Abstract
Historical DNA analysis of archival samples has added new dimensions to population genetic studies, enabling spatiotemporal approaches for reconstructing population history and informing conservation management. Here we tested the efficacy of Genotyping-in-Thousands by sequencing (GT-seq) for collecting targeted single nucleotide polymorphism genotypic data from archival scale samples, and applied this approach to a study of kokanee salmon (Oncorhynchus nerka) in Kluane National Park and Reserve (KNPR; Yukon, Canada) that underwent a severe 12-year population decline followed by a rapid rebound. We genotyped archival scales sampled pre-crash and contemporary fin clips collected post-crash, revealing high coverage (> 90% average genotyping across all individuals) and low genotyping error (< 0.01% within-libraries, 0.60% among-libraries) despite the relatively poor quality of recovered DNA. We observed slight decreases in expected heterozygosity, allelic diversity, and effective population size post-crash, but none were significant, suggesting genetic diversity was retained despite the severe demographic contraction. Genotypic data also revealed the genetic distinctiveness of a now extirpated population just outside of KNPR, revealing biodiversity loss at the northern edge of the species distribution. More broadly, we demonstrated GT-seq as a valuable tool for collecting genome-wide data from archival samples to address basic questions in ecology and evolution, and inform applied research in wildlife conservation and fisheries management.
Similar content being viewed by others
Introduction
Historical DNA analysis of archival samples has added new dimensions to population genetic studies, expanding our ability to reconstruct patterns and processes of evolution across time and space1. Generally defined as preserved hard or soft tissues collected within the past 200 years, data from archival samples have enabled comparisons of genetic diversity and changes in effective population size between past and present populations1,2. Furthermore these data can be used to include extirpated populations or extinct species within phylogenetic reconstructions3,4,5.
Calcified material from fish, such as scales, otoliths, and various types of bone, have been collected over the past century and effectively used to investigate individual growth history6,7 and response to climate-, population-, and fishing-related pressures8,9,10,11. In addition, microchemical analyses of these samples have been used to infer fish habitat use12,13, origin14,15, and exposure to pollutants16,17, as well as to reconstruct diet and trophic structure18,19,20,21,22. Such material also provides opportunities for recovering historical DNA. For example, mitochondrial DNA obtained from archival scales has been used to determine the genetic effects of historical stocking of Atlantic salmon (Salmo salar) in northern Spanish rivers23, while microsatellite data from archival and contemporary pectoral fin samples allowed investigation of the temporal genetic consequences of river fragmentation for lake sturgeon (Acipenser fulvescens) in Ontario, Canada24. More recently, studies have employed single nucleotide polymorphism (SNP) genotyping of archival samples to examine temporal stability of Atlantic cod (Gadus morhua) at the northern range margin around Greenland25,26 and assess spatiotemporal genetic changes in Atlantic salmon populations across the Baltic Sea27. These studies and others have clearly demonstrated the value of historical DNA analysis and temporal population genomics for directly investigating population history and informing fisheries management.
While archival DNA holds great promise for fisheries management, such historical samples can pose challenges as they may contain low DNA quantity, poor DNA quality28,29, and a high percentage of exogenous DNA30. A range of approaches have been employed for successfully obtaining SNP genotypic data from archival fish samples, including TaqMan® genotyping assays31, Illumina GoldenGate assays25,26, SNP chips27, and shotgun whole genome re-sequencing32. One promising approach that has yet to be employed with archival samples is Genotyping-in-Thousands by sequencing (GT-seq)33. GT-seq is a multiplex amplicon sequencing method that uses species-specific primer probes to simultaneously target hundreds of loci for up to thousands of individuals, while also reducing amplification of non-target species DNA. Data collection can be conducted using a single library, making preparation simple and cost-effective compared to other methods34. Though amplification success can vary during multiplex PCR35, GT-seq has been found to amplify consistently with low genotyping error rates33. Moreover, GT-seq has been effective in obtaining genotypic data from low quality samples, such as those typically obtained through minimally or non-invasively collected starting materials36. Here, we test the effectiveness of GT-seq for genotyping archival samples using as a case study kokanee, the freshwater resident form of sockeye salmon (Oncorhynchus nerka), in Kluane National Park and Reserve (KNPR), Yukon, Canada.
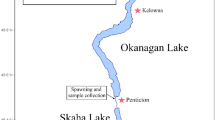
Kokanee in and around KNPR represent the northernmost wild populations documented in Canada (Fig. 1), which have experienced large population size fluctuations and extirpations over the past 50 years (Fig. 2). Kokanee within KNPR currently inhabit a connected set of waterbodies including Sockeye, Louise, and Kathleen (Mät’àtäna Män) Lakes (Fig. 1), with spawning in this system primarily occurring in Sockeye Creek and along the north shore of Sockeye Lake. An unconnected population that historically resided just outside of KNPR in Frederick Lake (Fig. 1) is believed to be extirpated since the late 1980’s37. Spawning numbers in KNPR have historically averaged ~ 3600 spawners each year, but a severe and prolonged population decline occurred between 2002–2014, reaching a low of just 20 observed spawners in 2009. The population rebounded in 2015 and 2016, with ~ 5550 spawners observed in those years, and has since fluctuated between ~ 400–3000 observed spawners annually (Fig. 2). The genetic consequences of this period of decline and subsequent rebound remain unknown. A recent study investigating the diversity, demographic history, and structure of the contemporary kokanee population in KNPR revealed a pattern of heterozygous excess, a possible signature of a past bottleneck event38. Population bottlenecks, in general, can lead to decreased genetic diversity and increased susceptibility to extirpation39,40. Genotypic data from archival samples collected pre-crash would allow for the direct investigation of the genetic consequences of population decline in this system.
