Abstract
Gammarus roeselii Gervais, 1835 is a morphospecies with a wide distribution range in Europe. The Balkan Peninsula is known as an area of pre-Pleistocene cryptic diversification within this taxon, resulting in at least 13 Molecular Operational Taxonomic Units (MOTUs). The morphospecies diversified there during Neogene and has probably invaded other parts of the continent very recently, in postglacial or even historical times. Thus, the detailed goals of our study were to (1) identify which lineage(s) colonized Central-Western Europe (CWE), (2) determine their possible geographical origin, (3) verify, whether the colonisation was associated with demographic changes. In total, 663 individuals were sequenced for the cytochrome oxidase I (COI) barcoding fragment and 137 individuals for the internal transcribed spacer II (ITS2). We identified two MOTUs in the study area with contrasting Barcode Index Number and haplotype diversities. The Pannonian Basin (PB) appeared to be a potential ice age refugium for the species, while CWE was colonised by a single lineage (also present in PB), displaying low genetic diversity. Our results suggest that G. roeselii is a relatively recent coloniser in CWE, starting demographic expansion around 10 kya.
Similar content being viewed by others
Introduction
The current distribution of animal and plant species and their phylogeographic lineages have been largely shaped by the past geological and climatic events1. For example, the last Pleistocene glacial period and the subsequent climate warming, has had severe effects on the genetic diversity and distribution of numerous species in Europe2. At the time of the Last Glacial Maximum (LGM) at ca. 26–15 kya, the northern parts of the continent were covered by the Eurasian Ice Sheet Complex3. The cold temperature and the reduced water availability forced most of the local fauna and flora to retreat into isolated southern ice-age refugia. The most prominent were the three main peninsulas in the Mediterranean region: the Iberian, the Apennine and the Balkan Peninsula, where prolonged isolation led to the divergence of phylogeographic lineages4. The post-LGM climatic warm-up was associated with northward expansions often combined with a series of founder events, leading to a general geographic pattern of genetic diversity reduction from south to north in the case of many species1,4,5. Apart from the three main refugia, several extra-Mediterranean refugia have been recognized, e.g., the Alps, the Carpathian Arch, and the Pannonian Basin (PB)6,7,8,9,10.
The majority of studies on the topic of diversification associated with glacial refugia and postglacial colonizations focuses on terrestrial taxa2, 6. In freshwaters, the dynamics can be often different compared to terrestrial landscapes. In the former, the habitats could be either patchier and isolated, promoting divergence, or on the opposite, could be interconnected promoting exchange. For example, pre-Pleistocene tectonic movements and subsequent changes in watersheds, with isolated drainages, possibly impacted the rate of species diversification in many Eurasian freshwater taxa11,12,13,14,15,16. However, on the opposite, present distribution and population connectivity in freshwater fauna is largely affected by the postglacial climate warming, as well as by human impact5,17,18,19. The construction of large navigable waterways (canals, channels) has been a primary anthropogenic factor heavily affecting dispersal of species both in historical and recent times20,21,22.
Gammarid amphipods can be considered as challenging models for phylogeographic studies in fresh waters. Members of this group are permanently aquatic and have very limited ability to survive drought23. The natural dispersal abilities of amphipods between isolated water bodies are minimal; although recent studies documented local (small-scale) distribution of amphipod haplotypes across watersheds24,25. In addition, rare cases of ectozoochory by birds26 or by mammals27 are also documented. On the other hand, gammarids breed fast, as they usually have 1–3 generations per year28. Potentially, a few gravid females can establish a population29. Many species can survive adverse cold climatic conditions, including severe winters under ice cover30. For that reason, the location of their glacial refugia, timing of lineage diversification, and dynamics of post-glacial colonisation are hard to predict. On one end of the spectrum, old isolated water basins (e.g. ancient lakes) can hold endemic lineages and species31,32,33. Recent phylogeographic studies have indicated high molecular cryptic diversity within several amphipod morphospecies, with numerous Molecular Operational Taxonomic Units (MOTUs) representing phylogenetic species of pre-Pleistocene divergence13,34. This is especially true for morphospecies with wide geographic distribution ranges, such as Gammarus balcanicus Schäferna, 192313 and Gammarus fossarum Koch, 183635. On the other end of the spectrum, artificially connected large river systems allow for homogenization of freshwater fauna21,36 and mixing the genetic pools of previously isolated populations37. Fish restocking might also be a vector of amphipod introduction38,39. Amphipod crustaceans are listed among the richest in the number of invasive species successfully colonizing large water bodies (rivers and lakes) in Europe40,41. The prominent examples are Dikerogammarus villosus (Sowinsky, 1894)40, Dikerogammarus haemobaphes42 and Pontogammarus robustoides (G.O. Sars, 1894)39.
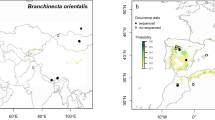
Gammarus roeselii has a wide but very peculiar distribution in Europe (Fig. 1). In the Balkans, G. roeselii can be found in various types of water bodies, from mountain streams, through rivers, to lakes11. Outside the Balkans, this species is associated mainly with the network of rivers and canals (Fig. 1). Already several decades ago, some studies suggested that G. roeselii originated from the Balkan Peninsula and its expansion outside the Balkans i.e., into the Pannonian Basin (PB) and Central-Western Europe (CWE) took place in historical times and progressed through artificial canals, possibly with the unintentional help of humans transferring water plants20,43. Currently, in PB and CWE, G. roeselii is considered as an old, well-established alien species17. This alien-exotic status is reinforced by recent reports of further expansion in France44 and Italy45. Surprisingly, G. roeselii was described by Gervais in 1835 from Coulanges-sur-Yonne (locus typicus), located 200 km south-east of Paris on the Yonne river. At that time, the place was not connected to the western part of G. roeselii's distribution46, thus presence of the species in those waterbodies was explained by human translocation with aquatic plants20. Jażdżewski20, as well as Jażdżewski and Roux43 suggested that presence of G. roeselii in the Oder River in Poland may be a result of both (1) the eastward colonisation through the Oder-Spree and Oder-Havel canal systems and (2) crossing the rather low watershed between the Oder and the Morava River (a Danube tributary) in the Czech Republic. In addition, the authors did not exclude that part of the current distribution of G. roeselii outside the Balkans could be associated with a natural post-glacial expansion, possibly of a single lineage, especially in the lower part of the Danube basin. In agreement with this hypothesis, Moret and colleagues47, based on 16S mtDNA, as well as Copilaş-Ciocianu and colleagues48, based on COI, suggested that only one phylogeographic lineage is present in Hungary, Czech Republic, but also France. On the other hand, Grabowski et al.11 recently identified 13 pre-Pleistocene MOTUs in the Balkan Peninsula showing that G. roeselii is characterised by a high level of cryptic diversity within this area.
Geographic range of Gammarus roeselii. Rebuilt after Jażdżewski (1980)20, Jażdżewski & Roux (1988)43, Copilaş-Ciocianu et al.96 and our own data. CWE = Central-Western Europe, PB = Pannonian Basin, BP = Balkan Peninsula. Map created with QGIS 3.4.5 (https://www.qgis.org/fr/site/).
The aim of our present study was to examine the phylogeography of G. roeselii in PB and CWE in relation to the 13 pre-Pleistocene MOTUs recently identified by Grabowski et al.11 in the Balkan Peninsula. To complete this goal, we: (1) hypothesised that PB could host further cryptic diversity acting as an additional glacial refugium, (2) tested how many lineages are present in PB and if this area could have served as a source for the colonisation of CWE.
