Abstract
Deleted in lung and esophageal cancer 1 (DLEC1) is a tumour suppressor gene that is downregulated in various cancers in humans; however, the physiological and molecular functions of DLEC1 are still unclear. This study investigated the critical role of Dlec1 in spermatogenesis and male fertility in mice. Dlec1 was significantly expressed in testes, with dominant expression in germ cells. We disrupted Dlec1 in mice and analysed its function in spermatogenesis and male fertility. Dlec1 deletion caused male infertility due to impaired spermatogenesis. Spermatogenesis progressed normally to step 8 spermatids in Dlec1−/− mice, but in elongating spermatids, we observed head deformation, a shortened tail, and abnormal manchette organization. These phenotypes were similar to those of various intraflagellar transport (IFT)-associated gene-deficient sperm. In addition, DLEC1 interacted with tailless complex polypeptide 1 ring complex (TRiC) and Bardet–Biedl Syndrome (BBS) protein complex subunits, as well as α- and β-tubulin. DLEC1 expression also enhanced primary cilia formation and cilia length in A549 lung adenocarcinoma cells. These findings suggest that DLEC1 is a possible regulator of IFT and plays an essential role in sperm head and tail formation in mice.
Similar content being viewed by others
Introduction
Spermatogenesis is a complex process in which spermatogonial stem cells differentiate into haploid sperm1. Spermatogonia proliferate by mitosis and differentiate into spermatocytes, which undergo two consecutive meiotic divisions to produce round spermatids. These round spermatids further undergo dynamic morphological and structural changes, such as elongation of the nucleus, condensation of chromatin, and formation of a flagellum, to form mature sperm. Many genes expressed in a spatially and temporally specific manner regulate these changes2,3. However, the molecular function of each gene is still unclear.
In the later phase of spermatogenesis, two microtubule-based structures, axoneme and manchette, are involved in sperm flagella formation and head shaping4. The axoneme is a core structure in the flagellum composed of nine outer doublet microtubules and two central microtubules (9 + 2 structure) attached with many structural components, such as an inner dynein arm (IDA), an outer dynein arm (ODA), a radial spoke, and the nexin–dynein regulatory complex (N-DRC)5. This structure is highly conserved in the motile cilia of somatic cells, except in nodal cilia, which have a 9 + 0 structure similar to non-motile primary cilia. During spermatogenesis, the axonemal microtubules act as a platform for intraflagellar transport (IFT)4. IFT is a bidirectional intracellular trafficking system necessary for cilia/flagella formation. Since cilia/flagella do not contain ribosomes, the proteins and membrane necessary to build a flagellum are transported from the base of the flagellum to the tip by the motor protein kinesin (anterograde transport), while unnecessary proteins are returned to the base by dynein (retrograde transport)6,7. IFT-A, IFT-B, and Bardet–Biedl syndrome proteins (BBSome) play an important role in IFT. These are large protein complexes comprising many subunits and are involved in the recognition of cargo and loading on the motor protein8. IFT-A and IFT-B bind to dynein and kinesin, respectively, and are involved in retrograde and anterograde transport, respectively. BBSome binds to cargo proteins and act as mediators of cargo loading onto the IFT complex. However, the regulation of the formation of these complexes is still unclear.
The manchette is a microtubule-/F-actin-containing skirt-like structure that transiently appears around the caudal region of nuclei and flagella in elongating spermatids. Although findings about the nucleation site and the direction of microtubules are inconsistent9,10, microtubules extend from the perinuclear ring (a portion that corresponds to the skirt waist and exists just below the acroplaxome marginal ring) into the cytoplasm and/or basal body of the spermatid. During spermatid differentiation, the manchette moves toward the tail neck region, with constriction of the perinuclear ring, which contributes to sculpting of the sperm head in mice. Similar to the axoneme, proteins are actively transported on manchette microtubules via intramanchette transport (IMT)4. IMT shares similar molecular components as IFT, including motor proteins and IFT protein complex.
Deletion of IFT-/IMT-associated genes significantly affects sperm morphogenesis and male fertility. Lehti et al.11 reported that Kif3a deletion in the germ cell lineage severely impairs flagella formation, nuclear remodelling, and manchette organization in mice. Similarly, several IFT complex subunit-deficient male mice (IFT2012, IFT2513 IFT2714 and IFT14015) also shows infertility due to impaired spermatogenesis with sperm head and flagella malformation.
Recessive mutation in the oligotriche locus (olt) causes male fertility in the mouse16. Homozygous mutant (olt/olt) mice show alopecia in the inguinal region and male infertility due to impaired spermatogenesis. In olt/olt mice, spermatogenesis is impaired after meiosis, and the number of sperm at step 13 or later is significantly lower compared to wild type (WT) mice. Additionally, olt/olt mice do not have flagella in the seminiferous lumen, indicating impaired flagellum development. Runkel et al.17 reported that olt/olt mice have a 234 kbp deletion in distal chromosome 9. This region contains six genes: carboxy-terminal domain small phosphatase-like protein (Ctdspl), villin-like (Vill), 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta-1 (Plcd1), Dlec1, acetyl-coenzyme A acyltransferase 1B (Acaa1b), and solute carrier 22a member 14 (Slc22a14). Of these, Plcd1 and Acaa1b are not required for male fertility in mice, indicating that Ctdspl, Vill, Dlec1, and Slc22a14 are candidate genes responsible for male infertility. Slc22a14, a member of the organic anion/cation transporter family, plays a critical role in male infertility18. However, spermatogenesis in Slc22a14-knockout (KO) mice is essentially normal. Therefore, a gene essential for male fertility other than Slc22a14 should be present in the olt locus; however, no reports showing that Ctdspl or Vill is involved in spermatogenesis or flagellar formation are available.
DLEC1 (also called DLC1) was originally identified as a gene that is downregulated in human lung and esophageal cancers19. Decreased DLEC1 expression is found in various cancers; hypermethylation of the DLEC1 promotor region causes this decreased expression20,21,22,23,24. Introduction of the DLEC1 expression vector into cancer cells or treatment with 5-aza-2′-deoxycytidine, which induces DNA demethylation, inhibits cell proliferation and/or malignancy23,25,26, so DLEC1 is believed to act as a tumour suppressor. FAP81, an ortholog gene of Dlec1 in Chlamydomonas reinhardtii, is found as a flagellar protein using proteomic analysis of isolated flagella27. Zhao et al.28 found that FAP81 is a novel central apparatus protein in Chlamydomonas, and Fu et al.29 showed that FAP81 is required for flagellar motility and assembly of the C1e-c complex that attaches to the central microtubule pair. In addition, DLEC1 has plural ASPM-SPD-2-Hydin (ASH) domains that are involved in protein–protein interactions and are often found in cilia/flagella or centrosome-associated proteins in mammalian cells30,31. These findings suggested that DLEC1 is involved in cilia/flagella formation or function, in addition to inhibition of tumorigenesis.
This study investigated the expression profiles of Dlec1, Vill, and Ctdspl for infertility in olt/olt male mice, with an emphasis on Dlec1. We disrupted Dlec1 in mice and analysed its function in spermatogenesis and male fertility.