Kokanee spawning counts in Kluane National Park and Reserve, 1973–2020 (Wong, pers. comm) illustrating the annual fluctuations in spawning numbers and severe period of decline from 2002–2014. Data were accessed under the Open Government Licence – Canada (https://open.canada.ca/data/en/dataset/4107dc81-79c0-406e-a1e6-301509d9e506).
Here, we employed GT-seq to collect targeted SNP genotypic data from population-level, archival scale samples in Sockeye, Kathleen, and Frederick Lakes collected pre-crash between 1973–1981 and analyzed them relative to data from the contemporary Sockeye Lake post-crash population as well as from a current hatchery stock historically sourced in KNPR. We used the resulting genotypic data to: (1) assess the effectiveness of GT-seq for genotyping archival scales; (2) quantify pre- and post-crash genetic diversity in KNPR; and (3) investigate spatial and temporal population structure in KNPR and Frederick Lake.
Methods
Sample collection and DNA extraction
Archival kokanee scale samples from Sockeye Lake were collected July 26–28, 1973 (n = 50) and July 15–August 26, 1975 (n = 13) as a part of a limnological survey of KNPR41. Samples from Kathleen Lake were also collected July 13–22, 1973 (n = 22) during the same survey, and during subsequent creel surveys conducted May 26–June 22, 1980 (n = 18), and June 1 – July 23, 1981 (n = 47)42. Samples from spawning kokanee in Frederick Lake were collected by angling July 30–31, 1981 (n = 12)42. All samples were dried and stored in envelopes. Although carefully labelled and stored, these samples were largely forgotten until unearthed at the KNPR Warden Office, Yukon, Canada, during an office move in 2012 (Wong, pers. comm.). Whole genomic DNA from dry scale samples was extracted following the protocol in43 within a dedicated historical DNA laboratory at the University of British Columbia Okanagan.
Previously extracted genomic DNA38 from Sockeye Lake and Sockeye Creek kokanee (total n = 46) originally sampled August 25–30, 2019 was used to represent the post-crash population. Also included was previously extracted kokanee DNA from the Whitehorse Rapids Fish Hatchery (n = 29) sampled August 31–September 18, 201938; this population was originally established from eggs and milt collected in Sockeye Creek in 1991–1994, 1999 and 2000 (Wong, pers. comm., LaRocque, pers. comm.). Genomic DNA from two archival and two contemporary samples were subject to Genomic DNA ScreenTape® analysis on an Agilent Tapestation 4150 to assess DNA quality and quantity.
GT‐seq library preparation and genotyping
GT-seq libraries were constructed following33 as modified in36. Within library (n = 7 within plates, n = 4 between plates) and between library (n = 3) duplicates were included to allow for estimation of genotyping error rates. Each individual was prepared in two panels using separate sets of previously designed primer pools targeting ~ 100 base pair fragments, one including 288 SNPs44 and the other containing 342 SNPs45. PCR1 products were diluted 1:20 before use in PCR2. PCR2 products were quantified using PicoGreen™ (Molecular Probes, Inc.), normalized manually to the concentration of the sample with the lowest concentration, and pooled. Pooled samples were purified using MinElute PCR Purification columns (Qiagen) and eluted into 24 μL nuclease-free water. Libraries were sequenced using a Mid Output Reagent Kit (300 cycles) on an Illumina MiniSeq within the Ecological and Conservation Genomics Laboratory at the University of British Columbia Okanagan.
Raw sequence data were genotyped using the GT-seq pipeline (https://github.com/GT-seq/GT-seq-Pipeline). Individuals and SNP loci with > 30% missing data were removed using PLINK v1.90b6.1746. Monomorphic loci and those that had been previously identified as outlier loci44,45 were removed using VCFTOOLS47. Forty loci identified as duplicates between the two panels were also removed.
