Abstract
The high-mobility group box-1 (HMGB1) protein is implicated in the development of various cancers and their proliferation. According to its function, HMGB1 shuttles between the cell nucleus and cytoplasm, assisting with nucleosome stabilization and gene transcription, or localizing in the cell membrane for outgrowth. The clinicopathologic and prognostic significance of these different subcellular locations and their correlation has been unclear in colorectal cancer (CRC). We found significantly higher rates of nuclear HMGB1 expression in CRC and colorectal adenoma tissue samples (84.0% and 92.6%, respectively) than in normal colorectal tissue (15.0%) and a significantly higher rate of positive cytoplasmic HMGB1 expression in CRC tissue (25.2%) compared with colorectal adenoma (11.8%) and normal colorectal tissue (0.0%). Positive cytoplasmic HMGB1 expression was associated with high-grade CRC, a poor prognosis, and was negatively correlated with strongly positive nuclear HMGB1 expression in CRC tissue specimens (r = – 0.377, P = 0.000). CRC patients with strongly positive nuclear HMGB1 expression had a better survival prognosis than other CRC patients. Preventing nuclear plasma translocation of HMGB1 may be a new strategy for CRC management.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is one of the most common types of cancers globally, and is ranked amongst the top three malignancies in terms of morbidity and mortality1,2. High-mobility group box-1 (HMGB1) protein, also known as amphoterin or HMG1, was discovered in calf thymus and named according to its high electrophoretic mobility in polyacrylamide gels3. The biological function of HMGB1 depends on its subcellular localization and expression4, whereby HMGB1 can play a paradoxical role in promoting or suppressing cancer during the progression of malignant tumors3,5,6,7,8,9,10. Inside the nucleus, HMGB1 is a highly conserved chromosomal protein engaged in DNA repair, transcription and genome stability3,5. Post-translational modifications, such as phosphorylation, acetylation and methylation, enables HMGB1 to be translocated into the cytoplasm and actively released outside the cell by means of secretory lysosomes or organelles, where HMGB1 promotes inflammation, immunity, cell proliferation, autophagy, apoptosis and metastasis3,5.
The function and molecular mechanisms of HMGB1 remain unclear in cancer. In CRC pathogenesis, the role of HMGB1 depends on the redox status of the protein11. While some studies have detected the expression of HMGB1 protein in CRC12,13,14,15, research on the subcellular localization of HMGB1 in CRC is only available from individual reports, both of which involve only small samples of CRC tissue16,17. Also, the research has failed to clarify any association between different subcellular locations of HMGB1 and their clinicopathologic prognostic significance.
We performed an immunohistochemical (IHC) analysis of HMGB1 expression in CRC and colorectal adenoma tissue samples obtained from a cohort of Chinese patients. We examined the expression of HMGB1 in different subcellular locations (nucleus and cytoplasm) and analyzed the association between them. This study aimed to determine the clinicopathologic and prognostic significance of different subcellular locations of HMGB1.
Results
Subcellular localization and HMGB1 expression in colorectal tissue
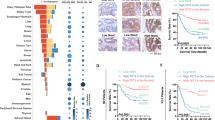
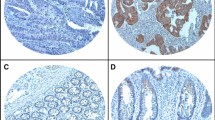
In normal colorectal tissue, HMGB1 protein was localized in the nucleus, while some cytoplasmic localization was observed in CRC and colorectal adenoma tissue samples (Fig. 1). Nuclear HMGB1 expression was generally positive in normal colorectal, adenoma and CRC tissue samples, with rates of 95.0% (19/20), 98.5% (67/68) and 99.5% (367/369), respectively. Strongly positive nuclear HMGB1 rates in normal colorectal, adenoma and CRC tissue samples were 15.0% (3/20), 92.6% (63/68) and 84.0% (310/369), respectively (P < 0.01, Table 1). Rates of positive cytoplasmic HMGB1 expression were 11.8% (8/68) in colorectal adenoma tissues and 25.2% (93/369) in CRC tissues (P < 0.01, Table 2).
Immunochemical analysis of HMGB1 expression and subcellular localization in colorectal tissue. (A–C) Normal colorectal tissue, not strongly nuclear positive/cytoplasmic negative. (D–F) Colorectal adenomas, strongly nuclear positive/cytoplasmic negative. (G–I) Low-grade CRC, strongly nuclear positive/cytoplasmic negative. (J–L) High-grade CRC, not strongly nuclear positive/cytoplasmic positive. (M–O) Negative control, nuclear negative/cytoplasmic negative.