Results
DLEC1 is expressed in spermatids and spermatozoa
Toward the identification of the gene responsible for male infertility in olt/olt mice, we investigated the expression profiles of candidate genes (Dlec1, Vill, and Ctdspl) in various mouse tissues using reverse transcription-polymerase chain reaction (RT-PCR). As shown in Fig. 1A, Dlec1 and Vill were expressed in the brain, lungs, kidneys, and testes, while Ctdspl showed a broad expression pattern. We investigated the expressions in W/Wv mouse testes which lack germ cells because of the mutation on c-kit32 to elucidate whether these genes are specifically expressed in germ cells. Expression of Ctdspl and Vill were observed in W/Wv mouse testes, and expression of Dlec1 was not detected, indicating that only Dlec1 among these genes is specifically expressed in germ cells in the testes (Fig. 1B).
Expression analysis of mouse DLEC1. (A–C) Analysis of Dlec1, Vill, and Ctdspl expression using RT-PCR. Expression in (A) various mouse tissues, (B) WT and W/WV mutant mouse testes, and (C) testes during first-wave spermatogenesis. The numbers indicate days after birth. (D) DLEC1 expression during postnatal testicular development was examined by western blotting. The numbers indicate days after birth. (E) DLEC1 expression in testes, cauda epididymis, and cauda epididymal sperm. 1% Triton X-100-containing lysis buffer (T) or 0.1% SDS-containing RIPA buffer (R) was used to solubilize the tissue and use for western blotting. Images of full-length gels and immunoblots are presented in the supplementary Fig. S6.
Next, we investigated Dlec1, Vill, and Ctdspl expression during first-wave spermatogenesis. Vill was expressed at PND (postnatal day) 5 and reached a maximum at PND 20–25. Ctdspl was expressed constantly during testicular development, while Dlec1 expression began at PND 20 and plateaued at PND 25 (Fig. 1C). DLEC1 protein expression also showed a similar pattern (Fig. 1D). DLEC1 expression was also detected in the cauda epididymis and cauda epididymal sperm (Fig. 1E). Since haploid spermatids first appear at 18 days after birth33, these results indicated that DLEC1 expression starts in round or early elongating spermatids. Interestingly, although DLEC1 could be solubilized in the testes with either 1% Triton X-100-containing lysis buffer or radioimmunoprecipitation assay (RIPA) buffer, it could be solubilized in the epididymis and sperm only with RIPA buffer, which has stronger protein solubilization ability (Fig. 1E).
Dlec1 is indispensable for spermatogenesis and male fertility
To investigate the physiological role of Dlec1, we developed Dlec1−/− mice using the Clustered Regularly Interspaced Short Palindromic Repeats and CRISPR-associated (CRISPR-Cas9) system (Supplementary Fig. S1A). Dlec1 deletion was confirmed by genome PCR (Supplementary Fig. S1B). Although we detected expression of Dlec1 messenger RNA (mRNA) that lacks exons 28–33 (~ 830 bp) in homozygous mutant mice (Supplementary Fig. S1C), this mRNA does not seem to be translated efficiently. The predicted short form of the protein (~ 157.1 kDa, DLEC1ΔC) in Dlec1−/− testes could be detected, but the expression level was approximately one-tenth expression of wild-type DLEC1 (Supplementary Fig. S1D). Although DLECΔC potentially acts as a dominant negative mutant because it lacks the third ASH domain (Supplementary Fig. S1E), the fertility and spermatogenesis of heterozygous mutant male mice was normal. Thus, we considered DLEC1ΔC does not play a substantial role in vivo. Validation of the anti-mouse DLEC1 antibody we developed is shown in Supplementary Fig. S2. In addition, we did not detect off-target effects for at least two potential off-target sites for each guide RNA (Supplementary Fig. S3).
Dlec1−/− mice were born at the expected Mendelian ratio, and visual examination showed their growth and behaviour to be normal. In contrast, the mating experiment showed that Dlec1−/− male mice are infertile. In WT pair mating, 75% of female mice delivered, with an average litter size of 9.2 ± 0.32 (Fig. 2A,B), while no pups were born when Dlec1−/− male mice were mated with WT female mice (Fig. 2A,B). The frequency of plug formation was the same for WT and KO mice. Although the gross appearance (Fig. 2C) and weight of testes (Fig. 2D) were comparable between WT and homozygous mutant mice, histomorphological analysis revealed that spermatogenesis is impaired in Dlec1−/− mice. Although we found spermatogonia, spermatocytes, and round spermatids in Dlec1−/− seminiferous tubules, mature sperm were hardly observed (Fig. 2E). The number of cauda epididymal sperm was extremely low in Dlec1−/− mice (Fig. 2E,F). Sperm collected from the cauda epididymides of Dlec1−/− mice showed an abnormal head shape and short flagellum (Fig. 2G) and were immotile (Fig. 2H). Impaired flagella formation in Dlec1−/− mice was also confirmed by immunofluorescence analysis using anti-acetylated tubulin antibody, which is known as a flagella marker (Fig. 2I).
Dlec1 is required for spermatogenesis and male fertility. Each male mouse was caged with two WT BDF1 female mice for 2 weeks, and the (A) pregnancy rate (number of pregnant females/number of females mated) and (B) average of litter size were measured (n = 6). (C) Appearance of testes and epididymides of WT and Dlec1−/− mice. (D) Testis weight per body weight of WT and Dlec1−/− mice (n = 6). No significant statistical difference between WT and Dlec1−/− mice was detected (p = 0.066). (E) Sections of testis (a–d) and cauda epididymis (e and f) were stained using H&E. *Seminiferous tubule where sperm flagella were observed. (c and d) Magnified areas of the boxes in (a) and (b), respectively. Scale bar = 100 µm. (F) Number of cauda epididymal sperm in WT and Dlec1−/− mice (n = 5). (G) Morphology of WT and Dlec1−/− sperm. Scale bar = 10 µm. (H) Percentage of motile sperm of WT and Dlec1−/− sperm (n = 5). Error bars indicate standard error. (I) The testis section was stained using anti-acetylated tubulin antibody (green) and DAPI (blue). No acetylated tubulin signal was observed in the seminiferous tubule lumen in Dlec1−/− testes. Scale bar = 100 µm.
We investigated spermatogenesis in Dlec1−/− mice more precisely. Although spermatogenesis was normal until steps 8 and 9 of spermatids, sperm head deformation became apparent after step 10 (Fig. 3A,B). The number of spermatids decreased as spermatogenesis proceeded in the cycle of the seminiferous epithelium, and at step 14 or later, we hardly observed any spermatids. This phenotype was very close to that of olt/olt mice17. We further investigated acrosome morphology of Dlec1−/− spermatids by staining with Alexa488-conjugated peanut agglutinin (Fig. 3C). The acrosome appeared to be normally formed until round spermatid in Dlec1−/− mice; however, it's shape gradually deformed as nuclei deformed during differentiation. Acrosomes were observed in the dorsal region of the sperm head in wild-type spermatids at step 11–12, but such polar localisation was not clearly observed in Dlec1−/− spermatids. The sperm head and flagella abnormality was confirmed by electron microscopy (Fig. 3D–K). Consistent with the results of light microscopy analysis, round spermatids of Dlec1−/− at stage VII look healthy (Fig. 3D,E, arrow). In WT testes, we observed many flagella cross sections in the same seminiferous tubule (Fig. 3D, arrowhead). In contrast, in Dlec1−/− testes, we observed many degenerated cells and irregular cellular components, such as nuclei, acrosomes, and flagella (Fig. 3E, arrowhead). In elongating spermatids, most of the nuclei were distorted or irregular in Dlec1−/− testes (Fig. 3F–I). We could not find an axonemal structure despite an extensive search. Instead, misaligned mitochondria were frequently observed around a fibre-like structure in Dlec1−/− elongating spermatids (Fig. 3J). Since these degenerated cellular components appeared to be incorporated into Sertoli cells (Fig. 3K), the decreased number of sperm in Dlec1−/− testes was probably because of phagocytosis by Sertoli cells.