Population genetic analyses
Observed (Ho) and expected heterozygosity (He), effective number of alleles (Ae), and Weir and Cockerham’s θ (999 permutations)48 were estimated as implemented in GenoDive49. Effective population size (Ne) was estimated using the linkage disequilibrium method as implemented in NeEstimator v.250; Ne was only calculated for populations with n ≥ 40, as the linkage disequilibrium method provides the most reliable results the closer that sample sizes approximate the true Ne51. Population structure was visualized using Principal Component Analyses (PCA) as implemented in PCADAPT52; the number of principal components (PC) retained was identified using a graphical approach based on the scree plot53 as recommended by52. The Bayesian clustering approach implemented in STRUCTURE v2.354 was used to infer population structure. Run length was set to 500,000 MCMC (Markov chain Monte Carlo) iterations following a burn-in of 100,000 using correlated allele frequencies under a straight admixture model. STRUCTURE was run with the number of clusters (K) varying between 1–9, with 10 replicates for each value of K. The most likely K was chosen by plotting the log probability (ln Pr(X|K)) of the data across the K values and choosing the value at which ln Pr(X|K) leveled and variance was minimized as recommended in54. Bar plots were generated and visualized using CLUMPAK55.
Results
Sample quality and GT-seq genotypic data processing
Genomic DNA from archival samples were of similar average quantity (archival = 31.5 ng/μL, contemporary = 37.5 ng/μL), but substantially poorer quality, compared to DNA from contemporary samples based on Genomic DNA ScreenTape® analysis (Fig. S1). Average genotyping of loci before filtering was 90.1% across all individuals. After filtering and the removal of outlier loci, monomorphic loci, and duplicate samples, a total of 271 loci were retained across 223 individuals, constituting our neutral dataset. Average read depth was 222.2 reads per loci. Retained individuals included those from Sockeye Lake 1973 (n = 49), Sockeye Lake 1975 (n = 10), Sockeye Lake 2019 (n = 40), Kathleen Lake 1973 (n = 21), Kathleen Lake 1980 (n = 17), Kathleen Lake 1981 (n = 47), Frederick Lake 1981 (n = 11), and the Whitehorse Rapids Fish Hatchery 2019 (n = 28). Genotyping error rates were < 0.01% within GT-seq library (both within and between plates) and 0.60% between GT-seq libraries. A total of 158 loci across 68 contemporary individuals (Sockeye Lake and Hatchery 2019) overlapped between the GT-seq and RADseq data38. The genotype discordance between these methods at these loci was 1.77%.
Population diversity and differentiation
Diversity metrics (Ho, He, Ae) from the archival samples were similar and stable across locations and sampling years in Sockeye (1973, 1975) and Kathleen Lakes (1973, 1980, 1981) (Table 1). A slight decrease in He and Ae was observed in the Sockeye Lake post-crash population (Table 1). There was a ~ 30% reduction in Ne in Sockeye Lake post-crash [2019: 86.8 (71.4–109.5)] compared to the pre-crash population [1973: 129.0 (104.1–166.8)], although confidence intervals overlapped (Table 1). Differentiation was low between sampling years in Sockeye and Kathleen Lakes, both pre-crash (θ ≤ 0.009), and pre-crash compared to post-crash (θ ≤ 0.011) (Table 2).
Frederick Lake exhibited substantially lower diversity metrics (Table 1) and significantly higher differentiation (θ > 0.450) from Sockeye and Kathleen Lakes (Table 2). Hatchery individuals also had lower observed heterozygosity than detected in both Sockeye and Kathleen Lakes across sampling years, though still higher than Frederick Lake (Table 1). The hatchery population was significantly differentiated from the Frederick Lake population (θ = 0.476; Table 2) and exhibited substantially higher θ values in comparisons with Sockeye and Kathleen Lakes across all sampling years (Table 2).
Two PCs were retained from the PCADAPT analysis of all populations across all sampling years; PC1 explained 12.5% of the genetic variation, while PC2 explained 3.7% (Fig. 3a). PC1 separated Frederick Lake from the rest of the individuals, while PC2 separated hatchery individuals from those sampled in Sockeye and Kathleen Lakes. When hatchery and Frederick Lake individuals were removed, there was no clear optimal number of PCs, so two were chosen for the purpose of comparison; both PCs explained very little of the genetic variation (PC1 = = 2.9%, PC2 = 2.7%). In this reduced analysis, there was no clear clustering by year or location for Sockeye or Kathleen Lake samples (Fig. 3b).
(a) PCA for individuals in Sockeye Lake, Kathleen Lake, Frederick Lake, and the Whitehorse Rapids Fish Hatchery (neutral, 271 SNPs) (PC1 = 12.5%, PC2 = 3.7%, genetic variation explained), showing a clear separation of Frederick Lake individuals and a partial separation of hatchery individuals from KNPR individuals. (b) PCA for individuals in Sockeye and Kathleen Lakes (neutral, 271 SNPs) (PC1 = 2.9%, PC2 = 2.7%, genetic variation explained) revealing no clear clustering by year or location.
Bayesian clustering analysis of the wild populations implemented in STRUCTURE found evidence for K = 2 (Table S1), separating the Frederick Lake population from those sampled in Sockeye and Kathleen Lakes, regardless of sampling year (Fig. 4); additional values of K did not reveal further structure within or among Sockeye and Kathleen Lakes, or among sampling years (Fig. 4).