The relationship between nuclear and cytoplasmic HMGB1 expression in CRC
According to levels of HMGB1 expression in the nucleus and cytoplasm, the 369 CRC patients were divided into four groups: nuclear HMGB1 expression was not strongly positive and cytoplasmic HMGB1 expression was negative in 22 patients; 254 exhibited strongly positive nuclear/cytoplasmic-negative HMGB1 expression; 37 were not strongly positive for nuclear HMGB1 expression and were cytoplasmic-positive; and 56 exhibited strongly positive nuclear/cytoplasmic-positive HMGB1 expression. Among those with cytoplasmic-positive CRC, the rate of strongly positive nuclear HMGB1 expression was 60.2% (56/93), which was significantly lower than that in the cytoplasmic-negative CRC cohort (92.0%, 254/276) (P < 0.01, Table 3). Spearman correlation analysis revealed a significantly negative correlation between cytoplasmic HMGB1 and nuclear HMGB1 expression in CRC tissue specimens (r = – 0.377, P = 0.000).
The relationship between nuclear and cytoplasmic HMGB1 expression and clinicopathologic characteristics of CRC patients
As shown in Table 4, positive cytoplasmic HMGB1 expression was associated with a poor tumor grade in the CRC cohort (P < 0.05). CRC patients aged ≥ 60 years (27.7%, 73/264) had a higher positive cytoplasmic HMGB1 rate than patients aged < 60 years (19.0%, 20/105), but the between-group difference was not significant (P = 0.086). No correlation was found between strongly positive nuclear HMGB1 expression and clinicopathologic characteristics of CRC patients.
High levels of nuclear or cytoplasmic HMGB1 expression predict opposite effects on survival in CRC
We analyzed levels of nuclear or cytoplasmic HMGB1 expression in relation to RFS and OS rates in patients with CRC. As shown in Fig. 2A, the 216 patients whose primary tumors exhibited strongly positive nuclear HMGB1 expression had a mean OS of 54.7 months (an estimated 5-year OS rate of 79.2%), while the 42 patients whose tumors did not exhibit strongly positive nuclear HMGB1 expression had a mean OS of 49.1 months (an estimated 5-year OS rate of 61.9%, P = 0.009). Similarly, CRC patients with strongly positive nuclear HMGB1 expression had a better RFS than those whose nuclear HMGB1 expression was not strongly positive, although the data did not support an unequivocal association (Fig. 2B, P = 0.054).
Associations between nuclear and cytoplasmic HMGB1 expression with survival of patients with colorectal cancer. (A,B). Associations of nuclear HMGB1 expression with overall survival (OS) (A) and relapse-free survival (RFS) (B) were analyzed in the colorectal cancer cohort. (C,D). Associations of cytoplasmic HMGB1 expression with OS (C) and RFS (D) were analyzed in the colorectal cancer cohort. P-values were calculated using the Mantel-Cox log-rank test.
The 68 patients whose primary tumors exhibited positive cytoplasmic HMGB1 expression had a mean OS of 51.2 months (an estimated 5-year OS rate of 67.6%), while the 190 patients whose tumors were negative for cytoplasmic HMGB1 expression had a mean OS of 54.7 months (an estimated 5-year OS rate of 79.5%, Fig. 2C, P = 0.041). Similarly, patients with cytoplasmic-positive HMGB1 expression experienced worse RFS. Patients whose primary tumors were HMGB1 cytoplasmic-positive had a mean RFS of 44.3 months (an estimated 5-year RFS rate of 57.4%), while patients whose tumors were HMGB1 cytoplasmic-negative had a mean RFS of 49.8 months (an estimated 5-year RFS rate of 70.0%, Fig. 2D, P = 0.042).
Discussion
Scant data are available in regard to the subcellular localization of HMGB1 in malignant tumors. Kostova and colleagues found that HMGB1 subcellular localization was associated with histologic differentiation of malignant tumors, as moderately differentiated carcinomas exhibit perinuclear localization of the protein, while poorly differentiated carcinomas show a tendency for non-specific nuclear localization18. Data about the subcellular localization of HMGB1 in CRC is only available from two individual reports, both of which involve small samples of CRC tissue and neither has clarified the correlation between different subcellular localization sites of HMGB1 and their clinicopathologic prognostic significance16,17.