Results
Mt-DNA: COI haplotypes, BIN and MOTU definition, divergence and diversification time-frames
Out of 711 individuals from the Pannonian Basin (PB) and the Central-Western Europe (CWE), 55 COI haplotypes were defined (Table 1; Fig. 2c) which were clustered in BOLD into twelve Barcode Index Numbers (BINs) (Fig. 2a,c). Three of these BINs (AAY1309, ADD4052, ADD6100) were known already from the four northernmost sites in the study by Grabowski et al.11, i.e., PB. Altogether, at the scale of Europe (including the Balkan Peninsula), a total of 34 BINs is now registered (Fig. 2a). The BI tree allowed to group these 34 BINs in 14 MOTUs (Fig. 2a), two of them, MOTUs O and C, being present in PB and only one, MOTU C, in CWE (Fig. 2b).
Phylogeny and distribution of Gammarus roeselii COI BINs: (a) Bayesian tree reconstructed from representative sequences of all COI BINs with MOTUs definition. MOTUs were defined after Grabowski et al.11. BINs produced in this study are indicated by frames. Values at nodes indicate the bootstrap values (above 0.5) for the parallelly reconstructed ML tree. (b) Map showing all sampling sites of the study. Colours indicate BINs of A. Triangles indicate sampling sites of sequences from Grabner et al.49. Black dots represent the sites from Grabowski et al.11, displaying also the COI MOTUs found in the Balkan Peninsula. (c & d) Haplotypic networks of COI and ITS2 markers, respectively. Colour codes indicate COI BINs. Stars on the ITS2 network indicate ITS2 haplotypes which were present in northern Italy. Map created with QGIS 3.4.5 (https://www.qgis.org/fr/site/).
MOTU O is new, includes only the BIN ADA0638, and it diverged in the late Miocene from its sister clade containing MOTUs F and G found in the northern part of the Balkan Peninsula (Fig. 2a,b).
MOTU C includes the three BINs from Grabowski et al.11, as well as eight new BINs (ADD6100, ACZ7565, ACZ7566, ADH3845, ADH3846, ADH3847, ADA0637 and ADD6734). The diversification of BINs within MOTU C occurred in the Pannonian Basin. It has started in the middle Pliocene and peaked during the Pleistocene. MOTU C is in a phylogenetic sister relationship with two MOTUs (A and B) found in the northern part of the Balkan Peninsula, from which it diverged in late Miocene (Fig. 2a,b, Supplementary Fig. S1).
Detailed geographic distribution of haplotypes and BINs in CWE
Outside the Balkan Peninsula the relative abundance and distribution of the different BINs are highly variable. Ten BINs are rare (1 to 17 individuals) and not found outside the Pannonian Basin. BINs, such as ADH3845, ADH3846 and ADH3847 from northern Croatia, ACZ7565 and ACZ7566 from the Apuseni Mountains in Western Romania, as well as ADD6734 from the North Hungarian Mountains, are restricted to single localities. The BINs ADA0637 and ACZ9504 include individuals from several localities, but close to each other geographically (Fig. 2b).
The BIN ADD6100 is moderately frequent (56 individuals, 7.88%) and relatively widespread, occurring in the area extending from the Pannonian Basin, westwards to the Alps and to northern Italy (Fig. 2b).
The most frequent BIN is AAY1309 (583 individuals, 82%), being present in the Pannonian Basin and almost all around CWE, from the catchment of the Somme River in France, through the Netherlands and Germany, the Alps in Austria, Czech Republic, the basin of Po River in northern Italy, and western Poland, reaching the Vistula River on the east (Fig. 2b).
The MJ haplotype network shows the presence of 6 haplotypes within BIN ADD6100, while BIN AAY1309 is more diverse, including 31 haplotypes, with higher genetic distances between them (Fig. 3a). BINs AAY1309 and ADD6100 co-occur only on three sites in the Pannonian Basin (Fig. 3b; Table 1). BIN ADD6100 is characterised by a higher haplotype diversity in the Pannonian Basin compared to northern Italy. Within BIN AAY1309, the most frequent haplotype, haplotype 1, and the rare peripheral haplotypes are generally frequent in the northern and western parts of CWE, while haplotype 12 and the surrounding haplotypes can be found more often in the central and southern areas (Fig. 3b). Some sites from the Pannonian Basin contain private and divergent haplotypes of BIN AAY1309 (haplotype 20, haplotype 60).
Haplotype distribution and frequencies of the two most widely distributed Gammarus roeselii COI BINs: (a) Haplotype network of BINAAY1309 and AAD6100. (b) Distribution and frequencies of haplotypes. Pie charts represent single localities. Site number (as in Table 1) and number of sequenced individuals per site are indicated. Colour codes for haplotypes are based on the haplotype network of the two BINs. Map created with QGIS 3.4.5 (https://www.qgis.org/fr/site/).
Mitochondrial versus nuclear haplotype relationships
The phylogenetic relationships based on COI and ITS2 haplotypes are illustrated with MJ haplotype networks (Fig. 2c,d).
In the COI network, the two most widely distributed BINs AAY1309 and AAD6100 consist, as already pointed out, of the highest number of haplotypes and individuals. Other BINs that have more localized distribution hold fewer haplotypes and are represented by peripheral haplotypes in the MJ network. BIN ADA0638, which forms MOTU O, is represented by only one haplotype, extremely divergent from all the others (Fig. 2c), as expected given its position in the BI tree (Fig. 2a).
The overall topology of the ITS2 network is largely incongruent with the COI-based one (Fig. 2d). As expected, the genetic distances between ITS2 haplotypes are smaller than in case of COI49,50, but at the same time, the topology and phylogenetic relationships are more complex. There is one haplotype represented by the highest number of individuals (ITS2 haplotype 2), while most of the other haplotypes are unique to single individuals (Fig. 2d). The grouping of COI haplotypes into BINs is not well reflected in the ITS2 network. Concerning MOTU C, the BINs AAY1309 and AAD6100 share the most frequent ITS2 haplotype (haplotype 2), while other ITS2 haplotypes corresponding to these BINs are scattered all around the network. Also the BINs AAY1309 and ADH3846 share one haplotype. Interestingly, MOTU O (BIN ADA0638) is characterized by one unique COI haplotype (haplotype 18) but its ITS2 haplotype is commonly observed and associated with an individual assigned to BIN AAY1309 of MOTU C. On the other hand, individuals of BIN ADA0637 are parted away from each other in the ITS2 network, while based on the COI sequences they are closely related. Only the individuals of the BINs ADA0637, ADD6734, ACZ7566, highly divergent on the COI network, show also the highest level of divergence on the ITS2 network. On the contrary, the BINs ADA0638 and ACZ7565 represent distant groups in the COI network, while in the ITS2 network both of these BINs have a central position, with few mutations separating them from the most frequent haplotype.
An interesting pattern can be observed while comparing the geographic distribution of the COI and ITS2 haplotypes to their position in the MJ networks. While in the case of COI, distant peripheral haplotypes are present only in the Pannonian Basin, the ITS2 haplotypes of the corresponding individuals are present both in the Pannonian Basin and in northern Italy. There are some relatively divergent ITS2 haplotypes observed only in northern Italy. These haplotypes can be found in different parts of the network, not forming a well-defined subgroup. At the same time, the COI haplotypes appearing in northern Italy are not unique to this region. One is the widely distributed haplotype 12, and the other one is haplotype 18, which also occurs in the Pannonian Basin (Table 1).
Demographic history
Although not univocal, the results of mismatch distribution analyses do not reject the scenario assuming demographic and spatial expansions for BINs AAY1309 and ADD6100 (Table 2; Supplementary Fig. S2). In the case of BIN AAY1309 a peak in the observed data can be detected at three pairwise differences. It is associated with the fact that this BIN contains two haplotypes (haplotype 1 and 12), each with a large number of individuals, divergent by three substitutions (Fig. 3a). The values of Tajima’s D neutrality test do not support the existence of population expansion of BIN AAY1309 neither in CWE, nor in the PB (Table 2). Fu’s F supports the expansion in CWE, but not in the PB (Table 2). In the case of BIN ADD6100 the values of both neutrality tests are negative, although non-significant (Table 2).