STRUCTURE plots for K = 2–4 showing proportion of cluster membership for each individual in Frederick, Kathleen, and Sockeye Lakes (neutral, 271 SNPs). Bayesian clustering analysis of the wild populations implemented in STRUCTURE found evidence for K = 2 (Table S1), separating the Frederick Lake population from those sampled in Sockeye and Kathleen Lakes. No structure was found temporally or spatially in or between Sockeye and Kathleen Lakes.
Discussion
Here we demonstrated GT-seq to be an effective approach for genotyping archival scale samples, exhibiting high coverage (> 90% average genotyping of loci before filtering across all individuals) and low genotyping error (< 0.01% within libraries, 0.60% between libraries) despite the substantially poorer quality of recovered DNA (Fig. S1). These data allowed for the spatial and temporal comparison of diversity metrics and population structure of kokanee in KNPR and surrounding areas, while showing the value of pairing GT-seq and archival DNA analysis more broadly for informing conservation strategies.
Spatial/temporal population structure and comparison of diversity metrics in KNPR
Overall, we found no evidence of population structure spatially or temporally among the connected lakes where kokanee reside in KNPR (Kathleen and Sockeye), as revealed by extremely low pairwise θ values (Table 2) and lack of separation in the PCA or Bayesian clustering analysis (Figs. 3 and 4). The absence of differentiation between Kathleen and Sockeye Lakes is consistent with observations that all kokanee in KNPR, regardless of where they live during other life stages, eventually spawn in the same locations in Sockeye Lake and Creek.
Diversity metrics (Ho, He, Ae) were similar temporally and spatially in Sockeye and Kathleen Lakes in pre-crash years, with a slight decrease in He and Ae observed in the post-crash population. There was a ~ 30% reduction in Ne in the post-crash population compared to the pre-crash population in Sockeye Lake, although there was some overlap of confidence intervals (Table 1). This finding, as well as the temporal clustering and lack of differentiation between pre- and post-crash populations, suggests that a large proportion of genetic diversity was retained in KNPR kokanee despite the well-documented population crash.
Several factors may affect the genetic outcomes of a demographic contraction, including the severity and duration of the bottleneck, the latter of which can have an outsized effect. Genetic theory predicts that the bulk of genetic diversity can be retained, even during a severe bottleneck, as long as it is not prolonged56,57,58,59. For example, populations of white-tailed eagles (Haliaeetus albicilla) in Europe and Peregrine falcons (Falco peregrinus) in North America underwent severe bottlenecks and rapid recoveries as a result of dichlorodiphenyl-trichloroethane (DDT)-containing pesticides and their subsequent bans. However, neither showed a significant loss of genetic diversity as a result of these demographic bottlenecks60,61. While these periods of decline lasted decades, the species’ long lifespans likely helped maintain this diversity; in the case of white-tailed eagles, the population bottleneck lasted 20–30 years, which was only equivalent to ~ 2 generations61. As kokanee in KNPR have a documented generation time of ~ 4–5 years42, it is likely the demographic contraction in this system only lasted ~ 2–3 generations. Though severe, this duration of the bottleneck may not have been long enough to significantly erode genetic diversity within the population.
Extirpated Frederick Lake
Our results revealed the extirpated Frederick Lake population to be genetically distinct from kokanee in KNPR, as evidenced by high levels of differentiation (θ > 0.450; Table 2) and patterns of clustering in the PCA and STRUCTURE analyses (Figs. 3A, 4). The two competing hypotheses of the origin of kokanee in KNPR and Frederick Lake have assumed that they originated from the same event, either by way of colonization from the (1) Alsek River or (2) Tatshenshini River to Klukshu Lake (Łu Ghą Män)42. Yet, the high differentiation between these populations may suggest that they originated from separate ancestral populations over multiple events. Further research that includes sampling of anadromous sockeye salmon populations in the Alsek River and Klushu Lake would be required to test the single/multiple origin hypotheses. Regardless, the extirpation of Frederick Lake kokanee represents a loss of biodiversity, which may be even more pronounced given its northerly distribution, as small, isolated populations at the periphery of a species range, may harbor unique traits not found in the core of a species range62,63.
Though it is difficult to conclude what led to the extirpation of the Frederick Lake population in the late 1980’s37, the population exhibited much lower diversity metrics in terms of Ho, He, and Ae than the population in KNPR, as well as the Whitehorse Rapids Fish Hatchery population (Table 1). There is general agreement that genetic diversity is vital to a population’s viability39,64. As a lack of diversity has been associated with the extinction or extirpation of many species or populations65,66, the low levels of genetic diversity seen in Frederick Lake kokanee may have contributed to the loss of this population; however, additional studies would be required to test this hypothesis.
Value of archival samples in conservation
Our study highlights the valuable insights archival samples can provide to fisheries conservation and management. For example, previous RADseq data showed the contemporary population in KNPR displayed a heterozygous excess38, which is one potential indicator of a genetic bottleneck67. Yet, genotypic data from archival samples in KNPR revealed that this heterozygous excess was likely a feature of the population, rather than a result of a recent bottleneck. If only contemporary data were available, conservation strategies, such as initiatives to propagate diversity through hatchery supplementation, may have been implemented, possibly in vain or with negative consequences38. That said, we did observe a slight decrease in some diversity metrics over time in KNPR, such as He and Ae, as well as a small, but not significant, reduction in Ne post-crash; these findings warrant continued monitoring to document trends, examine outcomes, and inform conservation planning moving forward.