Our study found that in normal colorectal tissue, colorectal adenoma and CRC samples, HMGB1 is generally positively expressed in the nucleus. In colorectal adenoma and CRC tissues, the rates of strongly positive nuclear HMGB1 expression were significantly higher than the rate in normal colorectal tissue, while the rate was slightly higher in colorectal adenoma than in CRC tissue, suggesting that the enhanced expression of nuclear HMGB1 occurs in the early stage of colorectal neoplastic lesions. In addition, the positive rates of cytoplasmic HMGB1 in normal colorectal tissue, colorectal adenoma and CRC showed a gradually increasing trend, with significantly higher expression of positive cytoplasmic HMGB1 in high-grade versus low-grade CRC tissue, suggesting that cytoplasmic HMGB1 expression is closely related to CRC tumor grade.
We also found a significantly lower strongly positive rate of nuclear HMGB1 expression in cytoplasmic-positive CRC than in cytoplasmic-negative CRC, and a significant negative correlation between cytoplasmic and nuclear HMGB1 expression. Moreover, we observed that in CRC or adenoma cells subjected to cytoplasmic staining, nuclear staining was either weak or not apparent. Thus, we have revealed an obvious nuclear plasma translocation phenomenon of HMGB1 protein in CRC.
We found that cytoplasm-positive HMGB1 expression is related to a poor prognosis in CRC. Interestingly, nuclear-positive HMGB1 expression had an opposite outcome on survival prognosis, as CRC patients with strongly positive nuclear HMGB1 expression had a better prognosis than other patients. Previous studies involving different cancers concluded that HMGB1 plays a paradoxical role in promoting or suppressing cancer3,5,6,7,8,9,10. This is the first time that HMGB1 has been found to play both positive and negative roles in the same study population. We believe that the enhancement of nuclear HMGB1 expression in colorectal adenoma and CRC lays the molecular basis for nuclear plasma translocation for the further progress of tumors in later stages of the disease. As nuclear HMGB1 is involved in a wide range of effects, such as DNA repair, transcription and genome stability3,5, it is understandable that its high expression would be protective in regard to survival in CRC. We speculate that high levels of nuclear HMGB1 expression activate many downstream genes or pathways, such as epidermal growth factor receptor (EGFR)19 and β-catenin20,21. We found a significantly positive correlation between strongly positive levels of nuclear HMGB1 and EGFR positive or β-catenin positive expression in CRC tissue specimens (Supplementary materials, Table S1–S4, Figure S1). Further research into the molecular mechanisms of HMGB1 nuclear plasma translocation in CRC is necessary. Preventing the nuclear plasma translocation of HMGB1 may be a potentially useful strategy for CRC treatment.
Materials and methods
Patients and tissue samples
Tissue samples were obtained from 369 Chinese patients with CRC and 68 patients with colorectal adenoma. All patients were undergoing primary surgical treatment at the Affiliated Dongyang Hospital of Wenzhou Medical University (Dongyang, Zhejiang, China) between 2008 and 2015. Twenty cases of adjacent normal colorectal tissue specimens were obtained from the above-mentioned 20 CRC samples post-surgery. Clinicopathologic characteristics were determined for all patients based on their medical records. CRC patients with a median age of 69 years (range 24–94), and patients diagnosed with colorectal adenoma with a median age of 68 years (range 33–88). The characteristics and distribution of CRC population are shown in Table 4. Pathohistologic and clinical diagnosis was made according to the World Health Organization classification of tumours of the digestive system22. CRC patients were staged according to the eighth edition of the AJCC cancer staging manual23. Follow-up information was available for 258 patients with CRC; the median follow-up was 60 months (range, 6–75 months). Study approval was granted by the Ethics Committee of the Affiliated Dongyang Hospital of Wenzhou Medical University. Written informed consent forms were signed by all patients or their guardians. All study methods satisfied the relevant guidelines and regulations issued by the Affiliated Dongyang Hospital of Wenzhou Medical University.
Tissue array preparation
We followed the methods described by Wang et al., 202024.
IHC analysis
IHC staining of paraffin-embedded tissue array sections was conducted using the Envision System (Dako, Glostrup, Denmark), as described previously25,26. Briefly, the sections were submerged in boiling sodium citrate (pH, 6.0) for 2 min in a pressure cooker. After being treated with 0.3% hydrogen peroxide for 10 min to block endogenous peroxidase, the sections were incubated with primary antibody for 1 h at room temperature. The sections were then incubated with secondary antibody (Dako) for 40 min at room temperature, followed by incubation with 3,3′-diaminobenzadine (Dako) at room temperature. The primary antibody were anti-HMGB1 rabbit monoclonal antibody (clone EPR3507, diluted at 1:1000, Abcam, Cambridge, England), anti-EGFR rabbit polyclonal antibody (clone 1005, diluted at 1:100, Santa Cruz Biotechnology, Santa Cruz, USA) and anti-β-catenin rabbit monoclonal antibody (clone D10A8; diluted at 1:400; Cell Signaling Technology, Boston, USA). The secondary antibody was Dako's HRP rabbit/mouse universal antibody (Dako, Glostrup, Denmark). The negative control was incubated with vehicle then with secondary antibody, without primary antibody. As positive control, we used the internal control in the colorectal tissue, to check for HMGB1 nuclear staining in the interstitial fibers and lymphocytes; a breast cancer tissue that is known to be EGFR-positive or β-catenin-positive serves as positive control for EGFR and β-catenin staining, respectively.
