Abstract
The Cancer/Testis Antigen (CTA) genes comprise a group of genes whose expression under physiological conditions is restricted to the testis but is activated in many human cancers. Depending on the particular expression pattern, the CTA genes are speculated to play a role in spermatogenesis, but evidence is limited thus far. Here, we reported patients with a hemizygous nonsense mutation in cancer-testis antigen 55 (CT55) suffering from male infertility with extreme disruption in sperm production, morphology, and locomotion. Specifically, the insufficiency of sperm individualization, excessive residue of unnecessary cytoplasm, and defects in acrosome development were evident in the spermatozoa of the patients. Furthermore, mouse models with depletion of Ct55 showed accelerated infertility with age, mimicking the defects in sperm individualization, unnecessary cytoplasm removal, and meanwhile exhibiting the disrupted cumulus-oocyte complex penetration. Mechanistically, our functional experiments uncovered CT55 as a new autophagic manipulator to regulate spermatogenesis via selectively interacting with LAMP2 and GABARAP (which are key regulators in the autophagy process) and further fine-tuning their expression. Therefore, our findings revealed CT55 as a novel CTA gene involved in spermatogenesis due to its unprecedented autophagy activity.
Similar content being viewed by others
Introduction
Spermatogenesis is a major physiological event in the seminiferous epithelium which includes four phases [1]: spermatogonial stem cells self-renew, and spermatogonia proliferate and differentiate into primary spermatocytes [2]; primary spermatocytes undergo meiosis to form round spermatids [3]; round spermatids transform to elongated spermatids and spermatozoa [4]; the sperm is released into the tubule lumen [1,2,3]. This is a normal condition, which only takes place in the testis. However, tumorigenesis is when the continuous and active cell renewal and proliferation take place in other tissues. Interestingly, the Cancer/Testis (CT) genes is restrictedly expressed in testis under physiological conditions, while being widely activated in various human cancers derived from different organs [4, 5]. Some of the CT genes encode tumor antigens that can be recognized by cytotoxic T lymphocytes and/or sera of cancer patients; these CT genes are referred to as CT antigens (CTAs) genes [4, 5].
Information regarding the functions of CTA genes in tumorigenesis is being explored, while only a few CTA genes have been described during spermatogenesis at a mechanistic level [6]. For example, Prame conditional KO (cKO) mice have smaller testes with reduced sperm count [7]. Spata19-cKO males exhibit the defective sperm motility associated with disorganized mitochondrial structure [8]. Male mice lacking Odf2, Ropn1 and Cabyr exhibit typical multiple morphological abnormalities of the sperm flagella (MMAF) phenotype [9,10,11]. Transgenic mice overexpressing Cep55 are sterile, showing the severe spermatogenic arrest [12]. However, only AKAP4, and PIWIL1 mutations are observed to cause human male infertility [13, 14]. Therefore, a clear link between CTA genes and male infertility requires urgent attention for better diagnosis and treatment of infertile patients; this needs the identification of more causative genes and further elucidation of the molecular mechanism.
In this study, we identified a hemizygous nonsense mutation in cancer-testis antigen 55 (CT55) in two infertile men. In addition to the sharp reduction in sperm count and motility and the aberrant sperm morphology, the insufficient sperm individualization and redundant cytoplasm removal were specifically outstanding in the patients with CT55 mutations. Mouse models with knocked out Ct55 showed an age-correlated infertility linking to impairments in sperm individualization, unnecessary cytoplasm removal and cumulus-oocyte complex (COC) penetration capability. Functionally, proteomics analysis revealed that the lack of CT55 impairs the autophagic process during spermatogenesis, and which might be linked to the downregulation of LAMP2 and GABARAP.
Materials and methods
Study participants
Two infertile siblings and their parents from a consanguineous family were recruited at the West China Second University Hospital, Sichuan University. Any other diseases or abnormalities that could induce decreased infertility were ruled out, such as androgenic or endocrine abnormalities, hypogonadotropic hypogonadism, cryptorchidism, varicocele, seminal ductal obstruction, testicular trauma, or tumors. 1000 healthy men who had undergone medical check-ups without evidence of any infertility and had fathered at least one child were enrolled as normal controls. The obstructive azoospermia patients provided the testicular puncture samples as the normal controls. The study was approved by the Ethical Review Board of West China Second University Hospital (IRB no.2019(040)), Sichuan University, and all the participants signed informed consents.
Whole-exome sequencing (WES) and Sanger sequencing
Genomic DNA was obtained from a peripheral blood sample of the proband using the QIAamp DNA Blood Mini Kit (QIAGEN, 51104). Exome captures were performed using the Agilent SureSelect Human All Exon V6 Kit. Subsequently, the Illumina HiSeq X system was used to sequence according to the manufacturer’s instructions. The average sequencing depth on targets was 100, and the ratio of the target fraction covered at minimum 10× was 99.7%. Reads were mapped to the reference genome (GRCh37/hg19) by the Burrows-Wheeler Aligner (BWA) software. ANNOVAR software was used for functional annotation with information from various databases such as dbSNP, 1000 Genomes Project, ExAC, GO, KEGG, and HGMD. The Genome Analysis Toolkit (GATK 3.7) was used to identify and quality-filter the variants. The c.465 T > A (p.Y155*) variant of CT55 identified by WES was further validated by Sanger sequencing in the two infertile siblings and their parents. The primers for PCR were listed in Table S1.
Tandem mass tagging (TMT) proteomics analysis
In this study, the spermatozoa used for proteomic analysis were obtained from the control with normal fertility and the proband carrying CT55 variant. Subsequently, the TMT proteomics analysis include whole-proteome preparation, trypsin digestion, TMT labeling, high performance/pressure liquid chromatography (HPLC) fractionation, liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis were conducted according to standard methods by Hangzhou Jingjie Biotechnology Co., Ltd (Hangzhou, China). Finally, the raw data were analyzed by Domain Annotation (InterProScan), GO Annotation (http://www.ebi.ac.uk/GOA/), and KEGG Pathway Annotation (KEGG online service tools KAAS mapper). Besides, STRING’s protein-protein interaction network analyses were conducted online (https://string-db.org/).
Generation of Ct55 knock out mouse models
All the animal experiments were performed in accordance with the recommendation of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The animal experiments were also approved by the Experimental Animal Management and Ethics Committee of West China Second University Hospital (IACUC no. 2021(070)). One-month C57BL/6 female mice were superovulated and then mated with male mice to generate zygotes. In brief, gRNAs were transcribed using T7 RNA polymerase and then purified with the MEGAclear kit (Life Technologies, AM1908) in vitro. Subsequently, the purified gRNAs and Cas9 proteins (NEB, M0646M) were microinjected into the cytoplasm of single-cell C57BL/6 J embryos to generate frameshift mutation by non-homologous recombination through the deletion of exons 2–4. Subsequently, the embryos were cultured and then transferred into the oviducts of the pseudopregnant mice at 0.5-day post coitum (dpc). Besides, the mutation in Ct55 was verified in the founder mice and their offspring by PCR. The sequences of gRNAs, the PAM site, and the primers for genotype identification were listed in Table S1. The male mice of 12 days, 8 weeks, 16 weeks, and 32 weeks were used in our study. The female mice of 8-10 weeks were used for fertility test in our study.
Total RNA isolation and real-time quantitative PCR (RT-qPCR)
We collected 50 mg testicular tissue samples, and extracted the total RNA was by Trizol reagent (Thermo Fisher, 15596026); then the RNA was converted to cDNA with SuperScript™ IV Reverse Transcriptase (Thermo Fisher, 18090010) according to the manufacturer’s instructions. RT-qPCR was conducted in 96-wells on an iCycler RT-PCR Detection System (Bio-Rad Laboratories) in a total volume of 10 μl that contained 1 μl cDNA template, 0.2 μl forward and reverse primer, 5 μl SYBR Green Mix (Sigma-Aldrich, KCQS00), and 3.6 μl DEPC-H2O. The PCR cycling conditions were as follows: pre-denaturation, 95 °C for 300 s and then 40 cycles of denaturation, 95 °C for 15 s, and annealing and extension at 60 °C for 30 s. The primers for qPCR were listed in Table S1.