Spermatogenesis is impaired at the elongating spermatid stage in Dlec1−/− mice. (A) Seminiferous epithelium in WT and Dlec1−/− mouse testis. Testis sections were stained using periodic acid solution and haematoxylin. Roman numerals indicate stages of the cycle. (B) Spermatids at each stage of spermatogenesis in WT and Dlec1−/− mice. Testis sections were stained as in (A). Numerals indicate the stages of spermatids. (C) Acrosome formation in WT and Dlec1−/− spermatids. Isolated spermatids (step 7–12) were stained with Alexa488-conjugated peanut agglutinin (PNA, green). (D–K) Electron microscopy of WT and Dlec1−/− testis cross sections. (D) Seminiferous tubule at stage VI in WT testis. Arrows and arrowheads indicate round spermatids and flagella cross sections, respectively. (E) Seminiferous tubule at stage VII in Dlec1−/− testis. Although round spermatids are normal (arrow), many degenerated cells and irregular cellular components were observed (arrowhead). (F–I) Elongating spermatids (step 11 and 12) in (F,H) WT and (G,I) Dlec1−/− testes at (F,G) stage XI and (H,I) stage XII. The head shape was deformed in Dlec1−/− spermatids. (J) Abnormal tail-like structure in Dlec1−/− spermatids. Misaligned mitochondria (arrowhead) and fibre-like structures (arrow) were observed. (K) Phagocytosis of degenerated spermatids by Sertoli cells. Digested cellular components in the phagosome (arrowhead) and incorporation of the fibre-like structure (arrow) were observed.
Dlec1−/− sperm has an abnormal manchette structure
Head deformation and impaired flagella formation in sperm are often accompanied by an abnormal manchette structure. We investigated the manchette structure by immunofluorescence analysis using anti-α-tubulin antibody. Although the shape of the Dlec1−/− sperm manchette was normal in round and early elongating spermatids (steps 8–9), it abnormally elongated as spermatogenesis proceeded (Step 10–12) (Fig. 4); 4′,6-diamidino-2-phenylindole (DAPI) staining confirmed the abnormal head shape of Dlec1−/− spermatids. Since these phenotypes of Dlec1−/− sperm were similar to those of various IFT-associated gene-KO sperm, such as Kif3a11, we believed Dlec1 deletion to affect those expressions. However, KIF3A, IFT25 (a component of the IFT-B complex), and IFT140 (a component of the IFT-A complex) expression in testes was not different between WT and Dlec1−/− mice (Supplementary Fig. S4).
Identification of DLEC1-interacting proteins
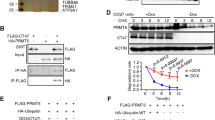
To determine the molecular function of DLEC1, we identified DLEC1-interacting proteins. Since commercially available or custom-made anti-DLEC1 antibodies are not suitable for immunoprecipitation of endogenous DLEC1, we introduced 3 × FLAG-tagged human DLEC1 (hDLEC1) into human embryonic kidney 293F (HEK293F) cells, and proteins that co-immunoprecipitated with DLEC1 were identified by liquid chromatography–tandem mass spectrometry (LC–MS/MS) (Supplementary Fig. S5 and Supplementary Table S1). In all, 67 proteins were identified, including many cilia-/flagella-associated proteins, such as α-tubulin, β-tubulin, and T-complex protein 1 (TCP-1) family proteins (Table 1). α- and β-Tubulin are the main structural components of axonemes in cilia and flagella. TCP-1 forms a complex called tailless complex polypeptide 1 ring complex (TRiC) or chaperonin-containing TCP-1 (CCT), which acts as a chaperon for tubulin and actin34. In addition to being a chaperon, TRiC enhances retrograde axonal transport in neurons35, forms a complex with BBS6, BBS10, and BBS12, and mediates BBSome assembly36. BBSome is a complex of seven BBS proteins (BBS1, BBS2, BBS4, BBS5, BBS7, BBS8, and BBS9) and is involved in the recognition and trafficking of cargo in or to cilia and flagella.
We confirmed the interaction of hDLEC1 with α- and β-tubulin in HEK293F cells by immunoprecipitation–western blotting (Fig. 5A). However, hDLEC1 did not bind to γ-tubulin efficiently (Fig. 5A). We also detected the interaction of hDLEC1 with TRiC subunits (TCP-1 α, TCP-1 β, TCP-1 γ, TCP-1 δ, TCP-1 ε, and TCP-1 η) (Fig. 5B). In addition, although LC–MS/MS could not identify BBSome subunits, hDLEC1 interacted with BBS2, BBS4, BBS5, and BBS6 (Fig. 5C).
DLEC1 interacts with tubulin, TRiC subunits, and BBSome subunits. 3 × FLAG-tagged human DLEC1 (FLAG-hDLEC1) or empty vector was expressed in HEK293F cells and immunoprecipitated using anti-FLAG antibody. SDS-PAGE was applied to the immunoprecipitates, followed by western blotting using (A) tubulin antibodies, (B) TRiC subunit antibodies, and (C) BBSome subunit antibodies. WCL, whole cell lysate. Images of full-length immunoblots are presented in the supplementary Fig. S7.
DLEC1 expression enhances ciliogenesis in A549 cells
The above results suggest that DLEC1 is involved in flagella formation through regulation of microtubule polymerization and/or intracellular vesicle trafficking in or to flagella. Initially, we investigated the effects of hDLEC1 expression on tubulin polymerization in HEK 293F cells using tubulin polymerization assay. DLEC1 expression did not affect tubulin polymerization (Fig. 6A,B). We also investigated the amount of polymerized tubulin in mouse testes and found that it was not different between WT and Dlec1−/− testes (Fig. 6C). This result is consistent with previous results that show that the manchette itself is formed in Dlec1−/− spermatids (Fig. 4). Next, we investigated the effect of expression of DLEC1 on primary cilia formation in A549 lung adenocarcinoma cells. These cells show ciliogenesis induced by serum starvation. We established a cell line stably expressing hDLEC1 (Fig. 6D) and immunostained cells with anti-ARL13B antibody, a marker of primary cilia (Fig. 6E). Quantification of results revealed that DLEC1 expression increased the percentage of ciliated cells (Fig. 6F). In addition, DLEC1 expression slightly but significantly increased ciliary length, although it is unclear whether the difference has functional meaning in the cilia (Fig. 6G). We also investigated intracellular localisation of DLEC1. DLEC1 appeared to be localized relatively uniformly in cytoplasm or as indistinct dot-like structures, but not to primary cilia (Fig. 6H). These results suggest that DLEC1 may regulate protein transport from cytoplasm to primary cilia rather than within primary cilia.