Assessment of HMGB1 staining
The entire tissue array section was scanned and scored separately by 2 pathologists. The intensity of nucleus and cytoplasm HMGB1 staining was assessed in colorectal tissue. Staining intensity was scored on a 4-point scale from 0 (negative) to 1 (weak), 2 (medium), or 3 (strong)27. In both the nucleus and cytoplasm, scores of 0 were negative and 1 to 3 were positive for HMGB1. A score of 2 to 3 in the nucleus was considered to be strongly positive. High levels of expression are expressed as strongly positive nuclear HMGB1 and positive cytoplasmic HMGB1 expression.
Patient follow-up
We followed the methods described by Wang et al., 202024.
Statistical analysis
Statistical analyses were conducted using SPSS version 19.0 (SPSS Inc, Chicago, IL, USA). Between-group differences were compared using a Pearson’s Chi-square test for qualitative variables. The correlation between nucleus and cytoplasm HMGB1 expression was assessed by Spearman’s correlation analysis. Patient RFS and OS rates were analyzed using the Kaplan–Meier method and compared using log-rank analysis. P < 0.05 was considered to be statistically significant.
Abbreviations
- IHC:
-
Immunohistochemistry
- CRC:
-
Colorectal cancer
- HMGB1:
-
High-mobility group box-1
- RFS:
-
Relapse-free survival
- OS:
-
Overall survival
References
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2019. CA: Cancer J. Clin. 69, 7–34 (2019).
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J. Clin. 68, 394–424 (2018).
Kang, R., Zhang, Q., Zeh, H. J. 3rd., Lotze, M. T. & Tang, D. HMGB1 in cancer: Good, bad, or both?. Clin. Cancer Res. 19, 4046–4057 (2013).
Youn, J. H. & Shin, J. S. Nucleocytoplasmic shuttling of HMGB1 is regulated by phosphorylation that redirects it toward secretion. J. Immunol. 177, 7889–7897 (2006).
He, S. J. et al. The dual role and therapeutic potential of high-mobility group box 1 in cancer. Oncotarget 8, 64534–64550 (2017).
Wang, K. et al. HMGB1-containing nucleosome mediates chemotherapy-induced metastasis of human lung cancer. Biochem. Biophys. Res. Commun. 500, 758–764 (2018).
Xia, J. et al. Inhibiting the cytoplasmic location of HMGB1 reverses cisplatin resistance in human cervical cancer cells. Mol. Med. Rep. 15, 488–494 (2017).
Kang, R. et al. Intracellular HMGB1 as a novel tumor suppressor of pancreatic cancer. Cell Res. 27, 916–932 (2017).
Amornsupak, K. et al. High ASMA(+) fibroblasts and low cytoplasmic HMGB1(+) breast cancer cells predict poor prognosis. Clin. Breast Cancer 17, 441–452 (2017).
Huang, C. Y. et al. HMGB1 promotes ERK-mediated mitochondrial Drp1 phosphorylation for chemoresistance through RAGE in colorectal cancer. Cell Death Dis. 9, 1004 (2018).
Cheng, K.J., Alshawsh, M.A., Mejia Mohamed, E.H., Thavagnanam, S., Sinniah, A., Ibrahim, Z.A. HMGB1: An overview of its versatile roles in the pathogenesis of colorectal cancer. Cell. Oncol. (2019).
Suren, D. et al. The role of high mobility group box 1 (HMGB1) in colorectal cancer. Med. Sci. Monitor 20, 530–537 (2014).
Zhang, W. et al. Increased HMGB1 expression correlates with higher expression of c-IAP2 and pERK in colorectal cancer. Medicine 98, e14069 (2019).
Hongo, K. et al. Immunohistochemical detection of high-mobility group box 1 correlates with resistance of preoperative chemoradiotherapy for lower rectal cancer: A retrospective study. World J. Surg. Oncol. 13, 7 (2015).