Lentivirus and plasmids transfection
HEK293T and K562 cells were obtained from the American Type Culture Collection (ATCC, CRL-11268, CRL-5844). The cell lines were identified and showed no mycoplasma contamination. The HEK293T cells were cultured in DMEM (Gibco, 11965092) and the K562 cells were cultured with RPMI 1640 Medium (Gibco, C11875500BT), which contains 10% fetal bovine serum (Sigma, F8318) and 0.1% penicillin/streptomycin in a 5% CO2 cell incubator at 37 °C.
The CT55 plasmids (RefSeq ID: NM_017863.2) were synthesized and cloned into the pCMV-MCS-3*Flag by Vigene. The mutant plasmid of CT55 was generated by the Fast Mutagenesis System (TransGen, FM111) following the instructions. The primers were listed in Table S1. The shCT55 lentiviral particles were synthesized from Vigene (DZ2456485), and the target sequence is listed in Table S1. The HEK293T cells were transfected with WT or mutant CT55 plasmid using jet PRIME regent (Polyplus, PT-114-15). The transfection of control or shCT55 lentiviral particles into K562 cells were enhanced by polybrene (Solarbio, H8761). 48 h after transfection, the cells were collected for further experimental analysis.
Western blot and Co-immunoprecipitation (Co-IP)
Cells or tissue samples were lysed for 30 min at 4 °C in RIPA lysis buffer (Beyotime, P0013B), which contains Protease Inhibitor Cocktail (Biomake, B14012). The concentrations of the total proteins were determined using the BCA Assay kit (Thermo, 23227); 20–30 μg total proteins were loaded into each lane and separated by electrophoresis in 10–12.5% SDS-PAGE gels and then blotted onto PVDF membranes (Millipore, ISEQ00010, IPVH00010). Subsequently, membranes were blocked in 5% fat free milk for 1 h at room temperature and then incubated with primary antibodies overnight at 4 °C, including CT55 (AVIVA SYSTEMS BIOLOGY, ARP70088_P050, 1:500), EMC4 (Abcam, ab184544, 1:10000), RPL27 (Abclonal, A13044, 1:1000), RPS6 (Abclonal, A6058, 1:2000), RPS7 (Abclonal, A6731, 1:2000), GABARAP (Proteintech, 18723-1-AP, 1:1000), LAMP2 (Proteintech, 66301-1-Ig, 1:2000), LC3 (Cell Signaling, 4108, 1:1000), p62 (Proteintech, 18420-1-AP, 1:5000), FLAG (Thermo Fisher, MA1-91878, 1:1000), SPAM1 (Abclonal, A2120, 1:1000), ACR (Bioss, bs-5151R, 1:1000), and GAPDH (Abcam, ab181602, 1:10000). Then the membranes were incubated with Goat anti-Mouse secondary antibody (Thermo Fisher, G-21040, 1:10000) or Goat anti-Rabbit secondary antibody (Thermo Fisher, 31460, 1:10000) for 1 h at room temperature and visualized with ECL HRP substrate (Millipore, WBKLS0500).
The extracted total proteins were incubated with 3 µg of target antibodies overnight at 4 °C. Subsequently, each sample was incubated with 50 μl of Protein A/G magnetic beads (Thermo Fisher, 88804) for 1 h at room temperature. The beads were then washed three times with 1× PBS and resuspended by 1×SDS loading buffer (Biosharp, BC502A) and denatured at 95 °C for 10 min. The prepared samples were then used for western blot assay as described above. All the original images of the protein bands were listed in Fig. S15.
Flagellar Beating analysis using SpermQ
The methods for analyzing flagellar beating using SpermQ software were previously described in detail [15]. First, the dark-field image sequences were briefly recorded with 200 frames per second (FPS) using a 10×objective (Nikon, Ti-U). Then the obtained image sequences were processed and analyzed by ImageJ and SpermQ to get the swing track of the flagella.
Immunofluorescence staining
The spermatogenic cells at different stages were fixed onto slides with 4% paraformaldehyde and then perforated with 0.5% Triton X-100 for 15 min. Subsequently, the slides were blocked in 5% BSA for 30 min at room temperature and then incubated with primary antibodies overnight at 4 °C; this included CT55 (AVIVA SYSTEMS BIOLOGY, ARP70088_P050, 1:100), GABARAP (Proteintech, 18723-1-AP, 1:100), LAMP2 (Proteintech, 66301-1-Ig, 1:100), EMC4 (Abcam, ab184544, 1:50), RPL27 (Abclonal, A13044, 1:50), RPS6 (Abclonal, A6058, 1:50), RPS7 (Abclonal, A6731, 1:50), LC3 (Cell Signaling, 4108, 1:50), p62 (Proteintech, 18420-1-AP, 1:50), α-TUBULIN (Thermo Fisher, A11126, 1:200), FLAG (Thermo Fisher, MA1-91878, 1:200), SPAM1 (Abclonal, A2120, 1:200), ACR (Bioss, bs-5151R, 1:100), PNA (Vector, RL-1072-5, 1:50) and CCZ1(Santacruz, sc-514290, 1:20). Next, the slides were incubated with Alexa Fluor 488 (1:1000, A21206, Thermo Fisher) or Alexa Fluor 594 (1:1000, A11005, Thermo Fisher) for 1 h at room temperature in the dark. Then, the slides were washed with 1× PBS twice and counterstained with 4, 6-diamidino-2-phenylindole (DAPI, D9542-1MG, Sigma-Aldrich). Lastly, the slides were observed under a laser scanning confocal microscope (Olympus 1000, Japan).
For immunofluorescence staining of testicular tissues, the prepared paraffin sections were dewaxed in dimethyl benzene for two consecutive 10 min and then hydrated in gradient declining alcohol (a serial dilution of 100%, 100%, 95%, 85%, 75%, and 50% alcohol). Then antigen repair was conducted in sodium citrate. Subsequently, after being washed by PBS twice for 5 min, the sections were blocked by goat serum at 37 °C for 30 min and then incubated with primary antibodies of CT55, PCNA (Proteintech, 60097-1-Ig, 1:200) and SYCP3 (Abcam, ab15093, 1:50) at 4 °C overnight. On the second day, the sections were incubated with Alexa Fluor 488 (1:1000, A21206, Thermo Fisher) or Alexa Fluor 594 (1:1000, A11005, Thermo Fisher) at room temperature for 1 h and then sealed with fluorescence decay-resistant medium containing 4’,6-diamidino-2-phenylindole (DAPI). The slides were observed under the fluorescence microscope (Zeiss, Ax10 Axio).
Spermatocyte spreading experiment
Seminiferous tubules were placed in hypotonic extraction buffer [30 mM tris (pH 8.2), 50 mM sucrose, 17 mM trisodium citrate dihydrate, 5 mM EDTA, 2.5 mM dithiothreitol (DTT), and 1 mM phenylmethylsulfonyl fluoride (PMSF)] for at least 25 min at room temperature to release the chromosome of spermatocytes. Next, cell suspension was prepared in 100 mM sucrose and then evenly spread on slides with 1% PFA (pH 9.2) containing 0.15% Triton X-100. Slides were humidified for 3 hours and then washed twice for 3 min in 0.4% Photoflo (Kodak) and air-dried at room temperature. Subsequently, slides were immediately used for immunofluorescence staining using primary antibodies of SYCP3 (Abcam, ab15097, ab97672, 1:100), γH2AX (Abcam, ab26350, 1:200), Rad51(Sigma-Aldrich, PC130, 1:100) and TEX12 (Proteintech, 17068-1-AP, 1:50).
