Abstract
Sucrose phosphate synthase (SPS), a key rate-limiting enzyme in the sucrose biosynthesis pathway in plants, is encoded by a multi-gene family. Until recently, the identification and characterization of the SPS gene family have been performed for dozens of plant species; however, few studies have involved a comprehensive analysis of the SPS family members in tropical crops, such as cassava (Manihot esculenta Crantz). In the current study, five SPS genes (MeSPS1, MeSPS2, MeSPS3, MeSPS4, and MeSPS5) were isolated from cassava, and their sequence characteristics were comprehensively characterized. These MeSPS genes were found distributed on five chromosomes (Chr2, Chr14, Chr15, Chr16, and Chr18). Phylogenetic analysis showed that the MeSPS protein sequences were clustered into three families, together with other SPS sequences from both dicot and monocot species (families A, B, and C). The spatio-temporal expression pattern analysis of MeSPS genes showed a tissue-specific and partially overlapping expression pattern, with the genes mainly expressed in source tissues during cassava growth and development. Correlation analysis revealed that the expression of MeSPS genes correlated positively with root starch content, indicating that the expression of MeSPS genes might accelerate the rate of starch accumulation in the roots of cassava plants.
Similar content being viewed by others
Introduction
Sucrose phosphate synthase (SPS; EC2.4.1.14) is a key rate-limiting enzyme in the sucrose biosynthesis pathway in plants1,2,3,4,5. SPS catalyses the reaction of uridine diphosphate glucose (UDPG) and fructose-6-phosphate to form sucrose-6-phosphate, which is subsequently dephosphorylated and hydrolyzed by sucrose-phosphate phosphatase (SPP; EC3.1.3.24), threreby releasing sucrose6,7,8. Previous studies demonstrated that SPS activity is significantly correlated with plant growth and development9,10,11, yield and quality of products12,13,14,15, and plant tolerance to abiotic stresses16,17. Further, the SPS protein was demonstrated to be encoded by a multi-gene family, with each family containing at least one distinct SPS isoform1,18. SPS genes present in dicotyledonous species are classified into three major families designated as families A, B, and C according to their phylogenetic evolution. In addition, two other subfamilies designated as subfamilies DIII and DIV) have been found in monocots19,20,21. The first SPS gene to cloned was obtained from maize22. To date, numerous distinct SPS isoforms have been identified and characterized for dozens of plant species, including Spinacia oleracea23, Saccharum officinarum24, Cucumis melo25, Ananas comosus26, Cerasus humilis27, and Litchi chinensis28 among others. Subsequently, the characterization and expression patterns of SPS gene sequences from these plant species have been explored29.
Cassava (Manihot esculenta Crantz) is a typical perennial crop of the Euphorbiaceae family. Owing to its starchy storage roots, cassava is one of the most important and widely cultivated staple food crops for feeding more than 800 million people in tropical and subtropical regions of Africa, Asia, South America, and the Pacific30. In cassava storage roots, sucrose is readily hydrolysed into fructose and UDPG, the latter being the immediate precursor of starch synthesis. To date, the physiological function of SPS isozyme was comprehensively illustrated in prior studies, but not yet about the molecular bases and expression patterns of SPS genes in cassava31,32. Thus, it is fundamental paramount for understanding of the SPS genes roles in regulating the sucrose biosynthesis and even in the starch accumulation pathway in cassava roots.
In the current study, members of the cassava SPS gene family were cloned and characterized from. Our study mainly focused on SPS gene cloning, chromosomal localization, exon–intron organization and structural analyses of conserved motifs. Additionally, we investigated the phylogenetic evolution and expression profile of these genes (in different tissues and at developmental stages). Further, the relationship between SPS gene expression and starch accumulation in the root was studied by the correlation analysis. Our results provide a solid foundation for a thorough understanding of the physiological role of SPS genes in regulating the starch accumulation in the storage roots of cassava.