Yao, X. et al. Overexpression of high-mobility group box 1 correlates with tumor progression and poor prognosis in human colorectal carcinoma. J. Cancer Res. Clin. Oncol. 136, 677–684 (2010).
Peng, R. Q. et al. Co-expression of nuclear and cytoplasmic HMGB1 is inversely associated with infiltration of CD45RO+ T cells and prognosis in patients with stage IIIB colon cancer. BMC Cancer 10, 496 (2010).
Li, Z. et al. Expression of high mobility group box-1 in colorectal cancer and its clinical significance. Zhonghua Wei Chang Wai Ke Za Zhi (Chin. J. Gastrointest. Surg.) 18, 616–619 (2015).
Kostova, N., Zlateva, S., Ugrinova, I. & Pasheva, E. The expression of HMGB1 protein and its receptor RAGE in human malignant tumors. Mol. Cell. Biochem. 337, 251–258 (2010).
Feng, L. et al. HMGB1 promotes the secretion of multiple cytokines and potentiates the osteogenic differentiation of mesenchymal stem cells through the Ras/MAPK signaling pathway. Exp. Ther. Med. 12, 3941–3947 (2016).
Wang, X. H., Zhang, S. Y., Shi, M. & Xu, X. P. HMGB1 promotes the proliferation and metastasis of lung cancer by activating the Wnt/beta-catenin pathway. Technol. Cancer Res. Treat. 19, 1533033820948054 (2020).
Jin, X. et al. High mobility group box 1 promotes aortic calcification in chronic kidney disease via the Wnt/beta-catenin pathway. Front. Physiol. 9, 665 (2018).
Bosman, F. T. C. F., Hruban, R. H. & Theise, N. D. WHO Classification of Tumours of the Digestive System 4th edn. (IARC Press, Lyon, 2010).
Amin, M.B., Greene, F.L., Edge, S.B., Compton, C.C., Gershenwald, J.E., Brookland, R.K., Meyer, L., Gress, D.M., Byrd, D.R., Winchester, D.P. The eighth edition AJCC cancer staging manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA: Cancer J. Clin. 67, 93–99 (2017).
Wang, C. Q. et al. High expression of both resistin and fascin-1 predicts a poor prognosis in patients with colorectal cancer. Biomed. Res. Int. 2020, 8753175 (2020).
Wang, C. Q. et al. Fascin-1 as a novel diagnostic marker of triple-negative breast cancer. Cancer Med. 5, 1983–1988 (2016).
Wang, C. Q. et al. EGFR conjunct FSCN1 as a novel therapeutic strategy in triple-negative breast cancer. Sci. Rep. 7, 15654 (2017).
Wang, C. Q. et al. Expression of HMGB1 protein in breast cancer and its clinicopathological significance. Zhonghua Bing Li Xue Za Zhi (Chin. J. Pathol.) 49, 57–61 (2020).
Acknowledgements
This work was supported by a grant from the National Natural Science Foundation of China (No. 81802660). We would like to thank Iona J. MacDonald from China Medical University for her English language revision of this manuscript.
Author information
Authors and Affiliations
Contributions
Y.W., Q.W. and Y.Z. performed the experiments. C.Q.W. and F.S. analyzed the data. C.Q.W. designed the experiments. B.F.H. and J.K.S. were responsible for management of the clinical samples. C.Q.W. wrote the manuscript. C.H.T. and H.C.J. revised the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, CQ., Huang, BF., Wang, Y. et al. Subcellular localization of HMGB1 in colorectal cancer impacts on tumor grade and survival prognosis. Sci Rep 10, 18587 (2020). https://doi.org/10.1038/s41598-020-75783-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-75783-2
This article is cited by
-
Musashi-2 potentiates colorectal cancer immune infiltration by regulating the post-translational modifications of HMGB1 to promote DCs maturation and migration
Cell Communication and Signaling (2024)
-
Dysfunctional TLR1 reduces the therapeutic efficacy of chemotherapy by attenuating HMGB1-mediated antitumor immunity in locally advanced colorectal cancer
Scientific Reports (2023)
-
Interleukin-1 alpha and high mobility group box-1 secretion in polyinosinic:polycytidylic-induced colorectal cancer cells occur via RIPK1-dependent mechanism and participate in tumourigenesis
Journal of Cell Communication and Signaling (2023)
-
Cell type-specific response of colon cancer tumor cell lines to oncolytic HSV-1 virotherapy in hypoxia
Cancer Cell International (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.