DLEC1 expression enhances primary cilia formation. (A) Upper panel: Tubulin polymerization assay. FLAG-hDLEC1 was introduced into HEK293F cells, and the cells were lysed with hypotonic buffer. After centrifugation, supernatants (S) and precipitates (P) were collected and used for western blotting. Nocodazole (Noc) and paclitaxel (Pac), which induce microtubule destabilization and stabilization, respectively, were used as a control experiment. Lower panel: DLEC1 expression was confirmed by western blotting. (B) Quantification results of tubulin polymerization assay shown in (A). P/S indicates ratio of signal intensity of precipitates (P) and supernatants (S). Data are presented as average ± standard error (n = 3). (C) Tubulin polymerization in WT and Dlec1−/− testicular cells was monitored as in (A). (D) Establishment A549 cells stably express human DLEC1. Empty vector or 3 × FLAG-tagged human DLEC1 expression vector were introduced into A549 cells and selected by puromycin. Expression of DLEC1 was monitored by western blotting using anti-human DLEC1 antibody. (E) Empty vector or hDLEC1 expressing A549 cells grown to confluent were serum-starved for 24 h and stained with anti-ARL13B antibody, a primary cilia marker. Scale bar = 10 μm. (F) Percentage of cells with cilia were quantified. Over 500 cells in one experiment were counted, and data were shown as average ± standard error (n = 3). Asterisk indicates a significant difference (Student’s t test, p < 0.05). (G) Ciliogenesis was induced as described in (E) and cilia length were measured. Data were counted from approximately 300 cilia from 3 independent experiments and shown in boxplots. Double asterisk indicates significant differences (Student’s t test, p < 0.01). (H) Intracellular localisation of hDLEC1 in A549 cells. 3 × FLAG-tagged hDLEC1 expressing A549 cells were immunostained with anti-ARL13B and anti-FLAG antibodies. Images of full-length immunoblots are presented in the supplementary Fig. S8.
Dlec1 deletion does not affect TRiC and BBSome expression, complex formation, and localisation
Next, we investigated the effect of Dlec1 deletion on TRiC and BBSome subunit expression, complex formation, and intracellular localisation. The expression of each TCP-1 and BBsome subunit in Dlec1−/− testis was comparable to that of WT testis (Fig. 7A). Complex formation of TRiC and BBSome in WT and Dlec1−/− testes were investigated using sucrose density gradient centrifugation. BBS2, BBS6, and TCP-1-α form a complex of ~ 670–880 kDa, 770–1000 kDa, and 770–1000 kDa, respectively, in the testes36, which was roughly similar to our result; however, there was no apparent difference in complex formation between WT and Dlec1−/− testes (Fig. 7B). Protein complex formation of TCP-1 alpha and BBS2 appears to be slightly different in KO mice, but results were not reproducible.
Expression, complex formation, and intracellular localisation of TRiC and BBSome is normal in Dlec−/− testis. (A) TRiC and BBSome subunit expression in WT and Dlec1−/− testes was monitored by western blotting. (B) Complex formation of TRiC and BBSome in WT and Dlec1−/− testes. Testis lysates were layered onto a 10–40% sucrose gradient and centrifuged at 28,000 rpm for 16 h. The fractions were recovered and western blotting applied. Positions of molecular weight standards are indicated at the top. (C) Intracellular localisation of TCP-1 α, BBS2, IFT25, and IFT140 in WT and Dlec1−/− round (upper) and elongating spermatids (lower). Scale bar = 10 μm. Images of full-length immunoblots are presented in the supplementary Fig. S9.
Intracellular localisation of TCP-1α, BBS2, IFT25, and IFT140 were investigated by immunofluorescence staining. Signals of these proteins in round spermatids were observed in the cytoplasm, and some were also seen in nuclei (Fig. 7C). These proteins in elongating spermatids were mainly localized posterior of nuclei (presumably manchette) in wild type. These staining patterns in Dlec1−/− spermatids were essentially equivalent to the wild type; although the cell morphology and cell nuclei were abnormal, the proteins appeared to be mainly localized to manchette. These results suggested that DLEC1 regulates functions of IFT-related protein but not its expression, complex formation, or intracellular localisation.
Discussion
This study revealed that Dlec1 is essential for spermatogenesis and male fertility in mice. Dlec1 is specifically expressed in germ cells in the testes, and Dlec1 expression increases during spermatogenesis. Although spermatogenesis in Dlec1−/− mice appears normal until step 8–9 spermatids, sperm head deformation is observed in subsequent stages of spermatogenesis. In addition, the tail of Dlec1−/− sperm is short, and we could not find normal axonemal structures. The number of mature sperm is extremely low compared to WT mice, and Dlec1−/− male mice are infertile. These abnormalities are closely similar to those of olt/olt mice. The oligotriche locus contains the Slc22a14 gene that plays a critical role in male fertility. Since spermatogenesis in Slc22a14−/− testis was normal, lack of the gene would not contribute to impaired spermatogenesis in olt/olt mice. In fact, spermatogenic phenotype of Slc22a14/Dlec1 double KO mice was the same as Dlec1 single KO mice (data not shown). Therefore, the lack of Dlec1 is probably responsible for infertility in olt/olt male mice, though careful investigation for the involvement of Ctdspl and Vill is necessary. In addition to sperm head and flagella malformation, Dlec1−/−sperm have an abnormally elongated manchette, which is frequently observed in mice sperm that lack microtubule-regulating protein, such as KATNB1 (a subunit of the microtubule-severing complex)37, leucine-rich repeats and guanylate kinase domain-containing isoform 1 (LRGUK-1)38, HOOK139, and cytoplasmic linker protein of 170 kDa (CLIP-170; the microtubule plus-end-tracking protein)40. Similarly, KO mice do not have IFT machinery proteins, such as KIF3A11, IFT2012, IFT2513, IFT2714, IFT14015, BBS241, BBS442, BBS643, and BBS744 show abnormal sperm head and flagella. These phenotype similarities suggest that DLEC1 is involved in the microtubule-associated function. In fact, we found that DLEC1 can interact with many microtubule-/IFT-associated proteins, such as α-tubulin, β-tubulin, TRiC subunits, and BBSome subunits in HEK 293F cells. Moreover, the expression of DLEC1 enhances primary cilia formation in A549 lung cancer cells. This result suggests that DLEC1 may regulate IFT through interacting proteins. However, since DLEC1 expression suppresses cancer cell proliferation and formation of cilia is induced by arrest of cell growth45, the possibility that DLEC1 indirectly increased cilia formation in A549 cells cannot be excluded. Moreover, DLEC1 is unlikely to be an essential role in the formation of primary cilia in vivo; Dlec1−/− mice did not show any ciliopathic disorder. Further, the functional significance of the interaction of DLEC1 and IFT-associated proteins need to be defined.