The results of the EBSP analysis based on well represented COI dataset also show different demographic histories according to the two aforementioned BINs and/or regions (Fig. 4). The EBSP analyses for BIN AAY1309 support demographic growth of population starting ca. 10 Ka (Fig. 4a) for CWE, but not for PB (Fig. 4b). Considering BIN ADD6100, a slight increase of the population size in the last 10K years can be seen (Fig. 4c). The COI + ITS2 dataset supports these results however, in case of AAY1309 from CWE the expansion starts ca. 20 kya (Supplementary Fig. S3).
Extended Bayesian Skyline Plots of the two most widely distributed COI BINs the study area: (a) BIN AAY1309 in CWE (b) BIN AAY1309 in PB and (c) ADD6100. The plots show the relative variation of effective population size in time in Ky. Elevation on graphs indicates population expansion. CWE = Central Western Europe; PB = Pannonian Basin. EBSP were generated using BEAST 2.4.8 (https://www.beast2.org/) and visualized using R software (https://www.r-project.org).
Discussion
The goal of the present study was to reveal the phylogeographic history within the CWE and PB part of the distribution of freshwater amphipod Gammarus roeselii. Although the study was centred on CWE and PB, it also included the Balkan Peninsula covering the whole distribution range of the species.
In our study, based on the samples from CWE and PB, we defined two COI MOTUs i.e. MOTU C (already identified in Grabowski et al.11) and a new MOTU, MOTU O. In contrast, Grabowski et al.11 found 13 COI MOTUs just in the Balkan Peninsula. MOTU C is a sister clade to MOTUs A and B, while MOTU O is a sister clade to MOTU F and G. The common ancestor of both groups of MOTUs existed most probably in the early Pliocene, i.e. ca. 5 Mya (Fig. 2a). It supports the hypothesis that G. roeselii originated and diversified predominantly in the Balkan Peninsula. The presence of other such old and divergent local lineages was not detected in any geographical region north of the Balkans. These findings, therefore, support the hypothesis by Jażdżewski and Roux43, suggesting the Balkan Peninsula as the possible area of origin for the species. G. roeselii diverged here in the early Miocene mostly due to dynamic geological changes of the region and following habitat isolation; the topic was thoroughly discussed by Grabowski et al.11.
The diversity observed in the CWE and PB area was low on the COI MOTU level (only two MOTUs). Within each MOTU, the level of BIN diversity was highly contrasted. MOTU O included only one BIN and was restricted to one location in Hungary. On the opposite, we detected 11 BINs in MOTU C. Based on our results, most of the BINs appeared during the last 2.5 My, in Pleistocene. While the Pannonian Basin (PB) harbours all the BINs’ diversity present within MOTU C, the northern part of the Apennine Peninsula was colonised by only two BINs AAY1309 and ADD6100 (Fig. 2b), and the rest of CWE was colonized exclusively by the BIN AAY1309 (Fig. 2b). This is in agreement with the results of another study, which identified only one phylogeographic lineage based on 16S mtDNA data from Hungary, Czech Republic and France47.
The fact that the PB contains most BIN diversity within MOTU C, suggests that the region could also be, following the Balkans, an area of lineage diversification. Contemporarily with the diversification of the Balkan MOTUs, the area of the PB was covered by the brackish Pannonian Lake. The lake had completely disappeared by the end of Pliocene, i.e. ca. 2.5 Ma51, and the newly formed hydrological network could be colonised by G. roeselii, which subsequently diversified into the observed BINs.
The distribution of genetic diversity in the CWE seems to be consistent with the postglacial pattern proposed by Hewitt1,2,52. At the end of the LGM after the ice retreated, numerous species belonging to a variety of taxa, such as insects, mammals or fishes, were documented to (re)colonize the newly available areas from southern ice age refugia. Several fishes have colonised post-glacial Europe migrating up the Danube, crossing the, yet unstable, Danube/Rhine watershed and expanding to the just forming river systems of CWE (ibidem). Such post-glacial (re)colonization by a single lineage (the so called “leading-edge colonization”), as e.g. in the case of the meadow grasshopper (Chorthippus parallelus (Zetterstedt, 1821)), results in relative genetic uniformity over vast areas in the recently ice-freed northern regions, if compared to the southern source populations, often with much smaller ranges1. Recently, such a pattern was evidenced for the European freshwater crustaceans, such as the waterlouse, Asellus aquaticus (Linnaeus, 1758)5, and the scud, Gammarus jazdzewskii53. In the case of G. roeselii, an overall loss of genetic diversity can be recognized in the northern regions of the continent. Population inhabiting CWE consists of individuals belonging to a single lineage (BIN AAY1309), while the PB hosts higher diversity in the much smaller geographic area (Fig. 2b). In this context, and considering the previously discussed levels of diversity, we propose the PB as a potential extra-Mediterranean ice age refugium for G. roeselii. The PB was free of ice during the LGM and had a rich and complex hydrological network3. Some studies have already reported the region to hold refugial populations of terrestrial and freshwater organisms during the last glacial maximum [See6 for a review, but also7, 8,10,54,55]. The results presented here strongly suggest that only one lineage (BIN AAY1309) was successful in expanding from the PB refugium after the ice-sheet retreated over the North European Plain, the Alps, the western part of France. Both, the star-like topology of the COI haplotype network (Fig. 2c) and the results of mismatch distribution analysis (Supplementary Fig. S2; Table 2), point out that the population of G. roeselii in CWE (excluding Italy) could have experienced a recent demographic and spatial expansion. The EBSP analysis suggests such expansion could start at ca. 10 Kya in this area (Fig. 4a). During this time the transition between the late Younger Dryas and the early Holocene was taking place. The Eurasian Ice Sheet complex had fully retreated from the CWE region and the climate had begun to warm up steadily56. From the hydrological point of view this era was characterised by an increased groundwater supply, followed by the general stabilisation of floodplains of rivers57,58. Although there are well identified climate attributes of this transition period, the Holocene hydrological systems has also been affected by human activity (e.g. animal and plant domestication, and the increasing influence of agricultural practices)59.
We also cannot exclude the possibility of anthropogenic factors playing a role in the dispersion of G. roeselii in more recent, historical times. The species was first recorded and taxonomically described from the vicinity of Paris by Gervais in 1835. Jażdżewski20, as well as Jażdżewski and Roux43 hypothesized that G. roeselii dispersed to this area from the middle Danube basin, with unintentional human help, possibly via the system of artificial canals built across various watersheds or with traded water plants. Concerning the artificial waterways, the possible dispersal route could be upstream the Danube, then through the Ludwig canal between the Danube and Rhine drainage systems, and then through the rich canal network in France. The importance of artificial canals for aquatic species dispersal has been stressed in many studies and for numerous alien and invasive species in Europe60. In addition, recent expansion, i.e. over the last fifty years, at the edge of the distribution was documented through temporal faunistic surveys in France20,43,44 but also, Italy45 and Poland61. The expansion of the species to smaller tributaries and upper reaches of rivers has been also recently reported in Germany62.
The past success of G. roeselii to invade CWE as well as the ongoing upstream expansion can be promoted by some important ecological attributes of the species. Generally, G. roeselii shows high phenotypic plasticity to a variety of biotic (population density), abiotic (e.g. temperature, flow velocity), and anthropogenic (thermally polluted water influx) factors62. An elevated tolerance to anthropogenic impact was reflected in the species’ habitat preference in comparison to G. fossarum63. At the same time, the high adaptive potential of the species is expressed through its ability to coexist with other native gammarids (e.g. G. pulex)64. Moreover, in thermal conditions specific to the middle and lower sections of the rivers, which are classified as summer-warm running waters, it is able to outcompete some native species such as G. fossarum65,66,67,68 or Echinogammarus stammeri (S. Karaman, 1931)45. The morphological variation of the dorsal spines was also documented as a possible adaptation to predation avoidance48. Such traits might very well facilitate the past and present expansion of G. roeselii. Although no direct evidence exists, considering the Balkan origin of the species, the present day climate warming may also facilitate the species’ expansion.