Results
Cloning and sequence characteristics of MeSPS genes
Results of agarose gel electrophoresis showed that the target fragments were effectively amplified via real-time PCR (RT-PCR). The full-length gels are shown in Supplementary Fig. S1. Five SPS genes, hereafter designated as MeSPS1, MeSPS2, MeSPS3, MeSPS4, MeSPS4, and MeSPS5, were isolated from cassava. The characteristics of these five SPS genes are summarized in Table 1. The results showed that the open reading frames (ORF) of the isolated MeSPS genes ranged from 2805 to 3198 bp in the length, while the predicted molecular weights (Mw) for the encoded proteins ranged between 104.12 and 119.46 kDa (934–1065 amino acids, their nucleotide and amino acid sequences are listed in Supplementary Data S1–S2). The theoretical isoelectric point (pI) of the MeSPS proteins ranged from 5.81 to 6.76. Furthermore, in order to ascertain the genomic organization of MeSPS genes, their locations and coordinates on the cassava chromosomes were mapped, and they were found to be distributed on chromosomes Chr14, Chr16, Chr15, Chr18, and Chr2, respectively (Table1; Fig. 1).
Structural analysis of exons–introns in MeSPSs indicated that MeSPS1 contains thirteen exons and twelve introns, MeSPS2 includes eleven exons and ten introns, MeSPS3 has twelve exons and eleven introns, and MeSPS4 and MeSPS5 are each composed of fourteen exons and thirteen introns. The length of exons differed among MeSPS genes (Fig. 2). To further analyse the diversity of the MeSPS proteins, conserved motifs were predicted using the MEME program (http://meme-suite.org/tools/meme) and Toolbox for Biologists (TBtools version 0.6) software (http://www.tbtools.com/). We distributed 12 putative conserved motifs (referred to as motifs 1–12) in the same direction in all MeSPS protein sequences, furthermore, most of these conserved motifs were located within the C-terminal region (Fig. 3). The average length of these motifs was 50 amino acids, except for motifs 9, 10, and 11, which contained 49, 43, and 29 amino acid residues, respectively (the features of these motifs are listed in Supplementary Table S1).
Conserved motif distribution in cassava MeSPS proteins. Motifs were analyzed by the MEME program and TBtools v0.6 software (Supplementary Table S1). Conserved motifs 1–12 are marked by different colours. The lengths of proteins and motifs can be estimated using the scale at bottom.
Multiple alignment and phylogenetic analyses
Multiple alignment analysis of the identified MeSPS sequences showed that the isolated MeSPS genes share a high sequence identity at both nucleotide (46.45–80.81%) and amino acid (47.90–91.09%) levels (Table 2). Among them, MeSPS4 and MeSPS5 showed the highest homology levels, with 80.81% and 91.09% at nucleotide and amino acid levels, respectively, compared with the other paralogs. Moreover, analysis of conserved domains indicated that three conserved domains—the sucrose synthase, glucosyl-transferase and resemble SPP domains, which have been catalogued as typical plant SPS domains—were also detected in all isolated MeSPS genes, using the SMART (http://smart.embl-heidelberg.de/) and the InterPro (http://www.ebi.ac.uk/interpro/search/sequence/) databases (Fig. 4). These results led to the suggestion that the five isolated MeSPS genes encode different SPS isozymes in cassava.
Multiple alignments of MeSPS deduced amino acid sequences. The alignment was performed using the multiple alignment program of DNAMAN v7.0. Consensus residues are in black, residues that are ≥ 75% identical among the aligned sequences are in pink, and residues that are ≥ 50% identical among the aligned sequences are in blue. Three characteristic functional domains were marked out. Black dotted lines indicate Sucrose-synth domain. Solid line boxes indicate Glycosyl transferase domain and black underlines indicate SPP-resemble domain.
To further illustrate the evolutionary relationship between MeSPSs and SPSs from other plant species, a total number of 80 full-length amino acid sequences from 36 species (comprising dicots and monocots, amino acid sequences summed in Supplementary Table S2) were selected to construct an unrooted phylogenetic tree using the MEGA (version X.0) software (https://www.megasoftware.net/). As shown in Fig. 5, all SPS proteins were clearly divided into four major families, hereafter designated as A, B, C, and D, with 100% bootstrap values. Families A, B, and C consist of SPS protein sequences from dicots and monocots plant species, whereas family D comprised only monocotyledonous sequences. The distribution of the seven cassava SPS proteins was the following: MeSPS1 and MeSPS2 belong to family A, MeSPS3 was assigned to family B, and MeSPS4 and MeSPS5 were clustered in family C.
Phylogeny of SPS proteins from cassava and other plant species (abbreviations and sequences of other plant SPS proteins are summarized in Supplementary Table S2). Four SPS gene families are distinguished by different colours. Numbers in the branch nodes are bootstrap values. Black dots represent SPS proteins of cassava.