While potential archival samples such as fish scales and otoliths are generally collected during surveys or studies, this material is sometimes only used to meet short-term objectives, with long-term preservation and storage given little priority. Although such samples are well preserved and recorded at some institutions, in others, standard operating procedures include retaining only the most recent five years of samples due to insufficient storage space68. Even where sufficient storage space exists, maintenance of archival material may be inconsistent or undervalued, leading to variable quality of samples69. In our case, the last remaining physical evidence of an assumed extirpated population was at risk of being discarded during an office move 40 years after collection. As the use of genomics in fisheries conservation and management continues to progress70,71, proper long-term storage and maintenance of archival samples, potentially through centralized storage facilities and collection management systems72,73, will be key in preserving their value for future studies.
Utility of GT-seq in studies using archival samples
Our study shows how GT-seq can be used to effectively and efficiently genotype degraded archival samples. Though diversity metrics in the historical GT-seq dataset were slightly lower than what was found using RADseq in the same contemporary individuals, similar trends, such as heterozygote excess, were observed in both38. These lower diversity metrics were most likely due to the inclusion of unique Frederick Lake individuals while calling SNPs rather than GT-seq itself. As a case in point, when Frederick Lake individuals were removed from the dataset, diversity metrics between the same individuals were similar between the RADseq and GT-seq datasets (Table 3).
While a range of methods have been used for genotyping archival DNA25,26,27,31,32, our study is the first to our knowledge to employ GT-seq, which has many advantages compared to other approaches. First, GT-seq only requires amplicon lengths of ~ 100 base pairs for effective genotyping33, positioning it as a suitable approach to apply to highly fragmented DNA that can be commonly obtained from archival samples of varying ages.
Second, GT-seq has a relatively low genotyping error rate33. High genotyping error is often a serious problem for historical samples, mainly due to nucleotide misincorporation during the amplification of archival DNA1,29. Estimates of genotyping error using more traditional methods, such as microsatellite fragment analysis, can range as high as 17–21%1,74,75. In our study there was a < 0.01% genotyping error rates within libraries and 0.60% error rate between libraries, while average genotyping of loci before filtering was 90.1% across all individuals, highlighting the ability of GT-seq to obtain high quality data with minimal error.
Third, GT-seq makes use of a straightforward protocol that is relatively cost-effective, allowing for the simultaneous genotyping of hundreds or thousands of individuals at hundreds of loci. Though panel design requires upfront development and investment, once optimized, multi-locus genotypes can be obtained for ~ $6.00 (USD) per sample34. When compared to RADseq, which costs ~ $30.00 per sample, or a targeted capture sequencing approach such as Rapture, which costs ~ $15.00 per sample, GT-seq is extremely cost-effective when processing hundreds or thousands of samples34. Moreover, GT-seq does not require highly specialized equipment for library preparation and uses simple scripts for bioinformatic processing of the recovered sequence data33,34. This reduces barriers to entry for research groups without advanced instrumentation or strong computational backgrounds to use this approach, while also standardizing methods across labs34.
Lastly, GT-seq can provide connectible data to monitor populations over time. Once designed, GT-seq panels can be employed by any lab or facility to directly target specific SNPs at specific locations in the genome. This is unlike more traditional markers, such as microsatellites, which are indirectly assayed using fragment analysis76. As such, GT-seq panels can be used not only as a means to compare genetic diversity and detect changes in effective population size and structure between populations past and present, but also to continually monitor these parameters into the future.
Taken together, these advantages make GT-seq a valuable tool for implementing genomics into conservation34. This approach has been previously shown to be effective for low quality DNA from minimally and non-invasively collected samples36. Here, we have demonstrated the utility of GT-seq for genotyping archival samples, further extending the temporal and spatial resolution this method can bring for addressing basic questions in ecology and evolution, as well as for informing applied research in wildlife conservation and fisheries management.
Data availability
All Illumina raw reads are available from the NCBI sequence read archive (BioProject ID: PRJNA769146). SNP genotypic data are deposited in DRYAD (https://doi.org/10.5061/dryad.qfttdz0j2).
References
Wandeler, P., Hoeck, P. E. & Keller, L. F. Back to the future: Museum specimens in population genetics. Trends Ecol. Evol. 22, 634–642 (2007).
Bi, K. et al. Unlocking the vault: Next-generation museum population genomics. Mol. Ecol. 22, 6018–6032 (2013).
Metcalf, J. L. et al. Historical stocking data and 19th century DNA reveal human-induced changes to native diversity and distribution of cutthroat trout. Mol. Ecol. 21, 5194–5207 (2012).
Mikheyev, A. S. et al. Museum genomics confirms that the Lord Howe Island stick insect survived extinction. Curr. Biol. 27, 3157–3161 (2017).