The decreased ITS2 polymorphism and the overall lower divergence compared to COI can be explained by a lower mutation rate for rDNA. This phenomenon was reported for other invertebrates, such as marine gastropods69 and the holoplanktonic scyphozoan Pelagia noctiluca (Forsskål, 1775)49. Intra-individual polymorphism of ITS2, observed in our dataset, was also reported in other arthropods70,71,72. However, the most striking and interesting feature while comparing ITS2 and COI haplotype networks was the incongruences between their topologies. Interestingly, a little pool of relatively divergent ITS2 haplotypes is restricted to the Po River basin in the northern Apennine Peninsula while others are shared between the Po basin and the PB (Fig. 2b and stars on 2d). In comparison, the only two COI haplotypes present in the Apennine Peninsula are those most common and of widest distribution in Europe. One possible scenario explaining this pattern could be that G. roeselii colonised the northern Apennine Peninsula from the PB, where they got into secondary contact with a local population, most probably originating from earlier colonisation event(s). Similar mixing between lineages through historical headwater river captures has been observed in freshwater fish, which share habitat preferences with G. roeselii, such as Phoxinus lumaireul (Schinz, 1840)73,74, Cobitis bilineata Canestrini, 1865 and Sabanejewia larvata (De Filippi, 1859)75. Apparently, the contact of different lineages of G. roeselii resulted in an asymmetric introgression that eventually wiped out the local maternal lineages. Similarly, the fact that MOTU O (BIN ADA0638) is characterized by one unique COI divergent haplotype (haplotype 18) but its ITS2 haplotype is commonly observed and associated with individual assigned to BIN AAY1309 of MOTU C could be explained by such asymmetric mating. Such a pattern was also suspected to have taken place in the Balkan Peninsula11. In both cases, it is hard to explain why the local females have not contributed to the present genetic pool of the Apennine population. Directional mate-choice has been mostly studied in vertebrates, although this phenomenon is commonly observed also in invertebrate reproduction. In case of the waterlouse Asellus aquaticus, males (which are generally bigger than females) favour females with larger body size. Such larger females were shown to ovulate faster and produce bigger broods76. In the case of Gammarus pulex assortative mating was evident, with significant and strong selection on male body size77. At the same time the phenomenon was proven to be influenced by the female moulting stage, increasing in magnitude with females closer to moulting78.
Conclusions
Our results supported the hypothesis that the Pannonian Basin was an important extra-Mediterranean glacial refugium for Gammarus roeselii and its divergence hotspot since the Pliocene. We also showed that G. roeselii MOTU C, and more precisely one BIN within this MOTU (BIN AAY1309), left the PB after the last glaciation, to colonise the rest of Central and Western Europe. We found also that the expansion of the G. roeselii MOTU C could have started already in the early Holocene and, as its distribution and genetic diversity patterns suggest, progressed in historical times aided by the construction of artificial waterways all over Europe. Finally, we presented an evidence suggesting asymmetric introgression events, which might be more common between invertebrate taxa, than previously thought.
Materials and methods
Overview of sampling strategy and dataset content
The present study focuses on the distribution of G. roeselii outside the Balkans, including 71 newly sampled localities as well as six localities from literature, i.e. four northernmost sites in the Pannonian Basin from Grabowski et al.11 and two sites in Germany from Grabner et al.79. All other sites located in the Balkans from Grabowski et al. 2017 were used to establish connections with phylogeographic lineages observed there11.
Sampling and taxonomic identification of new individuals
Individuals of Gammarus roeselii were collected at 71 localities during sampling campaigns in the years 2005–2017, covering the whole species range outside the Balkans (Figs. 1, 2b). The sampling included streams, rivers and lakes. Samples were collected with benthic hand nets via kick and sweep method61. The collected material was sorted at a site, and gammarids were immediately fixed in 96% ethanol. Individuals were identified to the species level under stereomicroscope, using the taxonomic keys46, 80,81. All animals have been stored in the permanent collection of the Department of Invertebrate Zoology and Hydrobiology, University of Lodz, Poland.
Laboratory procedures: DNA extraction, amplification and sequencing
DNA was extracted using the standard phenol–chloroform method82 from the muscle tissue of each individual from the newly sampled sites (1–12 individuals per site, altogether 663 individuals) (Table 1). The ca. 710 bp long part of the mitochondrial cytochrome oxidase I-subunit (COI) was amplified. Conditions of polymerase chain reaction (PCR) for the amplification of COI were set according to Grabowski et al.11. Following the definition of COI MOTUs and their geographic distribution (Fig. 2b; Table 1), representative individuals were chosen for amplification of the ca. 850 bp of the internal transcribed spacer 2 of the nuclear ribosomal operon (ITS2). PCR conditions for the amplification of ITS2 were set as following: pre-denaturation at 94 °C for 1 min followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 53 °C for 30 s, extension at 72 °C for 1 min 30 s, and final extension at 72 °C for 1 min 45 s (all primers used for amplification are specified in Supplementary Table S1). Prior to sequencing, the PCR products were cleaned using the EXO-FastAP method (Thermo Scientific). Sequencing was completed by Macrogen Europe—Amsterdam, The Netherlands, using BigDye terminator technology.
Assembling the dataset for further analysis: sequence edition and alignment
Altogether, within this study, we sequenced 663 individuals for COI and 137 individuals for ITS2 (Table 1). The identity of all the sequences was verified with the BLASTn tool (https://blast.ncbi.nlm.nih.gov/blast)83. Raw sequences were trimmed, edited and aligned using Geneious 11.1.5 software (https://www.geneious.com/)84. The translation frame in COI sequences containing no stop codons was found. Eighteen sequences from Grabowski et al.11 and 30 sequences from Grabner et al.79 supplemented the COI dataset, leading to a total of 711 sequences (Table 1). Due to the presence of polymorphic copies of ITS2, the chromatograms of sequences from some individuals contained double peaks. Ambiguous sites were selected and coded with IUPAC ambiguity code in Sequencher 4 (https://www.genecodes.com/)85. Phasing and reconstruction of the two alleles for each individual were completed in DnaSP 6 (https://www.ub.edu/dnasp/)86. Haplotypes for both markers were identified by DnaSP 6 software. All the new sequences have been deposited in GenBank: COI: MT324702–MT325364; ITS2: MT325365–MT325501. Relevant voucher information for 663 individuals is accessible through the public dataset “DS-GROEEUR” (https://doi.org/10.5883/DS-GROEEUR) in the Barcode of Life Data Systems (BOLD; https://v4.boldsystems.org).
BIN and MOTU delimitation
Clustering sequences into BINs was performed in BOLD (https://v4.boldsystems.org), where every uploaded sequence was compared to each other and to already available records and got assigned to an existing or a newly created Barcode Index Number (BIN)87. BINs are registered in BOLD and are unique identifiers of sequence clusters, which are created based on a graph analytical distance-based approach aiming to find discontinuities between groups87.
The MOTU framework was taken from the Grabowski et al.11 paper, to keep reference to our previous study dealing with the Balkan diversification hotspot. In the present study we extended the dataset by adding more sequences from PB and from CWE. In result, with the exception of one, all the new sequences extended the clade which we referred to as MOTU C (see next paragraph for the phylogeny reconstruction). This MOTU has become more diverse than in Grabowski et al.11 but, still, it holds its integrity against its sister clade, composed of MOTUs A and B. The remaining sequence diverged in the late Miocene from its sister clade containing MOTUs F and G (see below), thus decided to name it as another MOTU, MOTU O, more affiliated to the Balkan MOTUs than to MOTU C.