Expression patterns of MeSPS genes in different tissues and at developmental stages
In pursue of the physiological function of each gene in the MeSPS gene family, the spatio-temporal expression pattern of the isolated MeSPS genes were profiled in six cassava tissues—mature leaves (ML), the upper part of stem (US), the middle part of stem (MS), the lower part of stem (LS), tuber phloem (TP), and tuber xylem (TX)—at the root expansion stage (180 DAP, 2017) via real-time fluorescent quantitative PCR (RT-qPCR) analysis. Figure 6 indicated the expression pattern of MeSPS genes in each tissue. Overall, the expression of MeSPS genes revealed a tissue- and organ-specific pattern, where the five target genes showed a relatively low level of expression in US, MS, LS, TP, and TX, but had an abundant expression in the ML. Specifically, MeSPS3 revealed the highest level of expression in ML (i.e., more than 13-fold higher) compared with the other MeSPS genes. These findings suggested that cassava SPS genes were mainly expressed in source tissues rather than in transport or sink tissues.
The periodical variation of MeSPS expression in source (ML) and sink (TP and TX) tissues were also examined via RT-qPCR during four different growth periods: seedling stage (I), root formation stage (II), root expansion stage (III), and root maturation stage (IV) in 2017 and 2018. As shown in Fig. 7, all MeSPSs were relatively highly expressed (MeSPS3 reached near 40-fold) in source tissues (ML). These patterns are consistent with the expression profiles observed in a wide range of tissues (Fig. 6). Overall, MeSPS gene expression showed dynamic changes in both 2 years. In ML, MeSPS1 maintained a low and stab expression pattern during all growth stages in the 2 years. The expression patterns of MeSPS2 and MeSPS5 exhibited a similar trend (e.g., peaking at stage II and then gradually decreasing during the later stages of growth). In turn, MeSPS3 was first up-regulated and peaked at stage II in 2017, and at stage III in 2018. Meanwhile, MeSPS4 first increased to a peak at stage III and then gradually decreased at later stages in both 2 years (Fig. 7a,d). In TP, MeSPS1 showed a continuous down-regulation during the growth stages analysed, whereas MeSPS2 and MeSPS5 revealed a significant up-regulation to a peak at stage IV in both 2 years. In contrast, MeSPS3 was only weakly expressed across growth stages, and MeSPS4 was slightly up-regulated during the experimental period (Fig. 7b,e). Finally, as for TX, MeSPS1 and MeSPS2 showed a rapid increase from the beginning and peaked at stage IV in both 2 years. In turn, MeSPS3 showed a low level of expression in 2017, and a high expression level at stage IV in 2018. MeSPS4 was weakly expressed throughout the experimental period, while MeSPS5 reached a peak at stage II in 2017, and at stage III in 2018, but rapidly decreased at later stages in both years (Fig. 7c,f). Altogether, these results showed that none of the cassava tissues under study expressed only one MeSPS gene during cassava growth and development.
Physiological determination and correlation analysis
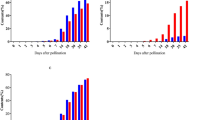
SPS activity and sucrose content in ML, SPS activity, sucrose content, and total starch content in TP and TX, were determined at four growth stages in 2017 and again in 2018. As shown in Fig. 8, SPS activity (Fig. 8a,d) and sucrose content (Fig. 8b,e) gradually increased in ML throughout growth in the two experimental years. Conversely, in TP, SPS activity in 2018 (Fig. 8d) and sucrose content (Fig. 8b,e) decreased gradually in both years. However, SPS activity in 2017 (Fig. 8a) increased gradually, and this trend was similar with that of the total starch content in both years (Fig. 8c,f). On the other hand, in TX, SPS activity in the 2 years (Fig. 8a,d) and sucrose content in 2018 (Fig. 8e) exhibited a similar behavior without any particular trend, whereas sucrose content gradually decreased in 2017 (Fig. 8b). While total starch content increased consistently throughout growth both in 2017 and 2018 (Fig. 8c,f).
Variation of SPS activity (a,d), sucrose content (b,e), and total starch content (c,f) in different tissues of cassava during growth. I, II, III, and IV represent seedling stage, root formation stage, root expansion stage, root maturation stage, respectively. ML, TP, and TX represent mature leaves, tuber phloem, and tuber xylem, respectively.