In this study, we did not obtain any direct evidence that DLEC1 is involved in regulating microtubule polymerization or IFT. DLEC1 expression does not affect tubulin polymerization, and IFT25, IFT140, and KIF3A expression is the same in WT and Dlec1−/− testes. Similarly, there is no difference in protein expression, complex formation, and intracellular localisation in TRiC and BBSome between WT and Dlec1−/−testes, indicating that DLEC1 is not involved in the regulation of microtubule polymerization and IFT machinery formation. One possible function of DLEC1 is the loading of cargo proteins on the IFT machinery. We identified some proteins other than IFT machinery as DLEC1-interacting proteins, such as heat shock proteins, glyceraldehyde-3-phosphate dehydrogenase (GADPH), and protein phosphatase 1B (formerly called PP2CB). These proteins are localised in the flagella of Chlamydomonas27,46 and mammalian sperm47. Therefore, DLEC1 might mediate the loading of these proteins on the IFT machinery, promoting flagella formation. Otherwise, DLEC1 itself could be a cargo protein transported by the IFT machinery into the flagella. Although intracellular DLEC1 localisation in sperm is not identified, FAP81 is localised to the central apparatus in Chlamydomonas flagella29. In addition, DLEC1 in sperm is more resistant to extraction by Triton X-100 compared to DLEC1 in testes that contain more immature germ cells. Since axonemal proteins, such as RSPH6A and acetylated tubulin, are fractionated to the Triton-insoluble fraction in mouse sperm48, our results might indicate that DLEC1 is transported from the cytosol to flagella as spermatogenesis proceeds. Regarding this, although we found exogenously expressed DLEC1 is not localized to primary cilia in A549 cells, it may not reflect the intracellular localisation in sperm because primary cilia do not have central pair of microtubules. Anyway, it is apparent that elucidation of the intracellular localisation of DLEC1 in sperm is critical for verifying our hypothesis. In this study, despite several attempts, we could not produce an antibody suitable for immunostaining. We also developed knock-in mice, which express FLAG-tagged DLEC1, and applied to immunohistochemical study, but specific signal was not detected in sperm. It may be due to low expression levels of endogenous DLEC1 is low. It is necessary to develop another tagged DLEC1 knock-in mice which enable higher sensitive detection.
Cryoelectron tomography shows that the Chlamydomonas FAP81 mutant lacks central apparatus projections C1e and C1c. Since the C1e-c subcomplex size is estimated to be 1.2 MDa29, the assembly of many proteins could be disturbed because of a lack of FAP81. In fact, the amount of FAP92 and FAP216 decreases to 37% and 0.07%, respectively, in the FAP81 mutant. Therefore, it is plausible that DLEC1 is involved in protein assembly in the central apparatus projection subcomplex of the mammalian sperm axoneme. However, one cannot simply apply the results in Chlamydomonas to mammalian sperm. The phenotypes of the FAP81 mutant and Dlec1−/− sperm are slightly different. The FAP81 mutant can swim but slowly, while Dlec1−/− sperm are immotile. In addition, FAP81 mutation does not affect the length of flagella in Chlamydomonas, and ortholog genes of FAP92 and FAP216 are not found in mammals28. Therefore, DLEC1 might play another role, in addition to the protein assembly of the central apparatus projection subcomplex.
In summary, we identified Dlec1 as an essential gene for spermatogenesis and male fertility in mice. In Dlec1−/− mice, the number of sperm is extremely low, accompanied by head and tail malformation. The human DLEC1 is a possible candidate for causative genes for idiopathic azoospermia or teratozoospermia. DLEC1 is located at 3p22-p21.3 in humans, which is frequently missing in various tumours, indicating that this chromosome region is more unstable compared to other regions. Azoospermic men have a 2.2-fold higher cancer risk compared to non-azoospermic men49. Since many reports suggest that DLEC1 acts as a tumour repressor, this study might provide a clue to elucidating the link between azoospermia and cancer.
Methods
cDNA synthesis, genomic DNA isolation, and PCR
Total RNA was isolated from mouse tissues using ISOGEN (Nippon Gene, Tokyo, Japan) and used as a template for reverse transcription. Complementary DNA (cDNA) synthesis was performed using ReverTra Ace (TOYOBO Co., Ltd., Osaka, Japan) according to the manufacturer’s instructions. Mouse genomic DNA was isolated from the tail tip using standard phenol/chloroform extraction. Polymerase chain reaction (PCR) was performed using KOD Fx Neo DNA polymerase (TOYOBO). Supplementary Table S2 lists the primers used for PCR.
Antibodies
Anti-mouse DLEC1 antibody was developed by immunization of recombinant mouse DLEC1 protein (1–300 aa) to rabbit. After immunization six times, serum was collected, and the antibody was affinity-purified by a column coupled with mouse DLEC1 recombinant protein. Immunization and serum collection were performed by Medical and Biological Laboratories Co., Ltd. Validation results for this antibody are provided in Supplementary Fig. S2. Supplementary Table S3 lists the other primary antibodies used and the concentration of use.
Dlec1−/− mice generation
Dlec1−/− mice were generated using the CRISPR-Cas9 system, as previously described50. Two target sequences for Dlec1 (5′-GGGGTGACCTTGACCGTAGTGGG-3′ and 5′-CCTCTGCACACGCTCGACACAGCC-3′) were selected to eliminate exons 28–33, and these DNA fragments were inserted into the plasmid vector containing guide RNAs and the T3 promoter. After linearization, the guide RNAs were transcribed in vitro using RNA polymerase. Similarly, Cas9 RNA transcripts were prepared using the Cas9 vector (Addgene ID: 48625). Fertilized eggs were collected from C57BL/6N mice, and two guide RNAs and Cas9 RNA were injected into the eggs, which were then transplanted into the oviducts of pseudopregnant ICR mice. The genotype of the pups was determined using genomic PCR. Considering the possibility of mosaicism in F0 mice, and to fix mutations, we used one heterozygous F1 mouse as the founder. All mice were 8 weeks to 8 months old.
Mice were purchased from Japan SLC Co. Ltd (Hamamatsu, Japan). All animal experiments were approved by the Institutional Committee for Experimental Animal Care and Use of Shizuoka University and the University of Tokyo, Japan, and carried out in accordance with the Act on Welfare and Management of Animals and the Guidelines for Proper Conduct for Animal Experimentation (Science Council of Japan).
Fertility evaluation
Ten-week-old male mice were mated with two 8-week-old B6D2F1 WT female mice for 2 weeks. The female mice were checked every day for a vaginal plug and separated if pregnant. Litter sizes were recorded on delivery.
Histological analysis of testes and sperm
Mouse testes and epididymides were dissected, fixed in Bouin’s solution, and embedded in paraffin. The paraffin blocks were sliced into 4-µm-thick sections, and the sections were then mounted on poly-l-lysine-coated glass slides. The slides were deparaffinized and stained using H&E (hematoxylin and eosin) or periodic acid solution. Images were captured using an IX-70 microscope (OLYMPUS, Japan) equipped with a digital camera (EOS6D; Cannon, Japan). To evaluate the morphology, number, and motility of sperm, the cauda epididymis was incised at a few places and sperm were released into TYH medium by gentle pushing. Subsequently, the sperm were smeared on glass slides. Sperm morphology was monitored under a microscope, the number of sperm was counted using a hemocytometer, and sperm motility was evaluated by visual observation under a stereomicroscope.