Time calibrated reconstruction of phylogeny
The time-calibrated phylogeny was reconstructed based on the COI sequences, using the Bayesian Inference (BI) in BEAST 2.4.8 (https://www.beast2.org/)88. One individual chosen at random from every BIN was used to build the tree (see previous paragraph for BIN definition). Dikerogammarus villosus (EF570297) and Pontogammarus robustoides (JF965990) sequences from GenBank (https://www.ncbi.nlm.nih.gov/genbank/) were used as an outgroup. The calibration points for the molecular clock were set the same as in the cross-validated calibration in Grabowski et al.11, with the initial rate set on 0.0113 substitutions per site per million years (My). The optimal model of evolution was selected using bModeltest (https://github.com/BEAST2-Dev/bModelTest/)89, clock model and tree prior were selected with the path sampling method in BEAST 2.4.8. The Tamura and Nei 1993 (TN93) model of nucleotide substitution with gamma-distributed rate heterogeneity (G) and a proportion of invariable sites (I) was chosen as the best-fitting model. The birth–death speciation process and lognormal relaxed molecular clock, were set as priors following Marginal Likelihood estimation through path sampling. The estimation was performed using BEAST software, the models were better for over 30 Bayes Factors in comparison to the next best-fitting model. Four MCMC runs were performed, each with 20 M iterations, sampled every 2000 iterations. The Effective Sampling Size (ESS) of each parameter was verified to be above 200 in Tracer 1.6 (https://beast.community/tracer)90. The outcomes of these runs were combined for tree construction in LogCombiner 2.4.888 with the first 25% burn-in iterations discarded. After summarizing all sampled trees, the produced maximum clade credibility tree (MCC) was annotated in TreeAnnotator 2.4.888, both programs being part of BEAST 2.4.8 package88 .
A maximum likelihood (ML) tree was constructed on the same dataset in MEGA7 software91 to give additional support to the topology of the Bayesian chronogram. The same model of evolution (TN93 + G + I) as in case of BI was used. To assess topology robustness 1000 bootstrap replicates were applied. These bootstrap values were reported on the chronogram (Fig. 2a).
Historical demography: haplotype diversity and distribution, population expansion
For COI and ITS2 median-joining (MJ) haplotypic networks were generated in PopArt 1.7 (https://popart.otago.ac.nz/index.shtml)92. The homoplasy level parameter was set to the default value (ε = 0). Networks were colour-coded based on the haplotypes belonging to BINs provided by BOLD.
The demography of the two most widely distributed BINs (BIN AAY1309 and ADD6100, see Results) was investigated. COI haplotype 20 and 60 were removed from the dataset, due to their divergent position on the haplotype network (Fig. 3a). Individuals from northern Italy were also removed due to the possible presence of introgression (see below) and geographical distinctness. Two BINs were run separately, additionally BIN AAY1309 was separated in two populations, first containing the individuals from CWE and the second containing those from PB. First, historical demographic and spatial expansion were examined by mismatch analyses. Additionally, the demographic expansion was tested with Tajima’s D93, and Fu’s F94 neutrality tests. All the analyses were performed in Arlequin 3.5.1.3 (https://cmpg.unibe.ch/software/arlequin35/)95. Finally, to infer the changes in the effective population size over time, Extended Bayesian Skyline Plot (EBSP) was generated in BEAST 2.4.888. The rate of 0.0113 substitution per site per My was used for COI. The preferred models of evolution for the aforementioned two BINs were set up with bModeltest89. EBSP was run separately for COI and COI + ITS2 datasets. The population scaling factor was set to 0.5 for COI and 2.0 for ITS2. Each run was 40 M MCMC long, sampled every 20,000 iterations. The ESS of each parameter was verified to be above 200 in Tracer 1.6. The plots were generated after removal of 10% burn-in using R software (https://www.r-project.org).
Data availability
All sequences with metadata are available in BOLD dataset DS-GROEEUR (https://doi.org/10.5883/DS-GROEEUR), GenBank accessions numbers for COI: (MT324702-MT325364) and ITS: (MT325365-MT325501).
References
Hewitt, G. M. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 58, 247–276. https://doi.org/10.1006/bijl.1996.0035 (1996).
Hewitt, G. M. Genetic consequences of climatic oscillations in the quaternary. Philos. Trans. R. Soc. B Biol. Sci. 359, 183–195. https://doi.org/10.1098/rstb.2003.1388 (2004).
Patton, H. et al. Deglaciation of the Eurasian ice sheet complex. Quat. Sci. Rev. 169, 148–172. https://doi.org/10.1016/j.quascirev.2017.05.019 (2017).
Hewitt, G. M. Postglacial re-colonisation of European biota. Biol. J. Linn. Soc. 68, 87–112 (1999).
Sworobowicz, L. et al. Revisiting the phylogeography of Asellus aquaticus in Europe: insights into cryptic diversity and spatiotemporal diversification. Freshw. Biol. 60, 1824–1840. https://doi.org/10.1111/fwb.12613 (2015).
Schmitt, T. & Varga, Z. Extra-Mediterranean refugia: the rule and not the exception?. Front. Zool. 9, 1–12. https://doi.org/10.1186/1742-9994-9-22 (2012).
Verovnik, R., Sket, B. & Trontelj, P. The colonization of Europe by the freshwater crustacean Asellus aquaticus (Crustacea: Isopoda) proceeded from ancient refugia and was directed by habitat connectivity. Mol. Ecol. 14, 4355–4369. https://doi.org/10.1111/j.1365-294X.2005.02745.x (2005).
Sworobowicz, L., Mamos, T., Grabowski, M. & Wysocka, A. Lasting through the ice age: the role of the proglacial refugia in the maintenance of genetic diversity, population growth, and high dispersal rate in a widespread freshwater crustacean. Freshw. Biol. https://doi.org/10.1111/fwb.13487 (2020).
Neumann, K. et al. Genetic spatial structure of European common hamsters (Cricetus cricetus)—a result of repeated range expansion and demographic bottlenecks. Mol. Ecol. 14, 1473–1483. https://doi.org/10.1111/j.1365-294X.2005.02519.x (2005).
Fussi, B., Lexer, C. & Heinze, B. Phylogeography of Populus alba (L.) and Populus tremula (L.) in Central Europe: secondary contact and hybridisation during recolonisation from disconnected refugia. Tree Genet. Genomes 6, 439–450. https://doi.org/10.1007/s11295-009-0262-5 (2010).
Grabowski, M., Mamos, T., Bącela-Spychalska, K., Rewicz, T. & Wattier, R. A. Neogene paleogeography provides context for understanding the origin and spatial distribution of cryptic diversity in a widespread Balkan freshwater amphipod. PeerJ 5, e3016. https://doi.org/10.7717/peerj.3016 (2017).
Hou, Z., Sket, B., Fiser, C. & Li, S. Eocene habitat shift from saline to freshwater promoted Tethyan amphipod diversification. Proc. Natl. Acad. Sci. 108, 14533–14538. https://doi.org/10.1073/pnas.1104636108 (2011).
Mamos, T., Wattier, R., Burzyński, A. & Grabowski, M. The legacy of a vanished sea: a high level of diversification within a European freshwater amphipod species complex driven by 15 My of Paratethys regression. Mol. Ecol. 25, 795–810. https://doi.org/10.1111/mec.13499 (2016).
Perea, S. et al. Phylogenetic relationships and biogeographical patterns in Circum-Mediterranean subfamily Leuciscinae (Teleostei, Cyprinidae) inferred from both mitochondrial and nuclear data. BMC Evol. Biol. 10, 1–27. https://doi.org/10.1186/1471-2148-10-265 (2010).