Correlation analysis was performed to determine the relationships among MeSPS gene expression level, sucrose biosynthesis and starch accumulation in ML, TP, and TX, respectively. Correlation coefficients are summarized in Table 3. Overall, all examined physiological indexes were significantly correlated with the expression level of one or more MeSPS genes in both years. Specifically, the expression level of MeSPS1 in TX was significantly and positively correlated with total root-starch content (both in TP and TX) both in 2017 and 2018. Similarly, the expression level of MeSPS2 and MeSPS5 in TX were positively correlated with total starch content in TX. Finally, the expression level of MeSPS3 in TP, and those of MeSPS3 and MeSPS4 in TX were also positively correlated with total starch content in TP. These findings clearly suggest that starch accumulation in the storage root of cassava might be promoted by the expression of MeSPS genes during development.
Discussion
SPS have been shown to play an important regulatory role in the sucrose biosynthesis pathway in higher plants. Recently, due to a growing number of high-quality whole genome sequences, various SPS genes from numerous plant species were cloned and classified. However, the prevalence and functional diversity of SPS gene family members are less well defined in tropical crops such as cassava, which is characterized by a high starch content in the roots. To our knowledge, this is the first report of gene structure, protein motifs, and protein multiple alignments of cassava SPS genes.
In the current work, five SPS isoforms were isolated from cassava, which were designated as MeSPS1–5, respectively. All MeSPS proteins harboured the glucosyl–transferase (N-terminal) and resemble the SPP (C-terminal) domains (Fig. 4), that allow the catalytic reactions in the sucrose biosynthesis pathway in higher plants20,33. The distribution of exons and introns varied among the MeSPS genes, which contained 11–14 exons and 10–13 introns (Fig. 2). Similar findings were reported for other crops, such as wheat8, maize20, and rice34, in which the number of exons and introns in the coding regions also ranged from 11 to 14, and from 10 to 13, respectively. These structures determined similar functions among SPS genes of dicot and monocot plant species distantly related in evolutionary history35,36,37.
A comprehensive phylogenetic analysis should help us gain novel insights into both the origins and evolutionary relationships among diverse members of gene family identified, as well as into their potential functions38. Based on the phylogenetic analysis of the five cassava MeSPS and other SPS proteins in this study, four major families were defined, among which, families A, B, and C include SPS proteins from dicots and monocots, whereas family D only includes monocot sequences (Fig. 5). These findings further corroborated SPS classifications previously reported, and lend support to the proposition that higher plant species may have at least three major families of SPS genes19,20,21. Interestingly, the phylogenetic tree constructed herein further revealed that most MeSPS proteins were more closely related to the proteins of RcSPS (R. communis), JcSPS (J. curcas), and HbSPS (H. brasiliensis), as these four species belong to the same Euphorbiaceae family. These findings are consistent with the conclusions by Bredeson et al.39.
In the recent decades, the expression of SPS genes has been comprehensively documented for a number of plant species, however, few detailed analyses have been conducted in cassava. In this study, the expression patterns of MeSPS genes in different cassava tissues and at different developmental stages were examined. The results showed that all MeSPS genes were mainly expressed in leaves, compared with other tissues, and there was at least one MeSPS gene predominantly expressed in source and sink tissues in both experimental years (Fig. 7), indicating that MeSPS genes exhibited a tissue-specific and partially overlapping expression pattern during cassava growth and development. Similar spatio-temporal expression of SPS genes in various other plants have been previously expounded. For instance, the expression of SPS genes from Citrus unshiu Marc., was found to be completely different in source and sink tissues. CitSPS1 was expressed to a higher extent in mature leaves and fruits than in young leaves, flowers, and immature fruits. In turn, CitSPS2 was expressed to a lower level in mature leaves than in flowers and mature fruits, whereas CitSPS3 was only detected in young and mature leaves40. Further, in rice, OsSPS1 was highly expressed in the youngest fully expanded leaves41, while PpSPS genes of peach (Prunus persica L.) were abundantly expressed in fruits rather than in leaves and phloem-enriched fractions during fruit development42. Lastly, in Cerasus humilis, ChSPS1 was expressed to higher levels in fruits than in leaves, and continuously increased during fruit development27. The temporal expression of MeSPS genes led us to hypothesize a functional collaboration between different SPS proteins in the metabolic pathways of cassava.