Histology hematoxylin-eosin (H&E) staining
Testicular and epididymal tissue samples from mice were fixed with 4% paraformaldehyde overnight, dehydrated in ethanol, embedded in paraffin, and sectioned at 5 μm. The sections were stained routinely with hematoxylin and eosin for histological examination.
Scanning electron microscopy (SEM) and transmission electron microscopy (TEM)
Fresh semen samples were washed three times with 1×PBS and then fixed on coverslips with 2.5% phosphate-buffered glutaraldehyde for 24 h (4 °C). The coverslips carrying spermatozoa were then dehydrated in gradient ethanol of 50%, 60%, 70%, 80%, 95%, and 100%. Subsequently, the specimens were dried and coated with gold particles using an ion sputter coater (Q150RS, RotaryPumped, Quorum Technologies, UK). The spermatozoa were finally observed under the SEM (Hitachi, Japan).
Semen and testicular samples were pretreated and fixed with 3% glutaraldehyde and osmic acid and then dehydrated with gradient ethanol. Samples were embedded with Epon 812, dodecenyl succinic anhydride, methylnaphthalic anhydride, dimethylaminomethylphenol, and rapid semithin sections were conducted to determine the location of seminiferous tubules. Subsequently, tultrathin sections (70–90 nm) were double-stained with lead citrate and uranyl acetate. Finally, the images were taken with TEM (TECNAI G2 F20, Philips) at 120 kV.
Computer-assisted semen analysis (CASA)
The parameters of sperm motility were quantified by a CASA system from Chengdu Panda Breeding Base. In brief, cauda epididymal sperm of mice were capacitated in TYH for 10 min and then transferred into a prewarmed counting chamber. Subsequently, the sperm motility was observed and recorded under an inverted microscope (IX73, Japan).
Isolation of human and mouse spermatogenic cells
Spermatogenic cells at different stages were obtained through cell diameter/density at unit gravity by the STA-PUT velocity sedimentation method as previously described [16]. Spermatogenic cells of adult male mice and humans were extracted and resuspended in 25 ml of 0.5% BSA solution and then filtered through an 80 mm mesh to remove cell aggregates. Subsequently, the cells were resuspended in a buffer containing 0.5% BSA and loaded in an STA-PUT velocity sedimentation cell separator (ProScience) for gradient separation. Finally, spermatogenic cells of different stages were isolated for further analysis.
In vitro fertilization (IVF)
The methods refer to the previously published literature [17]. Briefly, the oocyte-cumulus complexes (OCCs) were obtained from the oviductal ampulla of the superovulated mice (8–10 weeks) after hCG injection for 14 h and then placed in a 90 μl drop of TYH medium (Easy check, M2030) covered with mineral oil. Next, cauda epididymal sperm of male mice (C57BL6/J, 8–10 weeks) were capacitated by incubating 500 μl drops of TYH medium for 1 h in an incubator with 5% CO2 at 37 °C. Finally, a liquid drop of the capacitated sperm suspension was mixed with the 90 μl drops of TYH medium containing the OCCs to make a final sperm concentration of 1 × 105/ml. Subsequently, the mixtures were incubated at 37 °C under 5% CO2 for further observation and calculation of the fertilization and embryo development rates.
Cumulus cell dispersal assay
The OCCs in a 90 μl TYH drop were mixed with fresh capacitated epididymal sperm (1 × 105/ml) or sperm protein extracts (3 μg/10 μl). Methods for sperm protein extraction and concentration determination were described above. The mixtures were incubated at 37 °C under 5% CO2 for 30, 60, or 120 min, and the hyaluronidase (300 IU/ml, Sigma, 37326-33-3), apigenin (500 μM, TargetMOI, T2175), p-amino benzamidine (1 mM, ASTATECH, 7761-72-0) were added. The results were observed and recorded under a microscope (Nikon TS100).
Cumulus penetration assay
Superovulated OCCs were prepared as previously described. Cauda epididymal sperm were released and incubated in a 500 μl drop of TYH medium for 1 h to make the spermatozoa capacitated. Subsequently, the spermatozoa were incubated with 1 μg/ml Hoechst 33342 (Solarbio, C0031) for 10 min at 37 °C under 5% CO2. 10 μl of the capacitated sperm suspension was mixed with the OCCs in TYH medium (90 μl) to make the final sperm concentration of 1 × 105/mL, and then the mixture was incubated at 37 °C for 30 min under 5% CO2. After incubation, the OCCs were transferred to a fresh TYH medium drop of 100 μl, and fixed by the addition of 100 μl 1×PBS containing 8% paraformaldehyde and 1% PVP for 15 min. Next, the zona pellucida were visualized by staining with ZP3 antibody (Proteintech, 21279-1-AP, 1:200) at 4 °C overnight. The next day, the OCCs were incubated with Alexa Fluor 488 (1:1000, A21206, Thermo Fisher), the second antibody for 1 h at room temperature, then transferred to a confocal dish and observed under laser scanning confocal microscope (Olympus FV1000, Japan).
Sperm DNA fragment staining
The operation was conducted according to the Operators Manual of Sperm DNA fragment Staining Kit (Puhua Biotechnology Co., Ltd, SP001, China). Briefly, the coated slides with sperm were firstly immersed in reaction reagent A for 7 min and then immersed in reaction reagent B for 20 min. Subsequently, the slides were immersed in pure water for 2 min and then placed in gradient ethanol of 50%, 70%, 95% for 2 min. After drying in air, the slides were stained with prepared staining solution for 15 min at room temperature and then observed under an optical microscope (Zeiss, AX10 imager A2).
LC-MS/MS analysis
The immunoprecipitated mixtures were separated by 10% protein gels. Standard methods including in-gel protein extraction, trypsin digestion, LC-MS/MS analysis, and data processing were performed. Briefly, gel pieces were accordingly processed in acetonitrile, dithiothreitol, and iodoacetamide. Next, the mixtures were digested in trypsin at 37 °C overnight. Subsequently, the tryptic peptides were dissolved with EASY-nLC 1000 UPLC system, and then subjected to NSI source followed by tandem mass spectrometry (MS/MS) in Q ExactiveTM Plus (Thermo fisher) coupled online to the UPLC. Finally, the resulting MS/MS data were processed using Proteome Discoverer 1.3. Corresponding bioinformatics annotations were conducted according to requirements. The sample preparations and Liquid chromatography-Mass spectrum/Mass spectrum (LC-MS/MS) analysis were conducted by Hangzhou Jingjie Biotechnology Co., Ltd (Hangzhou, China).
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8.4.0 software and SPSS 17.0 software. All the data were presented as the means ± SEM. A p value of less than 0.05 was considered statistically significant. Statistical significance between two groups was calculated using an unpaired, parametric, two-sided student’s t test.
Results
Identification of a hemizygous nonsense mutation in CT55 in two infertile siblings
This study recruited two infertile siblings from a consanguine family (Fig. 1A). The routine semen analysis of the two patients showed extremely reduced sperm count, anomalous morphology of sperm head and flagellum, and significantly decreased sperm locomotion (Table 1). Papanicolaou staining and SEM observed notable sperm head deformities, which mainly manifested as round head, square head, irregular head, or double heads (Fig. 1B, C). Evident vacuoles and incomplete acrosome were exhibited in the sperm head by TEM (Fig. 1D). Moreover, numerous spermatozoa with curled or double flagella were observed (Fig. 1B, C). TEM showed sperm flagella with various abnormal ultrastructure, including swollen mitochondria, disordered or absent outer dense fibers (ODFs), missing or disorganized microtubules (MTDs), and inhomogeneous fibrous sheath (FS) (Fig. 1D). Notably, considerable wrapped spermatozoa with failed sperm individualization, or redundant cytoplasmic residue were observed in the patients under TEM (Fig. 1D). In addition, we detected the significantly increased sperm DNA fragmentation in the patients, suggesting the DNA damage in sperm heads of the patients (Fig. S1).