Poulakakis, N. et al. Historical DNA analysis reveals living descendants of an extinct species of Galápagos tortoise. Proc. Natl. Acad. Sci. 105, 15464–15469 (2008).
Farley, E. V. et al. Early marine growth in relation to marine-stage survival rates for Alaska sockeye salmon (Oncorhynchus nerka). Fish. Bull. 105, 121–130 (2007).
Pannella, G. Fish otoliths: daily growth layers and periodical patterns. Science 173, 1124–1127 (1971).
Matta, M. E., Black, B. A. & Wilderbuer, T. K. Climate-driven synchrony in otolith growth-increment chronologies for three Bering Sea flatfish species. Mar. Ecol. Prog. Ser. 413, 137–145 (2010).
Morrongiello, J. R., Sweetman, P. C. & Thresher, R. E. Fishing constrains phenotypic responses of marine fish to climate variability. J. Anim. Ecol. 88, 1645–1656 (2019).
Peyronnet, A., Friedland, K., Maoileidigh, N., Manning, M. & Poole, W. Links between patterns of marine growth and survival of Atlantic salmon Salmo salar, L. J. Fish Biol. 71, 684–700 (2007).
Smoliński, S. & Mirny, Z. Otolith biochronology as an indicator of marine fish responses to hydroclimatic conditions and ecosystem regime shifts. Ecol. Indic. 79, 286–294 (2017).
Brennan, S. R. et al. Shifting habitat mosaics and fish production across river basins. Science 364, 783–786 (2019).
Elliott, L. D., Ward, H. G. & Russello, M. A. Kokanee–sockeye salmon hybridization leads to intermediate morphology and resident life history: Implications for fisheries management. Can. J. Fish. Aquat. Sci. 77, 355–364 (2020).
Adey, E., Black, K., Sawyer, T., Shimmield, T. & Trueman, C. Scale microchemistry as a tool to investigate the origin of wild and farmed Salmo salar. Mar. Ecol. Prog. Ser. 390, 225–235 (2009).
Flem, B., Moen, V., Finne, T. E., Viljugrein, H. & Kristoffersen, A. B. Trace element composition of smolt scales from Atlantic salmon (Salmo salar L.), geographic variation between hatcheries. Fish. Res. 190, 183–196 (2017).
Limburg, K. E. et al. In search of the dead zone: Use of otoliths for tracking fish exposure to hypoxia. J. Mar. Syst. 141, 167–178 (2015).
López-Duarte, P. C. et al. Is exposure to Macondo oil reflected in the Otolith chemistry of marsh-resident fish?. PLoS ONE 11, e0162699 (2016).
Grønkjær, P. et al. Stable N and C isotopes in the organic matrix of fish otoliths: Validation of a new approach for studying spatial and temporal changes in the trophic structure of aquatic ecosystems. Can. J. Fish. Aquat. Sci. 70, 143–146 (2013).
MacKenzie, K. M. et al. Stable isotopes reveal age-dependent trophic level and spatial segregation during adult marine feeding in populations of salmon. ICES J. Mar. Sci. 69, 1637–1645 (2012).
Nonogaki, H., Nelson, J. A. & Patterson, W. P. Dietary histories of herbivorous loricariid catfishes: Evidence from δ 13 C values of otoliths. Environ. Biol. Fishes 78, 13–21 (2007).
Sirot, C. et al. Using otolith organic matter to detect diet shifts in Bardiella chrysoura, during a period of environmental changes. Mar. Ecol. Prog. Ser. 575, 137–152 (2017).
Trueman, C. N., MacKenzie, K. M. & Palmer, M. R. Stable isotopes reveal linkages between ocean climate, plankton community dynamics, and survival of two populations of Atlantic salmon (Salmo salar). ICES J. Mar. Sci. 69, 784–794 (2012).
Ciborowski, K. et al. Stocking may increase mitochondrial DNA diversity but fails to halt the decline of endangered Atlantic salmon populations. Conserv. Genet. 8, 1355–1367 (2007).
McDermid, J., Nienhuis, S., Al-Shamlih, M., Haxton, T. & Wilson, C. Evaluating the genetic consequences of river fragmentation in lake sturgeon (Acipenser fulvescens Rafinesque, 1817) populations. J. Appl. Ichthyol. 30, 1514–1523 (2014).
Therkildsen, N. O. et al. Spatiotemporal SNP analysis reveals pronounced biocomplexity at the northern range margin of Atlantic cod Gadus morhua. Evol. Appl. 6, 690–705 (2013).
Bonanomi, S. et al. Archived DNA reveals fisheries and climate induced collapse of a major fishery. Sci. Rep. 5, 1–8 (2015).
Östergren, J. et al. A century of genetic homogenization in Baltic salmon: Evidence from archival DNA. Proc. R. Soc. B 288, 20203147 (2021).
Hofreiter, M. & Shapiro, B. Ancient DNA: Methods and Protocols (Humana Press Incorporated, 2012).
Pääbo, S. et al. Genetic analyses from ancient DNA. Annu. Rev. Genet. 38, 645–679 (2004).