Recent reports demonstrated that SPS may participate in regulating carbon partitioning between starch and sucrose in source leaves22,43. For example, in tomato, SPS gene expression in transgenic plants revealed a specific role in starch mobilization, whereby diurnal starch content in NtSPSCi transgenic leaves was higher than in those of the wildtype44. SPS activity in cassava leaves was positively correlated with root starch content, implying that starch accumulation in the cassava root might be promoted by SPS activity35,45. Our results further support these observations. Indeed, we observed that the expression of one or more MeSPS genes was significantly and positively correlated with total starch content in the roots during development (Table 3), suggesting that the expression of MeSPS genes enhanced starch accumulation in the roots to a certain extent.
In conclusion, our study has expanded our understanding of the molecular bases and differential expression patterns of SPS genes, and their correlations with starch accumulation in cassava roots. However, the biological functions of these isoforms and their potential roles in sucrose transport and starch accumulation in cassava warrant further research.
Materials and methods
Materials
In the current study, cassava variety Radiation Selection 01(RS01) was used as plant experimental materials, as it is one of the most popular and widely cultivated cultivars in China because of its high root yield and high root starch content. All cassava seed-stems were cultivated at the experimental station of Guangxi University under conventional field management from March to December of 2017 and 2018. The three newest mature leaves (ML), from three healthy cassava plants of the same age, were sampled at 70 days after planting (DAP)—during the seedling stage, in 2017—for MeSPS gene cloning. To determine the physiological parameters and relative expression levels of MeSPS genes during cassava growth, ML and tubers from three healthy cassava plants were collected in 2 years at the seedling stage (70 DAP, except tubers), at the root formation stage (120 DAP), at the root expansion stage (180 DAP), and at the root maturation stage (210 DAP). Stem samples were also collected at 180 DAP for analysis of gene expression patterns in different cassava tissues in 2017; each stem sampled was separated into three parts (upper/top, middle and lower parts). Tubers were separated into two portions (phloem and xylem parts). All samples were immediately frozen in liquid nitrogen and stored at − 80 °C until used.
Cloning of MeSPS genes from Manihot esculenta
Total RNA from cassava leaves was extracted using the Quick Plant RNA Isolation Kit (Huayueyang) according to manufacturer instructions. First-strand cDNA was synthesized using the First Strand cDNA synthesis Kit (Takara), according to the protocol by the manufacturer, and then used for RT-PCR amplification using PrimeSTARMax DNA polymerase (Takara)46,47. All specific primers of target genes were designed using the Primer (version 5.0) software for cloning the ORFs based on the complete coding sequence (CDS) of the cassava genome (http://asia.ensembl.org/index.html) (primer sequences were listed in Supplementary Table S3).
The RT-PCR mixture was initially denatured at 94 °C for 5 min, and then ran for 35 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, elongation at 72 °C for 2.3 min with a final 7 min extension step at 72 °C. PCR products were detected by 1.2% agarose gel electrophoresis. Each PCR fragment was separately ligated into the pMD18-T cloning vector (Takara) and then transformed into E. coli DH5α competent cells to select positive clones.
Gene sequences analysis
Amino acid sequences of MeSPS genes were predicted by the online ORF finder tool of NCBI (https://www.ncbi.nlm.nih.gov/orffinder). The basic physical and chemical characteristics of these amino acid sequences, including protein length, molecular weight, and predicted isoelectric point, were calculated using the online ProtParam tool (http://www.expasy.org/tools/protparam.html). Multiple-sequence alignment of target amino acid sequences was performed by DNAMAN (version 7.0) software and conserved sequence domains were detected by the Simple Modular Architecture Research Tool (SMART, http://smart.embl-heidelberg.de/) and the InterPro database (http://www.ebi.ac.uk/interpro/search/sequence/)48. Analysis of exon–intron structures was visualized by the Gene Structure Display Server (GSDS) 2.0 program (http://gsds.cbi.pku.edu.cn/). Conserved motifs of MeSUS protein were analysed by the MEME program (http://meme-suite.org/tools/meme) and TBtools v0.6, http://www.tbtools.com/) software, according to the method of Bailey et al.49. The maximum number of motifs was set to 12, all other default parameters were normal. Chromosomal locations and coordinates of the MeSUS genes were mapped by the MG2C v2.0 program (http://mg2c.iask.in/mg2c_v2.0/) based on genomic information available from the cassava genome database (http://www.phytozome.net/cassava). The ClustalW algorithm was used to align the target protein sequences, and MEGA vX.0 software (https://www.megasoftware.net/), with the neighbour-joining (NJ) approach and 1000-replication bootstrap methods50, was used to construct the phylogenetic tree.