A The pedigree structure and segregation analysis of the infertile family. Squares represent male pedigree members, circles represent female pedigree members, solid symbols represent affected infertile members, hollow symbols represent unaffected members, black arrows indicate the proband. The Sanger sequencing (in the lower panel) of the family members indicated an X-linked variant in the siblings originated from their mother. The Papanicolaou staining (B) and SEM analysis (C) showed abnormal sperm morphology of the patients, including round head, irregular head, coiled flagella, and double heads or flagella were also visible. (scale bars,5 µm). D The TEM analysis exhibited the aberrant ultrastructure of the sperm head and flagellum from the two siblings. The vacuoles were visible in the morphologically abnormal sperm head of the patients compared with the normal control. (scale bars, 1 µm). In the mid-piece, the arrangement of ODF was disordered, the typical “9 + 2” microtubules were missing and the swollen mitochondria were observed (indicated by red arrow). In addition, central and peripheral microtubules were missing or disorderedly arranged in the principal and end pieces. Fibrous sheaths were also inhomogenous (indicated by blue arrow), and quite a number of spermatozoa with failed individualization and unnecessary cytoplasmic removal were found. CP central-pair microtubule, ODF outer dense fiber, MTD peripheral microtubule doublet. (scale bars, 200 nm).
WES was performed on this family. 111 and 90 rare clinically relevant variants were detected in the proband and his sibling respectively, while none of them were candidate causes of male infertility shared by the two siblings (Table S2). 10 and 16 rare functionally relevant homozygous or hemizygous variants were detected in the proband and his sibling respectively (Table S2). Among them, three homozygous or hemizygous variants are shared by the proband and the affected sibling (Table S2). Importantly, a hemizygous nonsense variant of c.465 T > A (p.Y155*) in CT55 caught our attention. This nonsense mutation was absent in various populations (Table 1), and was also not detected in our 1000 normal Chinese control males. Sanger sequencing confirmed this CT55 variant was inherited from their fertile mother (Fig. 1A). We further found the absence of CT55 expression of CT55 Y155* plasmid (Fig. S2A, B).
Importantly, a specifically high expression of CT55 in human testis compared with other organs (https://www.proteinatlas.org/ENSG00000169551-CT55/tissue) (Fig. S3A). In addition, CT55 expression in the testis was significantly increased from adolescence onwards and then stably maintained at a high level in humans (https://das.chenlulab.com/#/search) (Fig. S3B). Moreover, immunofluorescence staining of mouse testicular sections showed that CT55 was detected in various germ cells, with the high expression in late spermatids (Fig. S3C). Furthermore, we isolated germ cells in different steps from mouse and human testes. In particular, CT55 staining was mainly observed in the cytoplasm of spermatocyte and spermatogonia, and was also obviously found in acrosome or flagellum in early and late spermatids in mice and humans (Fig. S3D, E). Therefore, we reasoned that the causation of infertility in the two patients might be attributed to the loss of function mutation in CT55.
Disrupted acrosome biogenesis observed in the patients with CT55 deficiency
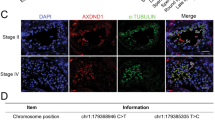
CT55 could be detected in the acrosome area of normal human spermatozoa, while the CT55 expression was missing in that of patients by immunofluorescence staining (Fig. 2A) and western blot (Fig. S2C). The PNA signals labelling acrosome were defective (Fig. 2A). The ultrastructure of the acrosome at different developmental stages in the testicular tissues of the patients and normal control was revealed by TEM. In the normal control, a concave Golgi complex formed over the round spermatid nucleus and released granules that fused to form an acrosomal vesicle (AV), with acrosomal granules (AG) that flattened over the area of the nuclear envelope underlined by the nuclear lamina (Fig. 2B). Elongating spermatids had highly condensed nucleus and formed flattened acrosome tightly attached to the nucleus (Fig. 2B). However, in the patients, round spermatids displayed missing Golgi apparatus or a more diffuse Golgi apparatus that did not have the typical arch over the nucleus in the Golgi phase (Fig. 2B). Besides, AGs were ectopic, absent or morphologically abnormal, accompanied by AVs to be too large or too small (Fig. 2B). These abnormalities in the developmental stages finally led to aberrant acrosomes in the maturation phase, which mainly manifested as small, incomplete, or structurally disordered acrosome (Fig. 2B). These results suggested that CT55 may play an important role in acrosome development in humans.
A In the spermatozoa of the patients, the expression of CT55 was completely missing, and the irregular staining of PNA indicated the incomplete acrosome. Three independent experiments were performed. (CT55, green; PNA, red; scale bars, 5 µm). B Numerous abnormalities at different developmental stages of acrosome were observed by TEM analysis, including missing or morphologically abnormal Golgi apparatus (G) (red arrows), too large or too small acrosomic vesicles (AV) (blue arrows), absent or ectopic acrosomic granules (AG) (yellow arrows), as well as the small or incomplete acrosome (AC) (green arrows) in maturation phase. N nucleus, NL nuclear lamina. (scale bars, 1 µm).
Damaged fertility of patients with CT55 deficiency can be rescued by intra-cytoplasmic sperm injection (ICSI) treatment
The ICSI treatment was implemented on these two infertile patients. Their female spouses had regular menstrual cycles and normal sex hormone concentrations (Fig. 3A). The detailed medical records of ICSI cycles were shown in Fig. 3A. For patient IV-1’s wife, 22 oocytes were retrieved, including seven immature oocytes and 15 metaphase II (MII) oocytes. After injection, 11 of 15 MII oocytes were fertilized and merogenetic, in which ten oosperm were successfully developed into blastocysts (Fig. 3B). For patient IV-3’s wife, all retrieved eight MII oocytes were injected, and seven were fertilized and successfully divided into blastocysts after 120 h (Fig. 3B). The detailed blastocyst quality rating for the two patients’ spouses was displayed in Fig. 3A. The vivid image of developmental processes from fertilization to blastocyst stage was exhibited in Videos S1 and S2. Finally, two embryos were transferred to the spouses of both patients respectively, and IV-3’s wife became pregnant, but the pregnancy of IV-1’s wife was not achieved. Therefore, CT55 variants might be linked to a good prognosis of ICSI therapy, and additional risk factors of ICSI cycles might lead to the failed outcome for patient IV-1’s wife.
Ct55 KO male mice show defects in acrosome development, sperm individualization and removal of unnecessary cytoplasm
We subsequently generated Ct55 KO mice to precisely define the onset of the disrupted spermatogenesis by the lack of CT55 (Fig. 4A). It was confirmed that Ct55 was null in KO mice (Fig. S4A, B, C). Furthermore, the Ct55 KO mice did not exhibit any gross abnormalities and survived to adulthood. The homozygous Ct55 KO female mice were fertile, while the numbers of both litters and pups were progressively decreased in hemizygous Ct55 KO male mice with various ages, which were totally infertile at the age around 32 weeks (Fig. 4B). The male Ct55 KO mice showed a similar weight of testis and epididymis compared with WT mice with different stages (Fig. S4D), and histological analysis of testes from Ct55 KO male mice showed no obvious abnormality when compared to WT mice with different ages by H&E staining (Fig. S5). Moreover, immunofluorescence staining showed the similar staining of PCNA (proliferating spermatogonia marker) and SYCP3 (spermatocyte marker) on the testicular sections of Ct55 KO and WT mice at different ages (Fig. S6). The normal meiosis process was confirmed in Ct55 KO mice using spermatocyte spreading assay (Fig. S7). Therefore, CT55 might not be involved in early spermatogenic process. However, spermatozoa progressively and evidently declined in different segments of epididymis from the KO mice compared to those of WT mice with increasing ages (Fig. S8). We then performed the phenotype analysis on the male mice with the age of 32 weeks. Strikingly, we examined the impaired movement characteristics of Ct55 KO spermatozoa using CASA technology (Table 2). Specifically, we observed the apparent motility declines in Ct55 KO spermatozoa, and the KO spermatozoa were almost immotile at the age about 32 weeks (Fig. 4C, Videos S3, S4). Vividly, SpermQ, a software for flagellar beating analysis, showed the lateral swing amplitude of flagella of the Ct55 KO mice was decreased compared with WT mice when the head was fixed by polylysine (Fig. 4D). Noticeably, the bunched or inseparable sperm flagella were distinct in Ct55 KO mice (Fig. 4E).