Saito, T. et al. Phylogeography of freshwater planorbid snails reveals diversification patterns in Eurasian continental islands. BMC Evol. Biol. https://doi.org/10.1186/s12862-018-1273-3 (2018).
Utevsky, S. & Trontelj, P. Phylogeography of the southern medicinal leech, Hirudo verbana: a response to Živić et al. (2015). Aquat. Ecol. 50, 97–100. https://doi.org/10.1007/s10452-015-9553-0 (2016).
Grabowski, M., Jażdżewski, K. & Konopacka, A. Alien crustacea in polish waters—Amphipoda. Aquat. Invas. 2, 25–38. https://doi.org/10.3391/ai.2007.2.1.3 (2007).
Kontula, T. & Väinölä, R. Postglacial colonization of Northern Europe by distinct phylogeographic lineages of the bullhead, Cottus gobio. Mol. Ecol. 10, 1983–2002. https://doi.org/10.1046/j.1365-294X.2001.01328.x (2001).
Mateus, C. S., Almeida, P. R., Mesquita, N., Quintella, B. R. & Alves, M. J. European lampreys: new insights on postglacial colonization, gene flow and speciation. PLoS ONE 11, 1–22. https://doi.org/10.1371/journal.pone.0148107 (2016).
Jażdżewski, K. Range extensions of some gammaridean species in European inland waters caused by human activity. 10–16 (1980).
Bij de Vaate, A., Jażdżewski, K., Ketelaars, H. A. M., Gollasch, S. & Van der Velde, G. Geographical patterns in range extension of Ponto-Caspian macroinvertebrate species in Europe. Can. J. Fish. Aquat. Sci. 59, 1159–1174. https://doi.org/10.1139/f02-098 (2002).
Panov, V. E. et al. Assessing the risks of aquatic species invasions via European inland waterways: from concepts to environmental indicators. Integr. Environ. Assess. Manag. 5, 110–126. https://doi.org/10.1897/IEAM_2008-034.1 (2009).
Väinölä, R. et al. Global diversity of amphipods (Amphipoda; Crustacea) in freshwater. Hydrobiologia 595, 241–255. https://doi.org/10.1007/s10750-007-9020-6 (2008).
Weiss, M. & Leese, F. Widely distributed and regionally isolated! Drivers of genetic structure in Gammarus fossarum in a human-impacted landscape. BMC Evol. Biol. 16, 153. https://doi.org/10.1186/s12862-016-0723-z (2016).
Weigand, A. M., Michler-Kozma, D., Kuemmerlen, M. & Jourdan, J. Substantial differences in genetic diversity and spatial structuring among (cryptic) amphipod species in a mountainous river basin. Freshw. Biol. 65, 1641–1656. https://doi.org/10.1111/fwb.13529 (2020).
Rachalewski, M., Banha, F., Grabowski, M. & Anastácio, P. M. Ectozoochory as a possible vector enhancing the spread of an alien amphipod Crangonyx pseudogracilis. Hydrobiologia 717, 109–117. https://doi.org/10.1007/s10750-013-1577-7 (2013).
Peck, S. B. Amphipod dispersal in the fur of aquatic mammals. Can. F. Nat. 89, 181–182 (1975).
Sainte-Marie, B. A review of the reproductive bionomics of aquatic gammaridean amphipods: variation of life history traits with latitude, depth, salinity and superfamily. Hydrobiologia 223, 189–227. https://doi.org/10.1007/BF00047641 (1991).
Rewicz, T., Grabowski, M., MacNeil, C. & Bącela-Spychalska, K. The profile of a ‘perfect’ invader—the case of killer shrimp, Dikerogammarus villosus. Aquat. Invas. 9, 267–288. https://doi.org/10.3391/ai.2014.9.3.04 (2014).
Vader, W. & Tandberg, A. H. S. Gammarid amphipods (Crustacea) in Norway, with a key to the species. Fauna Nor. 39, 12–25. https://doi.org/10.5324/fn.v39i0.2873 (2019).
Macdonald, K. S., Yampolsky, L. & Duffy, J. E. Molecular and morphological evolution of the amphipod radiation of Lake Baikal. Mol. Phylogenet. Evol. 35, 323–343. https://doi.org/10.1016/j.ympev.2005.01.013 (2005).
Grabowski, M., Wysocka, A. & Mamos, T. Molecular species delimitation methods provide new insight into taxonomy of the endemic gammarid species flock from the ancient Lake Ohrid. Zool. J. Linn. Soc. 181, 272–285. https://doi.org/10.1093/zoolinnean/zlw025 (2017).
Jabłońska, A., Wrzesińska, W., Zawal, A., Pešić, V. & Grabowski, M. Long-term within-basin isolation patterns, different conservation units, and interspecific mitochondrial DNA introgression in an amphipod endemic to the ancient Lake Skadar system, Balkan Peninsula. Freshw. Biol. 65, 209–225. https://doi.org/10.1111/fwb.13414 (2020).
Copilaş-Ciocianu, D. & Petrusek, A. The southwestern Carpathians as an ancient centre of diversity of freshwater gammarid amphipods: insights from the Gammarus fossarum species complex. Mol. Ecol. 24, 3980–3992. https://doi.org/10.1111/mec.13286 (2015).
Copilaş-Ciocianu, D. & Petrusek, A. Phylogeography of a freshwater crustacean species complex reflects a long-gone archipelago. J. Biogeogr. 44, 421–432. https://doi.org/10.1111/jbi.12853 (2017).
Leuven, R. S. E. W. et al. The river Rhine: a global highway for dispersal of aquatic invasive species. Biol. Invas. 11, 1989–2008. https://doi.org/10.1007/s10530-009-9491-7 (2009).
Kelly, D. W., Muirhead, J. R., Heath, D. D. & Macisaac, H. J. Contrasting patterns in genetic diversity following multiple invasions of fresh and brackish waters. Mol. Ecol. 15, 3641–3653. https://doi.org/10.1111/j.1365-294X.2006.03012.x (2006).
Panov, V. & Berezina, N. Invasive aquatic species of Europe. Distribution, impacts and management. Invas. Aquat. Species Eur. Distrib. Impacts Manag. https://doi.org/10.1007/978-94-015-9956-6 (2002).
Csabai, Z. et al. Mass appearance of the Ponto-Caspian invader Pontogammarus robustoides in the River Tisza catchment: bypass in the southern invasion corridor?. Knowl. Manag. Aquat. Ecosyst. https://doi.org/10.1051/kmae/2020003 (2020).
Rewicz, T., Wattier, R., Grabowski, M., Rigaud, T. & Bącela-Spychalska, K. Out of the Black sea: phylogeography of the invasive killer shrimp Dikerogammarus villosus across Europe. PLoS ONE 10, 1–20. https://doi.org/10.1371/journal.pone.0118121 (2015).
Rewicz, T. et al. The killer shrimp, Dikerogammarus villosus, invading European Alpine Lakes: a single main source but independent founder events with an overall loss of genetic diversity. Freshw. Biol. 62, 1036–1051. https://doi.org/10.1111/fwb.12923 (2017).
Jażdżewska, A. M. et al. Cryptic diversity and mtDNA phylogeography of the invasive demon shrimp, Dikerogammarus haemobaphes (Eichwald, 1841), in Europe. NeoBiota 57, 53–86. https://doi.org/10.3897/neobiota.57.46699 (2020).
Jażdżewski, K. & Roux, A. L. Biogéographie de Gammarus roeseli Gervais en Europe, en particulier répartition en France et en Pologne (1988).
Piscart, C. & Bollache, L. Crustacés amphipodes de surface : gammares d’eau douce.. Association Française de Limnologie, Introduction pratique à la systématique des organismes des eaux continentales de France (2012).