RT-qPCR analysis
Total RNA was extracted from each sample from each growth stages according to the protocol by the manufacturer (Huayueyang). The first-strand cDNA was synthesized using the instruction manual of First-Strand cDNA Synthesis Kit (Vazyme). Gene-specific primers for RT-qPCR were designed using the Primer 5.0 software (primer sequences were summed in Supplementary Table S3), using the housekeeping gene TBP-associated factor 15B (TAF15b) as an internal control for relative gene expression analysis51. The RT-qPCR reaction system contained 10 μl 2 × SYBR qPCR MasterMix (Vazyme), 2 μl template cDNA, 0.4 μl primers, and was made up to 20 μl with ddH2O. The amplification process was performed using the following protocol: 95 °C for 3 min, followed by 45 cycles at 95 °C for 10 s and 60 °C for 30 s. Three biological replicates were measured per reaction. Relative expression levels of the genes were calculated by the 2−ΔΔCt method. The error bar for each expression level was calculated based on the biological replicates using Microsoft Excel 2019.
Determination and correlation analysis of sucrose, starch contents and enzymatic activity
Sucrose and total starch contents were analysed according to the method described by Huang et al.52, while SPS activity was measured following the instruction manual of the Plant Enzyme-linked Immunosorbent Assay (ELISA) Kit (Jianglai). Three biological replicates were measured per sample.
Statistical analyses were performed using Pearson algorithm of IBM SPSS 19.0 (SPSS Science, Chicago, IL, USA) software. Correlation analysis was conducted between SPS activity, sucrose content, total starch content, and MeSPS expression levels at different developmental stages, in 2017, and 2018.
Data availability
All data generated or analysed during this study are included in this article and its supplementary information files.
References
Lunn, J. E. & MacRae, E. New complexities in the synthesis of sucrose. Curr. Opin. Plant Biol. 6, 208–214 (2003).
Strand, A. et al. Decreased expression of two key enzymes in the sucrose biosynthesis pathway, cytosolic fructose-1,6-bisphosphatase and sucrose phosphate synthase, has remarkably different consequences for photosynthetic carbon metabolism in transgenic Arabidopsis thaliana. Plant J. 23, 759–770 (2000).
Stitt, M., Lunn, J. & Usadel, B. Arabidopsis and primary photosynthetic metabolism-more than the icing on the cake. Plant J. 61, 1067–1091 (2010).
Seger, M. et al. Impact of concurrent overexpression of cytosolic glutamine synthetase (GS1) and sucrose phosphate synthase (SPS) on growth and development in transgenic tobacco. Planta 241, 69–81 (2015).
Chen, S. et al. Decreased sucrose-6-phosphate phosphatase level in transgenic tobacco inhibits photosynthesis, alters carbohydrate partitioning, and reduces growth. Planta 221, 479–492 (2005).
Huber, S. C. & Huber, J. L. Role and regulation of sucrose-phosphate synthase in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 431–444 (1996).
Winter, H. & Huber, S. Regulation of sucrose metabolism in higher plants: Localization and regulation of activity of key enzymes. Crit. Rev. Plant Sci. 19, 31–67 (2000).
Sharma, S. et al. Delineating the structural, functional and evolutionary relationships of sucrose phosphate synthase gene family II in wheat and related grasses. BMC Plant Biol. 10, 134 (2010).
Prioul, J. L. et al. From QTLs for enzyme activity to candidate genes in maize. J. Exp. Bot. 50, 1281–1288 (1999).
Whittaker, A. et al. Sucrose phosphate synthase activity and the co-ordination of carbon partitioning during sucrose and amino acid accumulation in desiccation-tolerant leaf material of the C4 resurrection plant Sporobolus stapfianus during dehydration. J. Exp. Bot. 58, 3775–3787 (2007).
Rocher, J. P. et al. Genetic variability in carbon fixation, sucrose-p-synthase and ADP glucose pyrophosphorylase in maize plants of differing growth rate. Plant Physiol. 89, 416–420 (1989).
Ishimaru, K., Ono, K. & Kashiwagi, T. Identification of a new gene controlling plant height in rice using the candidate-gene strategy. Planta 218, 388–395 (2004).
