366. https://doi.org/10.1165/rcmb.2018-0089OC (2019).
Avalos, C. R. et al. Quantitation of productively infected monocytes and macrophages of simian immunodeficiency virus-infected macaques. J. Virol. 90, 5643–5656. https://doi.org/10.1128/JVI.00290-16 (2016).
Martin, C. J. et al. Efferocytosis is an innate antibacterial mechanism. Cell Host Microbe 12, 289–300. https://doi.org/10.1016/j.chom.2012.06.010 (2012).
Hunt, P. W., Lee, S. A. & Siedner, M. J. Immunologic biomarkers, morbidity, and mortality in treated HIV infection. J. Infect. Dis. 214(Suppl 2), S44-50. https://doi.org/10.1093/infdis/jiw275 (2016).
Lien, E. et al. Elevated levels of serum-soluble CD14 in human immunodeficiency virus type 1 (HIV-1) infection: correlation to disease progression and clinical events. Blood 92, 2084–2092 (1998).
Maneglier, B. et al. Modulation of indoleamine-2,3-dioxygenase expression and activity by HIV-1 in human macrophages. Fundam. Clin. Pharmacol. 23, 573–581. https://doi.org/10.1111/j.1472-8206.2009.00703.x (2009).
Shive, C. L. et al. HIV-1 is not a major driver of increased plasma IL-6 levels in chronic HIV-1 disease. J. Acquir. Immune Defic. Syndr. 61, 145–152. https://doi.org/10.1097/QAI.0b013e31825ddbbf (2012).
Bunjun, R. et al. Effect of HIV on the frequency and number of mycobacterium tuberculosis-specific CD4+ T cells in blood and airways during latent M. tuberculosis infection. J. Infect. Dis. 216, 1550–1560. https://doi.org/10.1093/infdis/jix529 (2017).
Malnati, M. S. et al. A universal real-time PCR assay for the quantification of group-M HIV-1 proviral load. Nat. Protoc. 3, 1240–1248. https://doi.org/10.1038/nprot.2008.108 (2008).
Koyanagi, Y. et al. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science 236, 819–822 (1987).
Haltiner, M., Kempe, T. & Tjian, R. A novel strategy for constructing clustered point mutations. Nucleic Acids Res. 13, 1015–1025. https://doi.org/10.1093/nar/13.3.1015 (1985).
Cann, A. J. et al. Human immunodeficiency virus type 1 T-cell tropism is determined by events prior to provirus formation. J. Virol. 64, 4735–4742 (1990).
Collman, R. et al. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J. Virol. 66, 7517–7521 (1992).
Kim, F. M., Kolson, D. L., Balliet, J. W., Srinivasan, A. & Collman, R. G. V3-independent determinants of macrophage tropism in a primary human immunodeficiency virus type 1 isolate. J. Virol. 69, 1755–1761 (1995).
Doranz, B. J. et al. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85, 1149–1158 (1996).
Cavrois, M., Neidleman, J. & Greene, W. C. HIV-1 fusion assay. Bio. Protoc. 4, e1212. https://doi.org/10.21769/bioprotoc.1212 (2014).
Kwon, D. S., Gregorio, G., Bitton, N., Hendrickson, W. A. & Littman, D. R. DC-SIGN-mediated internalization of HIV is required for trans-enhancement of T cell infection. Immunity 16, 135–144 (2002).
Doitsh, G. et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 505, 509–514. https://doi.org/10.1038/nature12940 (2014).
Balazs, A. B. et al. Vectored immunoprophylaxis protects humanized mice from mucosal HIV transmission. Nat. Med. 20, 296–300. https://doi.org/10.1038/nm.3471 (2014).
Zhang, H. et al. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J. Virol. 78, 1718–1729. https://doi.org/10.1128/jvi.78.4.1718-1729.2004 (2004).
Acknowledgements
This work was supported in part by a National Institutes of Health Grant from the National Heart, Lung and Blood Institute, Award #U01HL121827 (to D.S.K. and B.D.M.) and F30HL134566-02 (to A.E.S.); from the National Institute of General Medical Sciences, Award #T32GM007753 (to A.E.S.); from the National Institute of Allergy and Infectious Diseases, Award #R01AI108538 (to A.M.T.); from the National Institute on Drug Abuse (NIDA) Avenir New Innovator Award #DP2DA040254 (to M.J.D. and A.B.B.); from the Burroughs Wellcome Fund (to D.S.K.); the MGH Transformative Scholars Program, the Charles H. Hood Foundation, and the Gilead Sciences Research Scholars Program in HIV (to M.J.D. and A.B.B.); the EDCTP2 program supported by the European Union (EU)’s Horizon 2020 program, #TMA2016SF-1535 (to W.A.B.); the South African National Research Foundation, Grants #96841 (to R.J.W.) and 86535 (to G.W.); the Wellcome Trust, Award #089832/Z/09/Z (to W.A.B.) and Awards #203135 and 104803 (to R.J.W.); the Francis Crick Institute, which receives support from Wellcome (FC0010218), Cancer Research UK (FC0010218), and UK Research and Innovation (FC0010218) (to R.J.W.) and separate Grants from Wellcome (FC0010218, 203135) (to R.J.W.); the European Union, Award #FP7-Health-F3-2012–305578 (to R.J.W.); and the Strategic Health Innovation Partnership, SAMRC (to R.J.W.) and the SATBBI grant (to G.W.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We would like to thank the Ragon Institute clinical team, including Daniel Worrall, Mamary Kone, Francis Ruzicka, and Sheila Ip, for recruiting and coordinating sample collection from the Boston bronchoscopy cohort, Antonella Lisanti-Park and Samantha Gates for assisting with sample processing, and Blandine Monel for kindly providing the BLaM-Vpr plasmid.
Author information
Authors and Affiliations
Contributions
Conceptualization, A.E.S., B.C., and D.S.K.; Methodology, A.E.S., S.B., and M.J.D.; Validation, A.E.S., S.N.L, A.H.L., and S.B.; Formal Analysis, A.E.S. and M.J.D.; Investigation, A.E.S., S.N.L., A.H.L., S.B., and M.J.D.; Resources, T.J.D., A.K.D., A.M.T., A.B.B., J.L.C., B.D.M., G.W., R.J.W., W.A.B., and D.S.K.; Data Curation, A.E.S., B.C., J.L.C., B.D.M., G.W., R.J.W., W.A.B., and M.J.D.; Writing—Original Draft, A.E.S., B.C., and D.S.K.; Writing—Review and Editing, A.E.S., S.B., A.K.D., J.L.C., B.D.M., G.W., R.J.W., W.A.B., B.C., and D.S.K.; Visualization, A.E.S., M.J.D., B.C., and D.S.K.; Supervision, B.C. and D.S.K.; Funding Acquisition, A.E.S., A.M.T., A.B.B., A.M.T., A.B.B., J.L.C., B.D.M., G.W., R.J.W., W.A.B., and D.S.K.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Schiff, A.E., Linder, A.H., Luhembo, S.N. et al. T cell-tropic HIV efficiently infects alveolar macrophages through contact with infected CD4+ T cells. Sci Rep 11, 3890 (2021). https://doi.org/10.1038/s41598-021-82066-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-82066-x
This article is cited by
-
HIV-1 Myeloid Reservoirs — Contributors to Viral Persistence and Pathogenesis
Current HIV/AIDS Reports (2024)
-
HIV-associated lung disease
Nature Reviews Disease Primers (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.