Immunofluorescence analysis of isolated testicular cells and tissue sections
Testes were dissected and then kept in a Petri dish containing phosphate-buffered saline (PBS). Seminiferous tubules were released by removing the tunica albuginea, put into a microtube, and cut into small pieces in PBS using scissors. Cells were mechanically dissociated by pipetting, filtered using a 70 µm nylon mesh, and washed twice with Dulbecco’s modified Eagle medium (DMEM) (D5796; Sigma-Aldrich, St. Loius, MO, USA). The cell suspension was placed on poly-L-lysine-coated glass cover slips in 35 mm dishes and incubated for 30 min in a CO2 incubator at 37 °C. Next, the cells were fixed in 3.8% paraformaldehyde (PFA) in PBS, permeabilized using 0.2% Triton X-100/PBS, and blocked using 5% skim milk/PBS for 1 h. Subsequently, the cells were incubated with a primary antibody for 1.5 h, washed with PBS, and incubated with Alexa Flour 488- or Alexa Flour 546-conjugated secondary antibody (Thermo Fisher Scientific, Waltham, MA, USA) for 1 h. Then, nuclei were stained using DAPI. Alexa488-conjugated peanut agglutinin (Thermo Fisher Scientific, Waltham, MA, USA) was used at 10 µg/mL for visualization of acrosome. Tissue sections were deparaffinized and heated for 5 min in citric acid buffer (pH 6.0) using a pressure cooker for antigen retrieval. Immunostaining was conducted similarly. Finally, fluorescence images were captured using an epifluorescence microscope (BX-60; OLYMPUS, Japan) equipped with a digital camera (DP-71; OLYMPUS, Japan or WRAYCAM-NOA630B; WRAYMER, Japan).
Cell culture and transfection
HEK293F and A549 cells were cultured in DMEM supplemented with 10% fetal bovine serum and penicillin–streptomycin–amphotericin B solution (FUJIFILM Wako Pure Chemical Corporation, Osaka Japan). Vector transfection was performed using Polyehylenimine Max (Polysciences, Inc., Warrington, PA, USA). The 3 × FLAG-tagged human DLEC1 expression vector was constructed by replacing the green fluorescent protein tag in the DLEC1 expression vector (Addgene ID: 90206) to the 3 × FLAG sequence. 3 × FLAG-tagged human DLEC1 was also subcloned into the pCX puro vector. This vector contains internal ribosome entry site sequences that allow the simultaneous expression of DLEC1 and puromycin resistance gene. 3 × FLAG-tagged hDLEC1 containing the pCX vector were transfected A549 cells for establishment of A549 cells stably expressing DLEC1. Cells were selected by 1 µg/mL puromycin for 2 weeks. Immunofluorescence analysis of A549 cells was performed like isolated testicular cells. Images were captured by a digital camera and cilia length was measured using Micro Studio software (WRAYMER, Japan).
Tubulin polymerization assay
Tubulin polymerization assay was performed, as previously described, with slight modifications51. Briefly, HEK293F cells transiently expressing human DLEC1 (transfection efficiency is 60–70%) or mouse testicular cells were cultured in a 35 mm dish, washed with prewarmed (37 °C) PBS, and lysed with hypotonic buffer [20 mM Tric-Cl (pH 6.8), 1 mM MgCl2, 2 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), 0.5% NP-40, 10 mM NaF, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride (PMSF)] supplemented with 1 μM paclitaxel at room temperature. The lysate was centrifuged at 17,000×g for 10 min at room temperature, and supernatants were put into new tubes and mixed with an equal volume of 2 × sample buffer. The precipitants were also solubilized with 2 × sample buffer, and the samples were incubated for 5 min at 100 °C. Finally, the amount of α-tubulin in each fraction was determined using western blotting.
SDS-PAGE and western blotting
Tissues or cells were lysed using 1% Triton X-100 lysis buffer [50 mM Tris–Cl (pH 7.4), 150 mM NaCl, 2 mM ethylenediaminetetraacetic acid (EDTA), 2 mM PMSF, 2 mM Na3VO4, 20 mM NaF, and 1% Triton X-100] or RIPA buffer [50 mM Tris–Cl (pH 7.4), 150 mM NaCl, 0.1% sodium dodecyl sulfate (SDS), 1% NP-40, 0.5% sodium deoxycholate, 2 mM PMSF]. When RIPA buffer was used, the lysate was sonicated to reduce viscosity. Next, the lysate was centrifuged for 20 min at 4 °C, and the supernatant was collected. For immunoprecipitation, cell lysates were incubated overnight with 1 µg anti-FLAG antibody and protein G-agarose beads with gentle agitation at 4 °C. Subsequently, the beads were washed several times. The lysate or beads were then mixed with sample buffer and incubated for 30 min at 37 °C or for 5 min at 100 °C. The proteins were separated with sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride (PVDF) membranes. Finally, western blotting was performed following standard procedures. HRP-conjugated light chain specific goat anti-mouse IgG (Jackson Laboratory, Bar Harbor, ME, USA) was used as secondary antibody to avoid the detection of antibody used for immunoprecipitation, if necessary.
Electron microscopy
Electron microscopy was performed, as previously described52. Briefly, mouse testes were fixed by perfusion with 4% PFA, followed by immersion fixation in 2.5% glutaraldehyde overnight at 4 °C. The testes were then postfixed in 1% osmium tetroxide and embedded in Epon resin 812. Ultrathin sections were cut and stained using uranyl acetate and lead citrate. Finally, the sections were observed under a JEOL JEM-1200 EX transmission electron microscope (JEOL Ltd., Tokyo, Japan).
Identification of DLEC1-interacting proteins
HEK293F cells transiently expressing 3 × FLAG-tagged hDLEC1 or an empty vector were lysed using 1% Triton X-100 buffer, and insoluble materials were removed by centrifugation. Then, 20 mg of lysates were mixed with 40 µL of anti-FLAG monoclonal antibody-conjugated agarose beads (Sigma-Aldrich) and incubated overnight with gentle rotation at 4 °C. The immunoprecipitants were washed several times and eluted with 2-mercaptoethanol-free 2 × sample buffer for 30 min at 37 °C. The suspension was centrifuged. Next, supernatants were collected, 2-mercaptoethanol was added, and the mixture was boiled for 5 min. Proteins were separated by SDS-PAGE and visualized using Coomassie Brilliant Blue (CBB) staining. The bands of proteins enriched by interaction with hDLEC1 were cut into small pieces. Then, proteins in the gel were reduced using 10 mM dithiothreitol (DTT) and S-alkylated cysteine residues with 55 mM iodoacetamide and digested using 10 ng/mL of sequence-grade trypsin (Promega Corporation, Madison, WI, USA) overnight at 37 °C. Peptides extracted from the gel were analysed using LC–MS/MS with a NanoFrontier eLD linear ion trap time-of-flight mass spectrometer (Hitachi High-Technologies Corporation, Japan) coupled with the NanoFrontier nLC (Hitachi High-Technologies). Briefly, trypsin-digested peptides were separated using a Capillary EX-Nano column (GL Sciences, Japan) and eluted with a linear gradient from 5 to 40% of solvent (98% acetonitrile and 0.1% formic acid) for 60 min at a flow rate of 200 nL/min. Next, the eluent was ionized using a nanoelectrospray ionization source equipped with a SilicaTip (New Objective, Woburn, MA, USA), and MS and MS/MS spectra were obtained in positive ion mode at a scan mass range of m/z 200–2000. To identify proteins, MS and MS/MS data were analysed using de novo sequencing and protein identification software PEAKS version 7.053. To avoid false positive, we selected proteins with two or more matched peptides and a higher − 10log (P-value) (≥ 50).