A The strategy for generating Ct55 KO mice using CRISPR-Cas9 by deleting exons 2–4 to induce a frameshift mutation. B WT and KO male mice were bred with the WT female mice, and the average litter sizes and pups were measured. The fertility of KO males was progressively reduced compared with WT control,and the KO male mice were totally infertile at the age about 32 weeks. Average litters means the average number of litters for each animal and the average pups means the average number of pups per litter. (n = 3 biologically independent male WT mice or KO mice were respectively mated with two WT female mice; two-sided student’s t test, *p < 0.05, error bars, s.e.m). C The percentage of the sperm motility in Ct55 KO mice and WT mice. (n = 3 biologically independent male WT mice or KO mice; two-sided student’s t test, *p < 0.05, error bars, s.e.m). D The swing amplitude of sperm flagella in Ct55 KO mice was reduced compared with WT mice analyzed by SpermQ. (n = 3 biologically independent WT mice or KO mice). E SEM analysis showing sperm aggregated into bundles in Ct55 KO mice. (n = 3 biologically independent WT mice or KO mice; scale bars, 1 µm).
Furthermore, some of the round spermatids displayed an absent or disordered Golgi apparatus (Fig. 5A). Besides, the AG was morphologically abnormal or with an abnormal number (missing or multiple) (Fig. 5A). These abnormalities in the developmental stage finally caused abnormal acrosome in the maturation phase (Fig. 5A). In addition, the abnormal cytoplasmic residue, impaired sperm individualization, incomplete FS, and aberrant mitochondria were found both in testicular spermatids and epididymal spermatozoa of Ct55 KO mice (Fig. 5B). Collectively, loss of CT55 might contribute to the progressive subfertility in mice.
A Abnormal acrosome development in Ct55 KO mice was shown by TEM analysis, including missing or abnormal Golgi apparatus (G) (red arrows) and ectopic or abnormal number AG (blue arrows), which induced abnormal acrosome morphology in the maturation phase (yellow arrows). (n = 3 biologically independent WT mice or KO mice; scale bars, 2 µm). B Impaired sperm individualization, excessive residual cytoplasm, incomplete FS, and dilated intermembrane spaces of mitochondria were observed in Ct55 KO mice. (n = 3 biologically independent WT mice or KO mice; scale bars, upper panel, 500 nm; lower panel, 1 µm).
Deficiency of CT55 impairs spermatogenesis via autophagic process
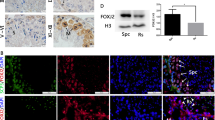
A proteomics approach was applied to the proband’ spermatozoa to determine the reason for the disruption of CT55-deficient sperm. A total of 6201 proteins were quantified, including 740 upregulated proteins and 1369 downregulated proteins (Fig. S9). Gene ontology (GO) enrichment analysis showed that the downregulated proteins were most relevant to biological processes or cellular components involved in spermatogenesis, such as spermatid differentiation, flagellated sperm motility, acrosomal vesicle, manchette, and so on (Fig. 6A). Moreover, these downregulated differential proteins formed a complex interaction network involved in acrosome function, autophagy, and spermatogenesis (Fig. 6B). Intriguingly, the cellular components of the ribosome and endoplasmic reticulum were found to be obviously and specifically enriched in semen from patient compared to normal control (Fig. 6C). A total of 47 ribosomal subunits and 10 endoplasmic reticulum-associated proteins were found to be significantly upregulated in the patient’s sperm (Fig. 6D). Western blot and immunofluorescence further verified the upregulation of ribosomal subunits and endoplasmic reticulum-associated proteins in the patient’s sperm and testes (Fig. 7A, B). The accumulation of cellular components of the ribosome and endoplasmic reticulum indicates the impaired function of autophagic removal of excessive cytoplasmic materials [18]. Accordingly, GO analysis revealed that the proteins involved in the autophagic process, such as phagolysosome assembly, regulation of autophagosome assembly, autophagosome maturation, and lysosome, were significantly reduced (Fig. 6B, E). Immunofluorescence staining consistently showed the increased signals of the ribosome and endoplasmic reticulum-associated protein in testes of Ct55 KO mice compared to WT mice (Fig. 7C), indicating the diminished autophagic process in Ct55 KO mice. The accumulation of p62 and LC3 was observed in Ct55 KO mouse testes (Fig. 7D). We further carried out LC-MS/MS analysis to search for the interacting partners of CT55 using the sperm lysate from fertile control. A total of 994 proteins were identified to interact with CT55. Specifically, GO enrichment analysis showed that most of the identified proteins were relevant to the biological processes of autophagy, spermatogenesis, and mitochondrion function, such as regulation of autophagy of mitochondrion, regulation of protein autophosphorylation, autophagosome maturation, spermatid development, regulation of flagellated sperm motility, mitochondrion organization, and mitochondrial transmembrane transport; the consistent cellular components were also observed, such as primary lysosome, sperm fibrous sheath, mitochondrial outer membrane, mitochondrial inter membrane (Fig. S10). Therefore, CT55 might play an important role in autophagy and spermatogenesis.
A Gene ontology analysis showed biological processes and cellular components in which the diminished proteins in the spermatozoa from the patient were remarkably involved in spermatogenesis. B The downregulated proteins formed a complex interaction network mostly associated with autophagy, acrosome, and spermatogenesis. C Gene ontology analysis indicated the enrichment of the upregulated cellular components was related to endoplasmic reticulum and ribosome. D List of the ribosome and endoplasmic reticulum proteins found in proteomics analysis that was upregulated in sperm from the patient without CT55 expression. E Gene ontology analysis showed the downregulated biological processes or cellular components involved in autophagy in the patient. F STRING analysis showed interactions between LAMP2/GABARAP and the proteins (downregulated in this study) related to spermatogenesis. G The heat map of the downregulated proteins involved in the fertility or acrosome biogenesis processes in the patient compared to normal control. Three independent experiments were performed.
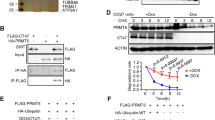
A The expression of autophagy-associated proteins LAMP2 and GABARAP was decreased, and the expression of endoplasmic reticulum and ribosome-associated proteins was increased in spermatozoa of the patient detected by western blot. Three independent experiments were performed. B Immunofluorescence staining confirmed the abnormal expression of LAMP2 and GABARAP, and the endoplasmic reticulum and ribosome-associated proteins in the testis from the patients. Three independent experiments were performed. (scale bars, 125 µm). C The proteins related to endoplasmic reticulum and ribosome were increased in the testes of Ct55 KO mice compared with WT mice by immunofluorescence. Three independent experiments were performed. (n = 3 biologically independent WT mice or KO mice; scale bars, 125 µm). D The increased expression of LC3 and p62 was observed in the testes of Ct55 KO mice by western blot and immunofluorescence. (n = 3 biologically independent WT mice or KO mice; scale bars, 125 µm). E The expression of LAMP2 and GABARAP was decreased in the spermatozoa of the patients by immunofluorescence. Three independent experiments were performed. (scale bars, 5 µm). F Western blot showed the downregulation of LAMP2 and GABARAP when knockout CT55 expression, or upregulation of LAMP2 and GABARAP by overexpressed CT55. Three independent experiments were performed. G No significant difference of LAMP2 and GABARAP expression detected by qPCR in cells transfected with shCT55 or WT-CT55. Three independent experiments were performed. (Two-sided student’s t-test; NS, not significant; error bars, s.e.m). H The increased ubiquitination of LAMP2 and GABARAP was observed by downregulation of CT55. Three independent experiments were performed. I The binding of CT55 between LAMP2 and GABARAP was detected in human testis by Co-IP analysis. Three independent experiments were performed. J The exogenous interaction of CT55 and LAMP2/GABARAP was observed in 293 T cells with overexpressed WT-CT55. Three independent experiments were performed.