Paganelli, D., Gazzola, A., Marchini, A. & Sconfietti, R. The increasing distribution of Gammarus roeselii Gervais, 1835: first record of the non-indigenous freshwater amphipod in the sub-lacustrine Ticino River basin (Lombardy, Italy). Bioinvas. Rec. 4, 37–41. https://doi.org/10.3391/bir.2015.4.1.06 (2015).
Karaman, G. S. & Pinkster, S. Freshwater gammarus species from Europe, North Africa and adjacent regions of Asia (Crustacea-Amphipoda) Part II. Gammarus roeseli-group and related species. Bijdragen tot de dierkunde 57, 207–260. https://doi.org/10.1163/26660644-05702005 (1977).
Moret, Y., Bollache, L., Wattier, R. & Rigaud, T. Is the host or the parasite the most locally adapted in an amphipod-acanthocephalan relationship? A case study in a biological invasion context. Int. J. Parasitol. 37, 637–644. https://doi.org/10.1016/j.ijpara.2006.12.006 (2007).
Copilaş-Ciocianu, D., Borza, P. & Petrusek, A. Extensive variation in the morphological anti-predator defense mechanism of Gammarus roeselii Gervais, 1835 (Crustacea:Amphipoda). Freshw. Sci. 39, 47–55. https://doi.org/10.1086/707259 (2020).
Miller, B. J., von der Heyden, S. & Gibbons, M. J. Significant population genetic structuring of the holoplanktic scyphozoan Pelagia noctiluca in the Atlantic Ocean. Afr. J. Mar. Sci. 34, 425–430. https://doi.org/10.2989/1814232X.2012.726646 (2012).
Brown, W. M., George, M. Jr. & Wilson, A. C. Rapid evolution of animal mitochondrial DNA. Genetics 76, 1967–1971. https://doi.org/10.1002/(sici)1097-4555(199706)28:6%3c433::aid-jrs125%3e3.3.co;2-5 (1979).
Kázmér, M. Birth, life and death of the Pannonian Lake. Palaeogeogr. Palaeoclimatol. Palaeoecol. 79, 171–188. https://doi.org/10.1016/0031-0182(90)90111-J (1990).
Hewitt, G. M. Post-glacial re-colonization of European biota. Biol. J. Linn. Soc. 68, 87–112. https://doi.org/10.1006/bijl.1999.0332 (1999).
Rudolph, K., Coleman, C. O., Mamos, T. & Grabowski, M. Description and post-glacial demography of Gammarus jazdzewskii sp. nov. (Crustacea: Amphipoda) from Central Europe. Syst. Biodivers. 16, 587–603. https://doi.org/10.1080/14772000.2018.1470118 (2018).
Copilaş-Ciocianu, D., Fišer, C., Borza, P. & Petrusek, A. Is subterranean lifestyle reversible? Independent and recent large-scale dispersal into surface waters by two species of the groundwater amphipod genus Niphargus. Mol. Phylogenet. Evol. 119, 37–49. https://doi.org/10.1016/j.ympev.2017.10.023 (2018).
Antal, L. et al. Phylogenetic evidence for a new species of Barbus in the Danube River basin. Mol. Phylogenet. Evol. 96, 187–194. https://doi.org/10.1016/j.ympev.2015.11.023 (2016).
Walker, M. J. C. Climatic changes in Europe during the last glacial/interglacial transition. Quat. Int. 28, 63–76. https://doi.org/10.1016/1040-6182(95)00030-M (1995).
Pawłowski, D. et al. The response of flood-plain ecosystems to the Late Glacial and Early Holocene hydrological changes: a case study from a small Central European river valley. CATENA 147, 411–428. https://doi.org/10.1016/j.catena.2016.07.034 (2016).
Notebaert, B. & Verstraeten, G. Sensitivity of West and Central European river systems to environmental changes during the Holocene: a review. Earth Sci. Rev. 103, 163–182. https://doi.org/10.1016/j.earscirev.2010.09.009 (2010).
Gibling, M. R. River systems and the anthropocene: a late pleistocene and holocene timeline for human influence. Quaternary 1, 21. https://doi.org/10.3390/quat1030021 (2018).
Gherardi, F. Biological invaders in inland waters: profiles, distribution, and threats. https://doi.org/10.1007/978-1-4020-6029-8 (2007).
Jazdzewski, K., Konopacka, A. & Grabowski, M. Recent drastic changes in the gammarid fauna (Crustacea, Amphipoda) of the Vistula River deltaic system in Poland caused by alien invaders. Divers. Distrib. 10, 81–87. https://doi.org/10.1111/j.1366-9516.2004.00062.x (2004).
Jourdan, J., Piro, K., Weigand, A. & Plath, M. Small-scale phenotypic differentiation along complex stream gradients in a non-native amphipod. Front. Zool. 16, 29. https://doi.org/10.1186/s12983-019-0327-8 (2019).
Mauchart, P., Bereczki, C., Ortmann-Ajkai, A., Csabai, Z. & Szivák, I. Niche segregation between two closely related Gammarids (Crustacea, Amphipoda)—native vs. naturalised non-native species. Crustaceana 87, 1296–1314. https://doi.org/10.1163/15685403-00003355 (2014).
Lagrue, C. et al. Interspecific differences in drift behaviour between the native Gammarus pulex and the exotic Gammarus roeseli and possible implications for the invader’s success. Biol. Invas. 13, 1409–1421. https://doi.org/10.1007/s10530-010-9899-0 (2011).
Pöckl, M. & Humpesch, U. H. Intra- and inter-specific variations in egg survival and brood development time for Austrian populations of Gammarus fossarum and G. roeseli (Crustacea: Amphipoda). Freshw. Biol. 23, 441–455. https://doi.org/10.1111/j.1365-2427.1990.tb00286.x (1990).
Pöckl, M. Effects of temperature, age and body size on moulting and growth in the freshwater amphipods Gammarus fossarum and G. roeseli. https://doi.org/10.1111/j.1365-2427.1992.tb00534.x (1992).
Pöckl, M. Reproductive potential and lifetime potential fecundity of the freshwater amphipods Gammarus fossarum and G. roeseli in Austrian streams and rivers. Freshw. Biol. 30, 73–91. https://doi.org/10.1111/j.1365-2427.1993.tb00790.x (1993).
Pöckl, M., Webb, B. W. & Sutcliffe, D. W. Life history and reproductive capacity of Gammarus fossarum and G. roeseli (Crustacea: Amphipoda) under naturally fluctuating water temperatures: a simulation study. Freshw. Biol. 48, 53–66. https://doi.org/10.1046/j.1365-2427.2003.00967.x (2003).
Aguilera-Muñoz, F., Lafarga-Cruz, F. & Gallardo-Escárate, C. Molecular analysis in Chilean commercial gastropods based on 16S rRNA, COI and ITS1-5.8S rDNA-ITS2 sequences. Gayana (Concepción) 73, 17–27. https://doi.org/10.4067/s0717-65382009000100003 (2009).
Alvarez, J. M. & Hoy, M. A. Evaluation of the ribosomal ITS2 DNA sequences in separating closely related populations of the Parasitoid Ageniaspis (Hymenoptera: Encyrtidae) article. Ann. Entomol. Soc. Am. https://doi.org/10.1603/0013-8746(2002)095 (2002).
Wesson, D. M., McLain, D. K., Oliver, J. H., Piesman, J. & Collins, F. H. Investigation of the validity of species status of Ixodes dammini (Acari: Ixodidae) using rDNA. Proc. Natl. Acad. Sci. U. S. A. 90, 10221–10225. https://doi.org/10.1073/pnas.90.21.10221 (1993).