Carpenter, M. L. et al. Pulling out the 1%: Whole-genome capture for the targeted enrichment of ancient DNA sequencing libraries. Am. J. Hum. Genet. 93, 852–864 (2013).
Smith, M. J. et al. Multiplex preamplification PCR and microsatellite validation enables accurate single nucleotide polymorphism genotyping of historical fish scales. Mol. Ecol. Resour. 11, 268–277 (2011).
Pinsky, M. L. et al. Genomic stability through time despite decades of exploitation in cod on both sides of the Atlantic. Proc. Natl. Acad. Sci. 118, (2021).
Campbell, N. R., Harmon, S. A. & Narum, S. R. Genotyping-in-Thousands by sequencing (GT-seq): A cost effective SNP genotyping method based on custom amplicon sequencing. Mol. Ecol. Resour. 15, 855–867 (2015).
Meek, M. H. & Larson, W. A. The future is now: Amplicon sequencing and sequence capture usher in the conservation genomics era. Mol. Ecol. Resour. 19, 795–803 (2019).
Andrews, K. R., De Barba, M., Russello, M. A. & Waits, L. P. Advances in using non-invasive, archival, and environmental samples for population genomic studies. (2018).
Schmidt, D. A., Campbell, N. R., Govindarajulu, P., Larsen, K. W. & Russello, M. A. Genotyping-in-Thousands by sequencing (GT-seq) panel development and application to minimally invasive DNA samples to support studies in molecular ecology. Mol. Ecol. Resour. 20, 114–124 (2020).
Buzzell, T. (Knowledge K., Director of Heritage, Lands and Resources), Champagne and Aishihik First Nations. Kokanee spawning. (2020).
Setzke, C., Wong, C. & Russello, M. A. Genome-wide assessment of kokanee salmon stock diversity, population history and hatchery representation at the northern range margin. Conserv. Genet. (in press) https://doi.org/10.1007/s10592-021-01418-2.
Frankham, R. Genetics and extinction. Biol. Conserv. 126, 131–140 (2005).
Luikart, G., Allendorf, F., Cornuet, J. & Sherwin, W. Distortion of allele frequency distributions provides a test for recent population bottlenecks. J. Hered. 89, 238–247 (1998).
Wickstrom, R. Limnological survey of Kluane National Park. Can. Wildl. Serv. Rep. Parks Can. Winn. 5, 352 (1978).
Wickstrom, R. Creel census, spawning enumeration and other studies of kokanee of the Kathleen drainage, Kluane National Park, Yukon Territory. 146 (1982).
Jensen, E. L. et al. Temporal mitogenomics of the Galapagos giant tortoise from Pinzón reveals potential biases in population genetic inference. J. Hered. 109, 631–640 (2018).
Chang, S. L., Ward, H. G. & Russello, M. A. Genotyping-in-Thousands by sequencing panel development and application to inform kokanee salmon (Oncorhynchus nerka) fisheries management at multiple scales. PLoS ONE In press.
Chang, S. L., Ward, H. G. & Russello, M. A. Genotyping-in-Thousands by sequencing panel to monitor kokanee-sockeye salmon (Oncorhynchus nerka) introgressive hybridization associated with a long-term reintroduction program. Mol. Ecol. Resour. Submitted.
Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Weir, B. S. & Cockerham, C. C. Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370 (1984).
Meirmans, P. G. genodive version 3.0: Easy-to-use software for the analysis of genetic data of diploids and polyploids. Mol. Ecol. Resour. 20, 1126–1131 (2020).
Do, C. et al. NeEstimator v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour. 14, 209–214 (2014).
England, P. R., Cornuet, J.-M., Berthier, P., Tallmon, D. A. & Luikart, G. Estimating effective population size from linkage disequilibrium: Severe bias in small samples. Conserv. Genet. 7, 303 (2006).
Luu, K., Bazin, E. & Blum, M. G. B. pcadapt: an R package to perform genome scans for selection based on principal component analysis. Mol. Ecol. Resour. 17, 67–77 (2017).
Jackson, D. A. Stopping rules in principal components analysis: A comparison of heuristical and statistical approaches. Ecology 74, 2204–2214 (1993).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Kopelman, N. M., Mayzel, J., Jakobsson, M., Rosenberg, N. A. & Mayrose, I. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 15, 1179–1191 (2015).
England, P. R. et al. Effects of intense versus diffuse population bottlenecks on microsatellite genetic diversity and evolutionary potential. Conserv. Genet. 4, 595–604 (2003).
Maruyama, T. & Fuerst, P. A. Population bottlenecks and nonequilibrium models in population genetics. I. Allele numbers when populations evolve from zero variability. Genetics 108, 745–763 (1984).
Maruyama, T. & Fuerst, P. A. Population bottlenecks and nonequilibrium models in population genetics. II. Number of alleles in a small population that was formed by a recent bottleneck. Genetics 111, 675–689 (1985).
Nei, M., Maruyama, T. & Chakraborty, R. The bottleneck effect and genetic variability in populations. Evolution 29, 1–10 (1975).