Autophagy has been implicated as a crucial component in spermatogenesis [19, 20], contributing to the acrosomal formation, organelle rearrangements, flagellum development, removal of unnecessary cytoplasm, survival of the spermatogenic cells, and remolding of the germ cells [21,22,23,24]. Strikingly, LAMP2 and GABARAP, which play important roles in autophagy, were significantly decreased in patient’s sperm, and these proteomics results were validated by western blot (Fig. 7A). LAMP2 is an essential factor for lysosome biogenesis during the autophagy process [25]. GABARAP is a member of ATG8 family and functions to sustain the growth of the autophagosome by activating the ULK complex and is crucial for autophagosome-lysosome fusion [26, 27]. Moreover, we observed the attenuated signals of LAMP2 and GABARAP in spermatozoa and the testicular tissues from the patients by immunofluorescence staining (Fig. 7B, E). Consistently, the reduced expression of LAMP2 and GABARAP was also confirmed in testes of Ct55 KO mice (Fig. S11A, B, C). We further isolated spermatogenic cells of human and mouse testes and then performed the immunofluorescence analysis of LAMP2 and GABARAP. LAMP2 is expressed in the cytoplasm (flagellum) and nucleus (head) of spermatocyte and different stages spermatids during spermatogenesis of humans and mice (Fig. S12). GABARAP is mainly localized in the cytoplasm or flagella and moderately expressed in the nuclei of various germ cells of humans and mice (Fig. S13). Importantly, LAMP2 and GABARAP showed robust interactions with the known proteins (downregulated in this study) related to spermatogenesis (Fig. 6F). These results thus strengthened the idea that CT55 plays a pivotal role in spermatogenesis by regulating autophagy process.
We next examined whether CT55 might regulate autophagy by interacting with LAMP2 and GABARAP. We first confirmed that CT55 directly mediated the expression of LAMP2 and GABARAP. Western blot revealed that CT55 knockdown reduced the level of endogenous LAMP2 and GABARAP, and the increased LAMP2 and GABARAP abundance was observed in the cells that overexpressed CT55 (Fig. 7F). The LAMP2 and GABARAP mRNA levels were not largely affected by CT55 depletion (Fig. 7G). The ubiquitin signals of LAMP2 and GABARAP were increased in shCT55 cells compared with control cells (Fig. 7H). To determine whether the effects on LAMP2 and GABARAP stability resulting from changes of CT55 expression were due to an interaction between the two proteins (LAMP2/CT55, GABARAP/CT55), an interaction between endogenous CT55 and LAMP2 or GABARAP was detected using human testicular lysates (Fig. 7I). Moreover, the exogenous Co-IP also showed the binding of CT55 and LAMP2 or GABARAP in 293 T cells (Fig. 7J). Together, our findings revealed that CT55 interacts with LAMP2 and GABARAP, suggesting CT55 might have a role in the maturation step of the autophagy pathway.
A cumulus-oocyte complex penetration defect observed in Ct55 KO male mice by IVF
We next analyzed the fertilizing ability of spermatozoa in Ct55 KO mice using an IVF assay. Intriguingly, the Ct55 KO spermatozoa fertilization rate with cumulus-intact oocytes was lower than that of WT spermatozoa (Fig. 8A). However, the Ct55 KO spermatozoa fertilization rate with cumulus-free oocytes had no significant difference compared to WT spermatozoa (Fig. 8A). The reduced fertilization rate resulted in the subsequent diminish in the percentages of 2-cell embryos and blastocyst in the group of Ct55 KO spermatozoa with cumulus-intact (Fig. 8A). Thus, we hypothesized that the capability of cumulus-oocyte complex (COC) penetration was impaired in Ct55 KO spermatozoa.
A IVF showed the zygotic formation rate, 2-cell embryo rate, and blastocyst rate were significantly decreased in Ct55 KO mice compared with WT mice when cumulus was intact. (n = 3 biologically independent WT mice or KO mice; scale bars, 200 µm; two-sided student’s t test; *p < 0.05; NS, not significant; error bars, s.e.m). B Time course analysis of cumulus cell dispersal was carried out in the presence of cauda epididymal sperm (or sperm protein extracts) of WT mice and Ct55 KO mice. After incubation, the OCCs were observed microscopically. The OCCs were also incubated without the sperm extracts or sperm as a control experiment (“free”). One mM p-amino benzamidine (pAB), an inhibitor sensitive to ACR, inhibits sperm entry into the OCCs. Hyaluronidase was used as the positive control. Apigenin (500 μM) is a hyaluronidase inhibitor. Delayed cumulus cell dispersal of epididymal sperm or sperm protein extract was observed in Ct55 KO mice compared to WT mice, and impairment could be rescued by hyaluronidase. (n = 3 biologically independent WT mice or KO mice; scale bars, 200 µm). C Penetration assays of epididymal sperm through the cumulus in vitro. Cauda epididymal sperm from WT and Ct55 KO mice were labelled by Hoechst 33342 (red dots). The sperm was then incubated with the OCCs for 30 min. Large and small white circles, respectively, indicated the cumulus matrix surface and the zonal pellucida (ZP, stained by ZP3, green) surface. (n = 3 biologically independent WT mice or KO mice; scale bars, 100 µm).
To ascertain that the impaired function of Ct55 affects the process of COC dispersion, we carried out a cumulus cell dispersal assay. A diminished capacity of the Ct55 KO sperm to penetrate the COC was observed after 30 min of incubation when compared to WT sperm (Fig. 8B). Furthermore, the oocytes nearly completely lost cumulus cells 60–120 min after the addition of the WT sperm, while cumulus cell dispersal was significantly delayed by the presence of Ct55 KO sperm (Fig. 8B). Consistent with the results of Ct55 KO sperm, the dispersion of cumulus cells was delayed by incubation with protein extracts of the Ct55 KO sperm compared to WT sperm extracts (Fig. 8B). Thus, the insufficient capacity of Ct55 KO sperm penetrating COC is another reason for the reduced fertility of Ct55 KO mice.
Proteomics analysis of spermatozoa from patient with CT55 mutation showed some decreased proteins involved in fertility or acrosome function (Fig. 6G). Among these proteins, we focused on ACR and SPAM1, which absence causes epididymal sperm to be defective in penetration through the cumulus matrix to reach the ZP [28, 29]. We further employed western blot and immunofluorescence to confirm the reduced expression of ACR and SPAM1 in patients’ sperm and testis tissues (Fig. S14A, B, C). We wondered if the defective COC penetration of Ct55 KO spermatozoa is related to the insufficient function of ACR and SPAM1. Strikingly, western blot and immunofluorescence results exhibited the downregulation of ACR and SPAM1 in Ct55 KO mice (Fig. S14D, E). SPAM1 is the major testicular hyaluronidase, which significantly increases cumulus penetration. As shown in Fig. 8B, COC dispersion was strongly inhibited in the presence of a Hyal inhibitor (300 μM apigenin) in both WT sperm (or sperm extracts) and Ct55 KO sperm (or sperm extracts) (Fig. 8B). Importantly, COC dispersal ability of the Ct55 KO sperm or Ct55 KO sperm protein extracts was restored by adding purified human sperm Hyal to the IVF medium (Fig. 8B). ACR is present in the acrosome as a matrix protein. Consistent with the delay of cumulus cell dispersal by the presence of Ct55 KO sperm or sperm protein extracts, the inhibitory effect of p-amino benzamidine, an inhibitor sensitive to ACR, was evident in cumulus cells with WT sperm or sperm protein extracts (Fig. 8B). Moreover, direct observation of the OCCs inseminated by Ct55 KO sperm showed the abundance of the mutant sperm on the OCC surface (Fig. 8C). Taken together, we concluded that the reduced fertility of male Ct55 KO mice is partly related to the defective COC penetration, which can be rescued by IVF treatment.