Tang, J., Toè, L., Back, C. & Unnasch, T. R. Intra-specific heterogeneity of the rDNA internal transcribed spacer in the Simulium damnosum (Diptera: Simuliidae) complex. Mol. Biol. Evol. 13, 244–252. https://doi.org/10.1093/oxfordjournals.molbev.a025561 (1996).
Palandačić, A., Bravničar, J., Zupančić, P., Šanda, R. & Snoj, A. Molecular data suggest a multispecies complex of Phoxinus (Cyprinidae) in the Western Balkan Peninsula. Mol. Phylogenet. Evol. https://doi.org/10.1016/j.ympev.2015.05.024 (2015).
Vucić, M., Jelić, D., Žutinić, P., Grandjean, F. & Jelić, M. Distribution of Eurasian minnows (Phoxinus : Cypriniformes) in the Western Balkans. Knowl. Manag. Aquat. Ecosyst. 419, 11. https://doi.org/10.1051/kmae/2017051 (2018).
Buj, I. et al. Peculiar occurrence of Cobitis bilineata Canestrini, 1865 and Sabanejewia larvata (De Filippi, 1859) (Cobitidae, Actinopteri) in the Danube River basin in Croatia. Fundam. Appl. Limnol. https://doi.org/10.1127/fal/2020/1272 (2020).
Manning, J. T. Male discrimination and investment in Asellus aquaticus (L.) and A. meridianus Racovitsza (Crustacea: Isopoda). Behaviour 55(1–2), 1–14 (1975).
Bollache, L. & Cézilly, F. Sexual selection on male body size and assortative pairing in Gammarus pulex (Crustacea: Amphipoda): field surveys and laboratory experiments. J. Zool. 264, 135–141. https://doi.org/10.1017/S0952836904005643 (2004).
Cornet, S., Luquet, G. & Bollache, L. Influence of female moulting status on pairing decisions and size-assortative mating in amphipods. J. Zool. 286, 312–319. https://doi.org/10.1111/j.1469-7998.2011.00882.x (2012).
Grabner, D. S. et al. Invaders, natives and their enemies: distribution patterns of amphipods and their microsporidian parasites in the Ruhr Metropolis, Germany. Parasites Vectors https://doi.org/10.1186/s13071-015-1036-6 (2015).
Karaman, G. S. & Pinkster, S. Freshwater gammarus species from Europe, North Africa and adjacent regions of Asia (Crustacea-Amphipoda) Part I. Gammarus pulex—group and related species (1977).
Karaman, G. S. & Pinkster, S. Freshwater gammarus species from Europe, North Africa and adjacent regions of Asia (Crustacea-Amphipoda). Part III. Gammarus balcanicus—group and related species (1987).
Hillis, D. M. & Moritz, C. Molecular Systematics (Sinauer Associates Inc., Sunderland, 1996).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410. https://doi.org/10.1016/S0022-2836(05)80360-2 (1990).
Kearse, M. et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. https://doi.org/10.1093/bioinformatics/bts199 (2012).
Sequencher version 5.4.6 DNA sequence analysis software, Gene Codes Corporation, Ann Arbor, MI USA https://www.genecodes.com.
Rozas, J. et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. https://doi.org/10.1093/molbev/msx248 (2017).
Ratnasingham, S. & Hebert, P. D. N. The barcode of life data system. Mol. Ecol. Notes 7, 355–364. https://doi.org/10.1111/j.1471-8286.2006.01678.x (2007).
Bouckaert, R. et al. BEAST 2: a software platform for bayesian evolutionary analysis. PLoS Comput. Biol. 10, 1–6. https://doi.org/10.1371/journal.pcbi.1003537 (2014).
Bouckaert, R. R. & Drummond, A. J. bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evol. Biol. 17, 1–11. https://doi.org/10.1186/s12862-017-0890-6 (2017).
Rambaut, A., Suchard, M. A., Xie, D. & Drummond, A. J. Tracer v1.6. Available at https://beast.bio.ed.ac.uk/Tracer (2014).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. https://doi.org/10.1093/molbev/msw054 (2016).
Leigh, J. W. & Bryant, D. POPART: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. https://doi.org/10.1111/2041-210X.12410 (2015).
Tajima, F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. (1989).
Fu, Y. X. New statistical tests of neutrality for DNA samples from a population. Genetics 143, 557–570 (1996).
Excoffier, L. & Lischer, H. E. L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x (2010).
Copilaş-Ciocianu, D., Grabowski, M., Parvulescu, L. & Petrusek, A. Zoogeography of epigean freshwater Amphipoda (Crustacea) in Romania: fragmented distributions and wide altitudinal variability. Zootaxa 3893, 243. https://doi.org/10.11646/zootaxa.3893.2.5 (2014).
Acknowledgements
We would like to thank Horia Bogdan, Krešimir Žganec, Radomir Jaskuła, Kamil Hupało and Michał Rachalewski for their help in fieldwork. Laboratory work and molecular analysis were supported by internal funds of the University of Lodz as well as, partly, by the Polish Ministry of Science and Education (Grant No. N N303 579439) and National Science Centre, Poland (Grant No. 2014/15/B/NZ8/00266), internal funds of the UMR-CNRS-Ub-6282 Biogéosciences and the Burgundy Region. TR and TM were supported by the Scholarship of the Polish National Agency for Academic Exchange (NAWA) at Bekker Programme (TR Project nb. PPN/BEK/2018/1/00162, TM Project nb. PPN/BEK/2018/1/00225) and TR, RW, KBS and MG by the Polish National Agency for Academic Exchange within Polonium Programme (PPN/BIL/2018/1/00138). Additional financing for HC’s stay in Poland was provided by the ERASMUS+ Mobility Grant (Grant No. 17/1/KA103/035116/SMP-063 and 18/1/KA103/0470013/SMS-040). The study was additionally supported by the New National Excellence Program of the Hungarian Ministry of Human Capacities (ÚNKP-18-2-I-PTE-146).
Author information
Authors and Affiliations
Contributions
K.B.S., M.G., R.W., T.M., T.R. designed the study. K.B.S., M.G., M.J., R.W., T.M., T.R. collected material. H.C., M.J., P.K. performed the experiments. K.B.S., M.G., R.W., T.M., T.R. designed data analysis. H.C., K.B.S., M.G., P.K., T.M., T.R. performed analyses on the data. H.C., K.B.S., M.J. prepared figures and/or tables. H.C., K.B.S., M.G., R.W., T.M., T.R. wrote the manuscript. H.C., K.B.S., M.G., M.J., R.W., T.M., T.R. revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Csapó, H., Krzywoźniak, P., Grabowski, M. et al. Successful post-glacial colonization of Europe by single lineage of freshwater amphipod from its Pannonian Plio-Pleistocene diversification hotspot. Sci Rep 10, 18695 (2020). https://doi.org/10.1038/s41598-020-75568-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-75568-7
This article is cited by
-
Infection with acanthocephalans increases tolerance of Gammarus roeselii (Crustacea: Amphipoda) to pyrethroid insecticide deltamethrin
Environmental Science and Pollution Research (2023)
-
Southernmost record of Rhachotropis aculeata (Lepechin, 1780) (Crustacea: Amphipoda: Eusiridae) in the Pacific Ocean with notes on the geographical genetic divergence
Marine Biodiversity (2023)
-
Molecular analysis reveals multiple native and alien Phoxinus species (Leusciscidae) in the Netherlands and Belgium
Biological Invasions (2022)
-
Cryptic survival and an unexpected recovery of the long-tailed mayfly Palingenia longicauda (Olivier, 1791) (Ephemeroptera: Palingeniidae) in Southeastern Europe
Journal of Insect Conservation (2022)
-
Current extinction rate in European freshwater gastropods greatly exceeds that of the late Cretaceous mass extinction
Communications Earth & Environment (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.