Tian, H. M. et al. Antisense repression of sucrose phosphate synthase in transgenic muskmelon alters plant growth and fruit development. Biochem. Biophys. Res. Commun. 393, 365–370 (2010).
Babb, V. M. & Haigler, C. H. Sucrose phosphate synthase activity rises in correlation with high-rate cellulose synthesis in three heterotrophic systems. Plant Physiol. 127, 1234–1242 (2001).
Haigler, C. H. et al. Transgenic cotton over-producing spinach sucrose phosphate synthase showed enhanced leaf sucrose synthesis and improved fiber quality under controlled environmental conditions. Plant Mol. Biol. 63, 815–832 (2007).
Zúñiga-Feest, A. et al. Light regulation of sucrose–phosphate synthase activity in the freezing-tolerant grass Deschampsia Antarctica. Photosynth. Res. 83, 75–86 (2005).
Yang, J., Zhang, J. & Wang, Z. Activities of starch hydrolytic enzymes and sucrose–phosphate synthase in the stems of rice subjected to water stress during grain filling. J. Exp. Bot. 52, 2169–2179 (2001).
Okamura, M. et al. Tissue specificity and diurnal change in gene expression of the sucrose phosphate synthase gene family in rice. Plant Sci. 181, 159–166 (2011).
Langenkämper, G. et al. Sucrose phosphate synthase genes in plants belong to three different families. J. Mol. Evol. 54, 322–332 (2002).
Castleden, C. K. et al. Evolution and function of the sucrose phosphate synthase gene families in wheat and other grasses. Plant Physiol. 135, 1753–1764 (2004).
Lutfiyya, L. L. et al. Phylogenetic and expression analysis of sucrose phosphate synthase isozymes in plants. J. Plant Physiol. 164, 923–933 (2007).
Worrell, A. C. et al. Expression of maize sucrose–phosphate synthase in tomato alters carbohydrate partitioning. Plant Cell. 3, 1121–1130 (1991).
Klein, R. R., Crafts-Brandner, S. J. & Salvucci, M. E. Cloning and developmental expression of the sucrose–phosphate–synthase gene from spinach. Planta 190, 498–510 (1993).
Sugiharto, B., Sakakibara, H., Sumadi, & Sugiyama, B. Differential expression of two genes for sucrose–phosphate synthase in sugarcane: Molecular cloning of the cDNAs and comparative analysis of gene expression. Plant Cell Physiol. 38, 961–965 (1997).
Wen, X. X., Zhang, W. Q., Feng, Y. Q. & Yu, X. Y. Cloning and characterization of a sucrose synthase-encoding gene from Muskmelon. Mol. Biol. Rep. 37, 695–702 (2010).
Zhang, X. M. et al. Cloning and expression of pineapple sucrose–phosphate synthase gene during fruit development. Afr. J. Biotechnol. 9, 8296–8303 (2010).
Wang, J., Du, J. J., Mu, X. P. & Wang, P. F. Cloning and characterization of the Cerasus humilis sucrose phosphate synthase gene (ChSPS1). PLoS One 12, e0186650. https://doi.org/10.1371/journal.pone.0186650 (2017).
Wang, D. et al. Identification and expression profile analysis of the sucrose phosphate synthase gene family in Litchi chinensis Sonn. Peer J. 6, e4379. https://doi.org/10.7717/peerj.4379 (2018).
McIntyre, C. L. et al. The identification and characterisation of alleles of sucrose phosphate synthase gene family III in sugarcane. Mol. Breed. 18, 39–50 (2006).
Adeyemo, O. S. et al. Overexpression of Arabidopsis FLOWERING LOCUST(FT) gene improves floral development in cassava (Manihot esculenta, Crantz). PLoS One 12, e0181460. https://doi.org/10.1371/journal.pone.0181460 (2017).
Luo, X. L., Pan, X. L. & Zhu, Y. M. The relationship between endogenous abscisic acid content and starch accumulation in root tuber of cassava. Chin. J. Trop. Crops. 39, 472–479 (2018).
Netrphan, S. et al. Differential expression of genes involved in sucrose synthesis in source and sink organs of cassava plants undergoing seasonal drought Stress. J. Agric. Sci. 4, 171–185 (2012).
Lunn, J. E. et al. Purification, molecular cloning, and sequence analysis of sucrose–6F-phosphate phosphohydrolase from plants. Proc. Natl. Acad. Sci. USA 97, 12914–12919 (2000).