Sucrose density gradient centrifugation
We prepared 12 mL of 10–40% sucrose density gradient in a polypropylene tube (331372; Beckman Coulter Inc., Brea, CA, USA) using a gradient maker (SANPLATEC Corp., Osaka, Japan). Testicular cell lysates were layered onto the sucrose gradient with a molecular weight marker (GE Healthcare, Chicago, IL, USA) and centrifuged at 28,000 rpm for 16 h using a SW41Ti rotor (Beckman Coulter) at 4 °C. Then, the fractions were collected from the bottom using a grass capillary connected with a peristaltic pump. Finally, the proteins were precipitated from each fraction using trichloroacetic acid, and the pellets were washed twice with ice-cold acetone, air-dried, and solubilized using 8 M urea.
References
Mäkelä, J. A. & Toppari, J. Spermatogenesis. In Endocrinology of the Testis and Male Reproduction (eds Simoni, M. & Huhtaniemi, I.) 1–39 (Springer, New York, 2017).
Shima, J. E., McLean, D. J., McCarrey, J. R. & Griswold, M. D. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol. Reprod. 71, 319–330. https://doi.org/10.1095/biolreprod.103.026880 (2004).
Schultz, N., Hamra, F. K. & Garbers, D. L. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc. Natl. Acad. Sci. U.S.A. 100, 12201–12206. https://doi.org/10.1073/pnas.1635054100 (2003).
Pleuger, C., Lehti, M. S., Dunleavy, J. E., Fietz, D. & O’Bryan, M. K. Haploid male germ cells—the Grand Central Station of protein transport. Hum. Reprod. Update https://doi.org/10.1093/humupd/dmaa004 (2020).
Porter, M. E. & Sale, W. S. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell Biol. 151, F37-42. https://doi.org/10.1083/jcb.151.5.f37 (2000).
Ishikawa, H. & Marshall, W. F. Intraflagellar transport and ciliary dynamics. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a021998 (2017).
Prevo, B., Scholey, J. M. & Peterman, E. J. G. Intraflagellar transport: mechanisms of motor action, cooperation, and cargo delivery. FEBS J. 284, 2905–2931. https://doi.org/10.1111/febs.14068 (2017).
Wingfield, J. L., Lechtreck, K. F. & Lorentzen, E. Trafficking of ciliary membrane proteins by the intraflagellar transport/BBSome machinery. Essays Biochem. 62, 753–763. https://doi.org/10.1042/ebc20180030 (2018).
Lehti, M. S. & Sironen, A. Formation and function of the manchette and flagellum during spermatogenesis. Reproduction 151, R43-54. https://doi.org/10.1530/rep-15-0310 (2016).
O’Donnell, L. & O’Bryan, M. K. Microtubules and spermatogenesis. Semin Cell Dev. Biol. 30, 45–54. https://doi.org/10.1016/j.semcdb.2014.01.003 (2014).
Lehti, M. S., Kotaja, N. & Sironen, A. KIF3A is essential for sperm tail formation and manchette function. Mol. Cell. Endocrinol. 377, 44–55. https://doi.org/10.1016/j.mce.2013.06.030 (2013).
Zhang, Z. et al. Intraflagellar transport protein IFT20 is essential for male fertility and spermiogenesis in mice. Mol. Biol. Cell 27, 3705–3716. https://doi.org/10.1091/mbc.E16-05-0318 (2016).
Liu, H. et al. IFT25, an intraflagellar transporter protein dispensable for ciliogenesis in somatic cells, is essential for sperm flagella formation. Biol. Reprod. 96, 993–1006. https://doi.org/10.1093/biolre/iox029 (2017).
Zhang, Y. et al. Intraflagellar transporter protein (IFT27), an IFT25 binding partner, is essential for male fertility and spermiogenesis in mice. Dev. Biol. 432, 125–139. https://doi.org/10.1016/j.ydbio.2017.09.023 (2017).
Zhang, Y. et al. Intraflagellar transporter protein 140 (IFT140), a component of IFT-A complex, is essential for male fertility and spermiogenesis in mice. Cytoskeleton (Hoboken) 75, 70–84. https://doi.org/10.1002/cm.21427 (2018).
Chubb, C. Oligotriche and quaking gene mutations. Phenotypic effects on mouse spermatogenesis and testicular steroidogenesis. J. Androl. 13, 312–317 (1992).
Runkel, F. et al. Alopecia and male infertility in oligotriche mutant mice are caused by a deletion on distal chromosome 9. Mamm. Genome 19, 691–702. https://doi.org/10.1007/s00335-008-9150-9 (2008).
Maruyama, S. Y. et al. A critical role of solute carrier 22a14 in sperm motility and male fertility in mice. Sci. Rep. 6, 36468. https://doi.org/10.1038/srep36468 (2016).
Daigo, Y. et al. Molecular cloning of a candidate tumor suppressor gene, DLC1, from chromosome 3p21.3. Cancer Res. 59, 1966–1972 (1999).
Kwong, J. et al. Candidate tumor-suppressor gene DLEC1 is frequently downregulated by promoter hypermethylation and histone hypoacetylation in human epithelial ovarian cancer1. Neoplasia 8, 268–278 (2006).
Navarro, A. et al. Genome-wide DNA methylation indicates silencing of tumor suppressor genes in uterine leiomyoma. PLoS ONE 7, e33284. https://doi.org/10.1371/journal.pone.0033284 (2012).
Wang, Z. et al. Epigenetic silencing of the 3p22 tumor suppressor DLEC1 by promoter CpG methylation in non-Hodgkin and Hodgkin lymphomas. J. Transl. Med. 10, 209. https://doi.org/10.1186/1479-5876-10-209 (2012).
Ying, J. et al. DLEC1 is a functional 3p22.3 tumour suppressor silenced by promoter CpG methylation in colon and gastric cancers. Br. J. Cancer 100, 663–669. https://doi.org/10.1038/sj.bjc.6604888 (2009).
Zhang, Q. et al. Aberrant promoter methylation of DLEC1, a critical 3p22 tumor suppressor for renal cell carcinoma, is associated with more advanced tumor stage. J. Urol. 184, 731–737. https://doi.org/10.1016/j.juro.2010.03.108 (2010).
Zhang, L. et al. DLEC1, a 3p tumor suppressor, represses NF-kappaB signaling and is methylated in prostate cancer. J. Mol. Med. (Berl) 93, 691–701. https://doi.org/10.1007/s00109-015-1255-5 (2015).
Li, L. et al. Epigenomic characterization of a p53-regulated 3p22.2 tumor suppressor that inhibits STAT3 phosphorylation via protein docking and is frequently methylated in esophageal and other carcinomas. Theranostics 8, 61–77. https://doi.org/10.7150/thno.20893 (2018).
Pazour, G. J., Agrin, N., Leszyk, J. & Witman, G. B. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170, 103–113. https://doi.org/10.1083/jcb.200504008 (2005).