Brown, J. W. et al. Appraisal of the consequences of the DDT-induced bottleneck on the level and geographic distribution of neutral genetic variation in Canadian peregrine falcons, Falco peregrinus. Mol. Ecol. 16, 327–343 (2007).
Hailer, F. et al. Bottlenecked but long-lived: high genetic diversity retained in white-tailed eagles upon recovery from population decline. Biol. Lett. 2, 316–319 (2006).
Allendorf, F. W. & Lesica, P. When are peripheral populations valuable for conservation?. Conserv. Biol. 9, 753–760 (1995).
Eckert, C., Samis, K. & Lougheed, S. Genetic variation across species’ geographical ranges: the central–marginal hypothesis and beyond. Mol. Ecol. 17, 1170–1188 (2008).
Markert, J. A. et al. Population genetic diversity and fitness in multiple environments. BMC Evol. Biol. 10, 205 (2010).
Menzies, B. R. et al. Limited genetic diversity preceded extinction of the Tasmanian tiger. PLoS ONE 7, e35433–e35433 (2012).
Spielman, D., Brook, B. W. & Frankham, R. Most species are not driven to extinction before genetic factors impact them. Proc. Natl. Acad. Sci. U. S. A. 101, 15261 (2004).
Cornuet, J. M. & Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144, 2001–2014 (1996).
Rivers, P. & Ardren, W. R. The value of archives. Fisheries 23, 6–9 (1998).
Vollmar, A., Macklin, J. A. & Ford, L. Natural history specimen digitization: challenges and concerns. Biodivers. Inform. 7, (2010).
Valenzuela-Quiñonez, F. How fisheries management can benefit from genomics?. Brief. Funct. Genomics 15, 352–357 (2016).
Price, M. H. H. et al. Genetics of century‐old fish scales reveal population patterns of decline. Conserv. Lett. 12, (2019).
Leadbetter, A. et al. A modular approach to cataloguing marine science data. Earth Sci. Inform. 13, 537–553 (2020).
Tray, E. et al. An open-source database model and collections management system for fish scale and otolith archives. Ecol. Inform. 59, 101115 (2020).
Nyström, V., Angerbjörn, A. & Dalén, L. Genetic consequences of a demographic bottleneck in the Scandinavian arctic fox. Oikos 114, 84–94 (2006).
Sefc, K. M., Payne, R. B. & Sorenson, M. D. Single base errors in PCR products from avian museum specimens and their effect on estimates of historical genetic diversity. Conserv. Genet. 8, 879–884 (2007).
Vieira, M. L. C., Santini, L., Diniz, A. L. & de Munhoz, C. F. Microsatellite markers: what they mean and why they are so useful. Genet. Mol. Biol. 39, 312–328 (2016).
Scott, W. & Crossman, E. Freshwater fishes of Canada. Bulletin 184 (1973).
Wong, C. Status of Ecological Integrity in Kluane National Park and Reserve 2017: Technical Compendium to the State of the Park Report (p. 66). Whitehorse, Yukon: Parks Canada. (2017).
Acknowledgements
We thank Champagne and Aishihik First Nations for taking care of the land where these kokanee travel. This work is part of a larger project undertaken by Parks Canada and Champagne and Aishihik First Nations with university partners aimed at the conservation of kokanee. We also thank Rolly Wickstrom, formerly Canadian Wildlife Service, and students, Clare Hawkin and Kim Beach, who worked with him collecting the historic samples in the 1970s and 1980s. Contemporary samples were collected by Parks Canada staff in Kluane National Park and Reserve and Lawrence Vano and Warren Kapaniuk at the Whitehorse Rapids Fish Hatchery. We thank Laura Grieve for generating Fig. 1 and Evon Hekkala and Danielle Schmidt for providing feedback on the manuscript. We are especially grateful to Lloyd Freese, formerly Parks Canada, who saw the value of the historic samples and kept them safely stored for decades. Funding for this work was provided by Parks Canada Conservation and Restoration Program Agreement # GC-1160 and the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant # RGPIN‐2019‐04621 to MAR.
Author information
Authors and Affiliations
Contributions
Conception: M.A.R. Study design: M.A.R., C.W., and C.S. Data collection: C.S. Data analysis: C.S. Interpretation of the data: C.S., M.A.R., and C.W. Drafting of the article: C.S. and M.A.R. Critical revision of the article for important intellectual content: C.S., M.A.R., and C.W.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Setzke, C., Wong, C. & Russello, M.A. Genotyping-in-Thousands by sequencing of archival fish scales reveals maintenance of genetic variation following a severe demographic contraction in kokanee salmon. Sci Rep 11, 22798 (2021). https://doi.org/10.1038/s41598-021-01958-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-01958-0
This article is cited by
-
Past population control biases interpretations of contemporary genetic data: implications for future invasive Sitka black-tailed deer management in Haida Gwaii
Biological Invasions (2023)
-
Genome-wide species delimitation analyses of a silverside fish species complex in central Mexico indicate taxonomic over-splitting
BMC Ecology and Evolution (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.