Hirose, T. et al. Analysis of gene-disruption mutants of a sucrose phosphate synthase gene in rice, OsSPS1, shows the importance of sucrose synthesis in pollen germination. Plant Sci. 225, 102–106 (2014).
Frugoli, J. A. et al. Intron loss and gain during evolution of the catalase gene family in angiosperms. Genetics 149, 355–365 (1998).
Lecharny, A. et al. Introns in, introns out in plant gene families: A genomic approach of the dynamics of gene structure. J. Struct. Funct. Genom. 3, 111–116 (2003).
Hu, L. F. & Liu, S. Q. Genome-wide identification and phylogenetic analysis of the ERF gene family in cucumbers. Genet. Mol. Biol. 34, 624–633 (2011).
Bredeson, J. V. et al. Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nat. Biotechnol. 34, 562–570 (2016).
Wang, Z. et al. Analysis of the sucrose synthase gene family in tobacco: Structure, phylogeny, and expression patterns. Planta 242, 153–166 (2015).
Komatsu, A., Takanokura, Y., Omura, M. & Akihama, T. Cloning and molecular analysis of cDNAs encoding three sucrose phosphate synthase isoforms from citrus fruit (Citrus unshiu Marc.). Mol. Genet. Genom. 252, 346–351 (1996).
Chávez-Bárcenas, A. T. et al. Tissue-specific and developmental pattern of expression of the rice SPS1 Gene. Plant Physiol. 124, 641–653 (2000).
Zhang, C. H. et al. Cloning and expression of genes related to the sucrose metabolizing enzymes and carbohydrate changes in peach. Acta Physiol. Plantarum. 35, 589–602 (2013).
Huber, S. C. & Huber, J. L. Role of sucrose–phosphate synthase in sucrose metabolism in leaves. Plant Physiol. 99, 1275–1278 (1992).
Chen, S., Hajirezaei, M. & Börnke, F. Differential expression of sucrose–phosphate synthase Isoenzymes in tobacco reflects their functional specialization during dark-governed starch mobilization in source leaves. Plant Physiol. 139, 1163–1174 (2005).
Cheng, Y. E. et al. Activities of related enzymes associated with high starch accumulation in cassava tuberous roots. Genom. Appl. Biol. 36, 376–381 (2017).
Yamamoto, K. et al. A novel TRB@/NOTCH1 fusion gene in T-cell lymphoblastic lymphoma with t (7; 9) (q34; q34). Eur. J. Haematol. 90, 68–75 (2013).
Yamanaka, K. et al. Rapid detection for primary screening of influenza a virus: Microfluidic RT-PCR chip and electrochemical DNA sensor. Analyst 136, 2064–2068 (2011).
Jones, P. et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014).
Bailey, T. L. et al. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic. Acids. Res. 34, W369–W373 (2006).
Kumar, S. et al. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Hyunjoo, E. et al. TAF15b, involved in the autonomous pathway for flowering, represses transcription of FLOWERING LOCUS C. Plant J. 93, 79–91 (2018).
Huang, T. W. et al. Comparison of growth vigor and leaf physiological features among different cassava varieties during seedling stage. J. South. Agric. 48, 51–56 (2017).
Acknowledgements
This work was supported by the National Grand Fundamental Research 937 Program (no. 2010CB126601), Open Sciences Fund Project of State Key Laboratory for Conservation and Utilization of subtropical Agro-Bioresources (nos. SKLCOSA-b201609, SKLCUSA-b201704, SKLCUSA-a201802), Guangxi Science and Technology Plan Project (no. AB18221127), the Key Project of the Natural Sciences Fund of Guangxi (no. 2010GXNSFD013025) and Innovation Project of Guangxi Graduate Education (no. YCBZ2018013).
Author information
Authors and Affiliations
Contributions
X.L. designed, initiated, and supervised the experiments. T.H., Z.S., Y.Z., Y.Y. and Z.F. performed the experiments and collected the data. T.H. analysed the data and drafted the manuscript. M.W. revised the manuscript. All authors read and approved the final manuscript for submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Huang, T., Luo, X., Wei, M. et al. Molecular cloning and expression analysis of sucrose phosphate synthase genes in cassava (Manihot esculenta Crantz). Sci Rep 10, 20707 (2020). https://doi.org/10.1038/s41598-020-77669-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-77669-9
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.