Zhao, L., Hou, Y., Picariello, T., Craige, B. & Witman, G. B. Proteome of the central apparatus of a ciliary axoneme. J. Cell Biol. 218, 2051–2070. https://doi.org/10.1083/jcb.201902017 (2019).
Fu, G. et al. Structural organization of the C1a-e-c supercomplex within the ciliary central apparatus. J. Cell Biol. 218, 4236–4251. https://doi.org/10.1083/jcb.201906006 (2019).
Ponting, C. P. A novel domain suggests a ciliary function for ASPM, a brain size determining gene. Bioinformatics 22, 1031–1035. https://doi.org/10.1093/bioinformatics/btl022 (2006).
Schou, K. B., Morthorst, S. K., Christensen, S. T. & Pedersen, L. B. Identification of conserved, centrosome-targeting ASH domains in TRAPPII complex subunits and TRAPPC8. Cilia 3, 6. https://doi.org/10.1186/2046-2530-3-6 (2014).
Russell, E. S. Hereditary anemias of the mouse: a review for geneticists. Adv. Genet. 20, 357–459 (1979).
Bellve, A. et al. Spermatogenic cells of the prepuberal mouse: isolation and morphological characterization. J. Cell Biol. 74, 68–85 (1977).
Spiess, C., Meyer, A. S., Reissmann, S. & Frydman, J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 14, 598–604. https://doi.org/10.1016/j.tcb.2004.09.015 (2004).
Chen, X. Q. et al. T-complex protein 1-ring complex enhances retrograde axonal transport by modulating tau phosphorylation. Traffic 19, 840–853. https://doi.org/10.1111/tra.12610 (2018).
Seo, S. et al. BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc. Natl. Acad. Sci. U.S.A. 107, 1488–1493. https://doi.org/10.1073/pnas.0910268107 (2010).
O’Donnell, L. et al. An essential role for katanin p80 and microtubule severing in male gamete production. PLoS Genet. 8, e1002698. https://doi.org/10.1371/journal.pgen.1002698 (2012).
Liu, Y. et al. LRGUK-1 is required for basal body and manchette function during spermatogenesis and male fertility. PLoS Genet. 11, e1005090. https://doi.org/10.1371/journal.pgen.1005090 (2015).
Mendoza-Lujambio, I. et al. The Hook1 gene is non-functional in the abnormal spermatozoon head shape (azh) mutant mouse. Hum. Mol. Genet. 11, 1647–1658. https://doi.org/10.1093/hmg/11.14.1647 (2002).
Akhmanova, A. et al. The microtubule plus-end-tracking protein CLIP-170 associates with the spermatid manchette and is essential for spermatogenesis. Genes Dev. 19, 2501–2515. https://doi.org/10.1101/gad.344505 (2005).
Nishimura, D. Y. et al. Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 101, 16588–16593. https://doi.org/10.1073/pnas.0405496101 (2004).
Mykytyn, K. et al. Bardet–Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc. Natl. Acad. Sci. U.S.A. 101, 8664–8669. https://doi.org/10.1073/pnas.0402354101 (2004).
Fath, M. A. et al. Mkks-null mice have a phenotype resembling Bardet–Biedl syndrome. Hum. Mol. Genet. 14, 1109–1118. https://doi.org/10.1093/hmg/ddi123 (2005).
Zhang, Q. et al. BBS7 is required for BBSome formation and its absence in mice results in Bardet–Biedl syndrome phenotypes and selective abnormalities in membrane protein trafficking. J. Cell Sci. 126, 2372–2380. https://doi.org/10.1242/jcs.111740 (2013).
Izawa, I., Goto, H., Kasahara, K. & Inagaki, M. Current topics of functional links between primary cilia and cell cycle. Cilia https://doi.org/10.1186/s13630-015-0021-1 (2015).
Bloch, M. A. & Johnson, K. A. Identification of a molecular chaperone in the eukaryotic flagellum and its localization to the site of microtubule assembly. J. Cell Sci. 108, 3541–3545 (1995).
Sasaki, M. et al. Disruption of the mouse protein Ser/Thr phosphatase 2Cbeta gene leads to early pre-implantation lethality. Mech. Dev. 124, 489–499. https://doi.org/10.1016/j.mod.2007.04.001 (2007).
Abbasi, F. et al. RSPH6A is required for sperm flagellum formation and male fertility in mice. J. Cell Sci. https://doi.org/10.1242/jcs.221648 (2018).
Eisenberg, M. L., Betts, P., Herder, D., Lamb, D. J. & Lipshultz, L. I. Increased risk of cancer among azoospermic men. Fertil. Steril. 100, 681–685. https://doi.org/10.1016/j.fertnstert.2013.05.022 (2013).
Fujii, W., Kawasaki, K., Sugiura, K. & Naito, K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res. 41, e187. https://doi.org/10.1093/nar/gkt772 (2013).
Giannakakou, P. et al. Paclitaxel-resistant human ovarian cancer cells have mutant beta-tubulins that exhibit impaired paclitaxel-driven polymerization. J. Biol. Chem. 272, 17118–17125. https://doi.org/10.1074/jbc.272.27.17118 (1997).
Ito, C. et al. Odf2 haploinsufficiency causes a new type of decapitated and decaudated spermatozoa, Odf2-DDS, in mice. Sci. Rep. 9, 14249. https://doi.org/10.1038/s41598-019-50516-2 (2019).
Ma, B. et al. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 17, 2337–2342. https://doi.org/10.1002/rcm.1196 (2003).
Bhowmick, R. et al. Photoreceptor IFT complexes containing chaperones, guanylyl cyclase 1 and rhodopsin. Traffic 10, 648–663. https://doi.org/10.1111/j.1600-0854.2009.00896.x (2009).
Li, Y., Zhao, L., Yuan, S., Zhang, J. & Sun, Z. Axonemal dynein assembly requires the R2TP complex component Pontin. Development 144, 4684–4693. https://doi.org/10.1242/dev.152314 (2017).
Acknowledgements
The authors thank Ryosuke Eba, Kenzo Kagomiya, and Kohei Ohishi for the technical assistance. This study was supported in part by MEXT (The Ministry of Education, Culture, Sports, Science and Technology) KAKENHI (Grant Number 15K07778).
Author information
Authors and Affiliations
Contributions
Y.O., M.N., T.Y., C.I., K.T., H.D., W.F., and K.Y. performed research; W.F. contributed generation of knockout mice; H.D. analysed data; C.I., K.T., H.D., W.F., and K.Y. contributed to interpretation of data; K.Y. designed research and wrote the paper; C.I., K.T., H.D., and W.F. participated in the manuscript revision.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Okitsu, Y., Nagano, M., Yamagata, T. et al. Dlec1 is required for spermatogenesis and male fertility in mice. Sci Rep 10, 18883 (2020). https://doi.org/10.1038/s41598-020-75957-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-75957-y
This article is cited by
-
Joubert syndrome causing mutation in C2 domain of CC2D2A affects structural integrity of cilia and cellular signaling molecules
Experimental Brain Research (2024)
-
Identifying pleiotropic variants and candidate genes for fertility and reproduction traits in Holstein cattle via association studies based on imputed whole-genome sequence genotypes
BMC Genomics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.