Discussion
As a CTA gene, CT55 has been detected in several cancers, such as lung, gastric, and cervical cancers [30]. Our previous study reported that Ct55 deficiency in mice suppresses the development of colitis-associated colorectal (CAC) cancer [31]. Actually, we have found the reduced fertility of Ct55 KO males in our previous study. But until we identified an X-linked nonsense mutation in CT55 in two infertile siblings with impaired spermatogenesis, we decided to explore the role of CT55 in male reproduction. Noticeably, mouse models with depletion of Ct55 showed a progressive reduction in fertility with age, showing the impairments in sperm individualization, unnecessary cytoplasmic removal and the insufficient capability of COC penetration. Mechanistically, our findings uncovered CT55 as a new autophagy regulator by selectively interacting with GABARAP and LAMP2 during spermatogenesis in humans and mice.
Vesicle function is essential for spermatogenesis. The acrosome is sequentially assembled from pro-acrosomal vesicles, and this process depends on the coordinated transport of vesicles to the nuclear membrane, where they ultimately adhere and spread to cover the anterior half of the nucleus. In addition, pro-acrosomal vesicles are involved in spermatid head shaping via the acroplaxome. Noticeably, the transport of vesicles plays a crucial role in the intra-flagellar transport (IFT), which is required for the assembly of the sperm tail accessory structures. Therefore, it is suggested that vesicle dysfunction will certainly lead to male infertility, specifically teratozoospermia and/or asthenozoospermia. However, limited vesicle-related genes have been identified in the involvement of human male sterility. Mutations in PICK1 gene have been implicated in globozoospermia [32]. SPATA16 is required for Golgi function, and mutations in this gene have been suggested to lead to sperm acrosome defects [33]. Intriguingly, we uncovered two patients with male infertility carrying a loss of function mutation in CT55, which is localized in a vesicle. Noticeably, humans lacking CT5 activity exhibited defective acrosome development, and mouse models without CT55 expression also showed in the impaired acrosome function. Additionally, the abnormalities of sperm morphology and movement were also evident in the patients and mice. Thus, our study revealed a novel vesicle-associated gene involved in male reproduction.
Autophagy is an evolutionarily conserved fundamental process that maintains cellular homeostasis by degrading and recycling cytotoxic components [34,35,36]. Accumulative evidence shows that autophagy functions in various physiological processes, such as acrosomal formation, organelle rearrangements, flagellum development, removal of unnecessary cytoplasm, spermatogenic cells survival, and remolding of the germ cells [21,22,23,24], to ensure successful spermatogenesis. The disruption of autophagy activity has been suggested to be closely related to male infertility with various phenotypes. Likewise, lacking ATG5 in mouse germ cells led to male subfertility with anomalous sperm differentiation [37]. The cytoplasm of Fbxo7-deficient mouse spermatids cannot be remodeled and eliminated correctly, causing spermatid death through wrongful phagocytotic cell processes [38]. Moreover, the deletion of Pacrg in mice led to male infertility resulting from the abnormality of the mid-piece of the sperm flagellum and improper removal of redundant cytoplasm [39]. The knockout Atg7 mice displayed severe sperm acrosome defects [23]. In our study, a nonsense mutation of CT55 was identified in two infertile patients, representing aberration in sperm morphology of acrosome and flagella, and also the sharp diminution in sperm count and motility. Deleting Ct55 in the mouse model led to the age-dependent male infertility, which resulted from the impairments in sperm individualization, unnecessary cytoplasmic removal, and ability of COC penetration. Noticeably, incomplete removal of ribosomes and endoplasmic reticulum was observed in the patient’ sperm, as well as the reduced expression of regulators (LAMP2 and GABARAP) of autophagy, which indicates the deficient autophagic activity. Consequently, these observations that the deficient sperm individualization, excessive cytoplasmic residue, and defective acrosome development in patients and Ct55 KO mice are attributed to the inactive autophagic process. We thus proposed that CT55 is a new regulator of autophagy during spermatogenesis.
Accumulating evidence indicates several diseases resulting from impaired autophagic process are age-dependent [40]. Importantly, a progressive impaired spermatogenesis has been observed in Tet1 deficiency mice with age, which is linked to reduced autophagic activity [41]. Specifically, the Tet1-/- males showed the progressive loss of germ cells and defects in meiosis with the increased age [41]. Intriguingly, here our Ct55 KO male mice similarly exhibited the accelerated infertility with increased age in the involvement of lacking the crucial role of CT55 in autophagy during spermatogenic process, showing the impairments of sperm individualization and unnecessary cytoplasm removal. Noticeably, our patients, who were 31-year-old and 33-year-old respectively (both the infertile mice and patients were at the same age), were totally sterile and presented the similar autophagic deficiency-related phenotype. Hence, CT55 maintains spermatogenesis with age, which might give useful information for the treatment of infertility related to pathological autophagic process.
In conclusion, based on our findings, CT55 is first identified as a novel autophagy regulator involved in spermatogenesis. Loss of CT55 impairs fertility in humans and mice, uncovering CT55 as a new CTA gene essential for the reproductive process. It is worthwhile to screen this gene in individuals with infertility. Autophagy is critical for both tumor progression and spermatogenesis. Although previous studies have implicated CT55 in cancer, its roles and the underlying mechanism are poorly understood. Therefore, our findings on CT55 in spermatogenesis via regulating autophagic activity could provide new insights into understanding its potential role in tumorigenesis. Further studies are also required to explore the importance of CT55 in the regulation of mammalian autophagy in other physiological and pathological processes.
Data availability
The data analysed during this study are included in this published article and the Supplementary data files. Additional supporting data are available from the corresponding authors upon reasonable request.
References
Hess RA, Renato de Franca L. Spermatogenesis and cycle of the seminiferous epithelium. Adv Exp Med Biol. 2008;636:1–15.
Cheng CY, Mruk DD. The biology of spermatogenesis: the past, present and future. Philos Trans R Soc Lond Ser B, Biol Sci. 2010;365:1459–63.
O’Donnell L, Nicholls PK, O’Bryan MK, McLachlan RI, Stanton PG. Spermiation: The process of sperm release. Spermatogenesis. 2011;1:14–35.
Boon T, Coulie PG, Van, den Eynde B. Tumor antigens recognized by T cells. Immunol today. 1997;18:267–8.
Chen YT, Old LJ. Cancer-testis antigens: targets for cancer immunotherapy. cancer J Sci Am. 1999;5:16–7.
Aurriere J, Goudenege D, Baris OR, Boguenet M, May-Panloup P, Lenaers G, et al. Cancer/Testis Antigens into mitochondria: a hub between spermatogenesis, tumorigenesis and mitochondrial physiology adaptation. Mitochondrion. 2021;56:73–81.
Lu C, Yang M, Rossi RM, Wang A, Feitosa WB, Diaz FJ, et al. Deletion of the mouse X-linked Prame gene causes germ cell reduction in spermatogenesis. Mol Reprod Dev. 2020;87:666–79.
Mi Y, Shi Z, Li J. Spata19 is critical for sperm mitochondrial function and male fertility. Mol Reprod Dev. 2015;82:907–13.
Donkor FF, Monnich M, Czirr E, Hollemann T, Hoyer-Fender S. Outer dense fibre protein 2 (ODF2) is a self-interacting centrosomal protein with affinity for microtubules. J Cell Sci. 2004;117:4643–51.
Fiedler SE, Dudiki T, Vijayaraghavan S, Carr DW. Loss of R2D2 proteins ROPN1 and ROPN1L causes defects in murine sperm motility, phosphorylation, and fibrous sheath integrity. Biol Reprod. 2013;88:41.
Young SA, Miyata H, Satouh Y, Aitken RJ, Baker MA, Ikawa M. CABYR is essential for fibrous sheath integrity and progressive motility in mouse spermatozoa. J cell Sci. 2016;129:4379–87.
Sinha D, Kalimutho M, Bowles J, Chan AL, Merriner DJ, Bain AL, et al. Cep55 overexpression causes male-specific sterility in mice by suppressing Foxo1 nuclear retention through sustained activation of PI3K/Akt signaling. FASEB J: Off Publ Federation Am Societies Exp Biol. 2018;32:4984–99.
Coutton C, Escoffier J, Martinez G, Arnoult C, Ray PF. Teratozoospermia: spotlight on the main genetic actors in the human. Hum Reprod update. 2015;21:455–85.
Gou LT, Kang JY, Dai P, Wang X, Li F, Zhao S, et al. Ubiquitination-Deficient Mutations in Human Piwi Cause Male Infertility by Impairing Histone-to-Protamine Exchange during Spermiogenesis. Cell. 2017;169:1090–104.e13.
Hansen JN, Rassmann S, Jikeli JF, Wachten D. SpermQ(-)A Simple Analysis Software to Comprehensively Study Flagellar Beating and Sperm Steering. Cells. 2018;8:10.
Alipour F, Jalali M, Nikravesh MR, Fazel A, Sankian M, Khordad E. Assessment of sperm morphology, chromatin integrity, and catSper genes expression in hypothyroid mice. Acta Biologica Hungarica. 2018;69:244–58.
Kawano N, Kang W, Yamashita M, Koga Y, Yamazaki T, Hata T, et al. Mice lacking two sperm serine proteases, ACR and PRSS21, are subfertile, but the mutant sperm are infertile in vitro. Biol Reprod. 2010;83:359–69.
Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16:495–501.
Ozturk N, Steger K, Schagdarsurengin U. The impact of autophagy in spermiogenesis. Asian J Androl. 2017;19:617–8.
Yin J, Ni B, Tian ZQ, Yang F, Liao WG, Gao YQ. Regulatory effects of autophagy on spermatogenesis. Biol Reprod. 2017;96:525–30.
Herpin A, Englberger E, Zehner M, Wacker R, Gessler M, Schartl M. Defective autophagy through epg5 mutation results in failure to reduce germ plasm and mitochondria. FASEB J: Off Publ Federation Am Societies Exp Biol. 2015;29:4145–61.
Shang Y, Wang H, Jia P, Zhao H, Liu C, Liu W, et al. Autophagy regulates spermatid differentiation via degradation of PDLIM1. Autophagy. 2016;12:1575–92.
Wang H, Wan H, Li X, Liu W, Chen Q, Wang Y, et al. Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res. 2014;24:852–69.
Zhuo C, Ji Y, Chen Z, Kitazato K, Xiang Y, Zhong M, et al. Proteomics analysis of autophagy-deficient Atg7-/- MEFs reveals a close relationship between F-actin and autophagy. Biochem Biophys Res Commun. 2013;437:482–8.
Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–30.
Joachim J, Tooze SA. Control of GABARAP-mediated autophagy by the Golgi complex, centrosome and centriolar satellites. Biol Cell. 2018;110:1–5.
Nguyen TN, Padman BS, Usher J, Oorschot V, Ramm G, Lazarou M. Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. The. J Cell Biol. 2016;215:857–74.
Yamagata K, Murayama K, Okabe M, Toshimori K, Nakanishi T, Kashiwabara S, et al. Acrosin accelerates the dispersal of sperm acrosomal proteins during acrosome reaction. J Biol Chem. 1998;273:10470–4.
Park S, Kim YH, Jeong PS, Park C, Lee JW, Kim JS, et al. SPAM1/HYAL5 double deficiency in male mice leads to severe male subfertility caused by a cumulus-oocyte complex penetration defect. FASEB J: Off Publ Federation Am Societies Exp Biol. 2019;33:14440–9.
Dong XY, Li YY, Yang XA, Chen WF. BJ-HCC-20, a potential novel cancer-testis antigen. Biochem cell Biol = Biochim et biologie cellulaire. 2004;82:577–82.
Zhao H, Pan WM, Zhang HH, Song Y, Chen J, Xiang Y, et al. Cancer testis antigen 55 deficiency attenuates colitis-associated colorectal cancer by inhibiting NF-kappaB signaling. Cell Death Dis. 2019;10:304.
Liu G, Shi QW, Lu GX. A newly discovered mutation in PICK1 in a human with globozoospermia. Asian J Androl. 2010;12:556–60.
Dam AH, Koscinski I, Kremer JA, Moutou C, Jaeger AS, Oudakker AR, et al. Homozygous mutation in SPATA16 is associated with male infertility in human globozoospermia. Am J Hum Genet. 2007;81:813–20.
Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev cell developmental Biol. 2011;27:107–32.
Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41.
Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–67.
Huang Q, Liu Y, Zhang S, Yap YT, Li W, Zhang D, et al. Autophagy core protein ATG5 is required for elongating spermatid development, sperm individualization and normal fertility in male mice. Autophagy 2021;17:1753–67.
Rathje CC, Randle SJ, Al Rawi S, Skinner BM, Nelson DE, Majumdar A, et al. A Conserved Requirement for Fbxo7 During Male Germ Cell Cytoplasmic Remodeling. Front Physiol. 2019;10:1278.
Wilson GR, Wang HX, Egan GF, Robinson PJ, Delatycki MB, O’Bryan MK, et al. Deletion of the Parkin co-regulated gene causes defects in ependymal ciliary motility and hydrocephalus in the quakingviable mutant mouse. Hum Mol Genet. 2010;19:1593–602.
Leidal AM, Levine B, Debnath J. Autophagy and the cell biology of age-related disease. Nat Cell Biol. 2018;20:1338–48.
Huang G, Liu L, Wang H, Gou M, Gong P, Tian C, et al. Tet1 Deficiency Leads to Premature Reproductive Aging by Reducing Spermatogonia Stem Cells and Germ Cell Differentiation. iScience. 2020;23:100908.
Acknowledgements
We thank to the patients, their family members, and the normal controls for their support and contribution in this study. We thank to the Analytical and Testing Center of Sichuan University for the morphology characterization and the authors would be grateful to Guiping Yuan and Yi He for their help of TEM and SEM images.
Funding
This work was supported by The National Natural Science Foundation of China (82000514).
Author information
Authors and Affiliations
Contributions
YS designed and supervised the study experiments. Yihong Y, YW, QS and TZ collected the data and conducted the clinical evaluations of the participants. GZ and CJ performed most of the experiments and analyzed the data. Yanting Y, SD, Yushang Y, LY and ML participated in the experiments including immunofluorescence, western blotting and cell culture. HZ and XZ generated the CRISPR mice and conducted the fertility assessment. YS wrote the manuscript. All authors revised and approved the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All the animal experiments were performed in accordance with the recommendation of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and the animal experiments were approved by the Experimental Animal Management and Ethics Committee of West China Second University Hospital (IACUC no. 2021(070)). For human participants, the study was approved by the Ethical Review Board of West China Second University Hospital (IRB no. 2019(040)), Sichuan University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by D. Aberdam
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, G., Jiang, C., Yang, Y. et al. Deficiency of cancer/testis antigen gene CT55 causes male infertility in humans and mice. Cell Death Differ 30, 500–514 (2023). https://doi.org/10.1038/s41418-022-01098-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-01098-6
This article is cited by
-
Spem2, a novel testis-enriched gene, is required for spermiogenesis and fertilization in mice
Cellular and Molecular Life Sciences (2024)