Abstract
Mucin 1 (MUC1) is a transmembrane glycoprotein involved in tumorigenesis of diverse cancers. However, the role of MUC1 in glioblastoma (GBM) has not yet been fully explored. In this study, the anticancer mechanism of MUC1 suppression in GBM was investigated. The expression level of MUC1 was analyzed in human glioma and paired normal brain tissues. MUC1 was overexpressed in GBM and was negatively associated with overall survival. Moreover, we silenced MUC1 to investigate its effect in GBM cell lines and found that knockdown of MUC1 inhibited cell proliferation and resulted in cell cycle arrest at G1 phase. MUC1 silencing decreased the phosphorylation of RB1 and increased the expression of CDKN1B. Gene set enrichment analysis showed that a series of genes related to cell cycle, telomere maintenance and transforming growth factor Beta (TGF-β) signaling in epithelial mesenchymal transition (EMT) were influenced by MUC1 knockdown. Notably, the reduced TERT expression levels combined with impaired telomerase activity and the switching of telomere maintenance mechanism to alternative lengthening of telomeres (ALT) were observed after MUC1 knockdown. Our results support the role of MUC1 in oncological process in GBM which can be developed as a therapeutic target for cell cycle control and telomere maintenance mechanism.
Similar content being viewed by others
Introduction
Glioblastoma (GBM) is the most common primary malignant brain tumor in adults1. Despite standard treatments including surgery, radiotherapy and chemotherapy, the outcome of this malignant tumor remains dismal1,2,3. To date, several prognostic genetic/epigenetic biomarkers, such as isocitrate dehydrogenase (IDH) mutation, human telomerase reverse transcriptase (hTERT) promoter mutation, and O6-methylguanine DNA methyltransferase (MGMT) promoter methylation have been identified through extensive molecular and genetic studies for glioblastoma4. Among those biomarkers, hTERT promoter mutation is associated with expression of hTERT and elevation of telomerase activity (TA), which is one of the poor prognostic factor recently identified in gliomas5.
Mucin 1 (MUC1) is a single pass type I transmembrane protein with a heavily glycosylated extracellular domain6,7. Full length MUC1 compose with two subunits, N-terminal subunit (MUC1-N) and C-terminal subunit (MUC1-C). MUC1-C again consist of extracellular cellular domain (ECD), transmembrane domain (TMD), and cytoplasmic tail (CT)7. MUC1 is normally expressed at low levels on the glandular or luminal epithelial cells in breast, lung, gastrointestinal tract, pancreas, uterus and, prostate, and a lesser extent in hematopoietic cells7,8. However, aberrant glycosylation and overexpression of MUC1 has been described in most of human epithelial cancers and even in hematological malignancies7,8. Evidences showed that MUC1 may act as an oncogene related to tumor formation and progression in many cancers9,10,11. Moreover, MUC1 is reported to be associated with the cancer invasiveness and metastasis, neo-angiogenesis, drug resistance, and poor prognosis12,13,14,15. Thus, MUC1 is thought to be the universal player that acts in various steps of oncogenesis.
Epithelial-mesenchymal transition (EMT) is well known oncogenic process accelerating invasiveness and metastasis of cancer cells16. MUC1 CT upregulates the EMT inducers directly as well as indirectly by modulating the expression of miRNAs that control gene expression related to EMT11,17. Transforming Growth Factor Beta (TGF-β) is a cytokine with a dichotomous role in tumorigenesis18. TGF-β1 plays a role as a tumor suppressor which induces apoptosis or cell suicide by SMAD signaling pathway at early stage of oncogenesis19,20. However, in the late stage of aggressive and invasive tumors, TGF-β signaling stimulate tumor progression by its pleiotropic activities on the cancer cells which include induction of EMT, migration, invasion, and tumor metastasis18,21,22. MUC1 is reported as a key inducer of EMT and is partially responsible for the functional switch of TGF-β from a tumor suppressor to a tumor promoter during EMT in multiple cancers23. However, the role of MUC1 has not been fully elucidated in GBM, although only sporadic reports mentioned the MUC1 in relation to the maintenance of aggressive of gliomas24.
In this study, we examined the anticancer effect of MUC1 suppression in GBM. Our work reveals a role of MUC1 in GBM oncogenesis involving cell cycle control and telomere maintenance mechanism, which can be developed as a potential prognostic marker and therapeutic target in GBM.
Results
MUC1 is significantly overexpressed in GBM tissue
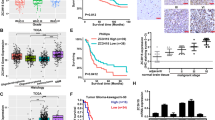
In order to study the mechanisms of differential gene expression in glioma tumorigenesis, we performed gene expression profiling by RNA sequencing data of paired normal brain and glioma tissue of 30 glioma patients. Among the differentially expressed genes identified, MUC1 was one of the significantly upregulated genes (p-value < 0.05, log2FC ≥ 2) in glioma tissue (Fig. 1A). However, the role of MUC1 in glioma cells has been rarely studied. The upregulation of MUC1 was a universal phenomenon in gliomas regardless of their WHO grades, although only high-grade gliomas including GBM showed statistical significances (Fig. 1B).
MUC1 is significantly overexpressed in gliomas. (A) Identification of differentially expressed genes from the RNA-seq data of paired normal brain and glioma tissue of 30 glioma patients. Volcano plot showing MUC1 as one of the significant upregulated gene. (B) Comparison of MUC1 mRNA expression in normal brain tissue and glioma (WHO grade II, III, and IV) tissues. MUC1 is significantly overexpressed in all glioma samples compared to normal brain samples. (C) Comparison of MUC1 RNA expression in glioblastoma (GBM) and lower grade glioma (LGG) data from TCGA (The Cancer Genome Atlas) with normal brain data from GTEx (Genotype-Tissue Expression). MUC1 is highly expressed in both GBM and LGG. (D) Overall survivals in GBM patients in TCGA dataset classified by MUC1 expression. The prognosis is significantly poor in MUC1 overexpressed patients.
Using GBM and lower grade glioma (LGG) data from TCGA (The Cancer Genome Atlas) and normal brain data from GTEx (Genotype-Tissue Expression), we could confirm that the MUC1 was overexpressed in glioma tissue compared with normal brain (Fig. 1C). Moreover, there was significant difference in overall survival between the groups of low- and high-expression level of MUC1 in TCGA dataset of GBM (Fig. 1D).
Knockdown of MUC1 inhibits the proliferation of GBM cells
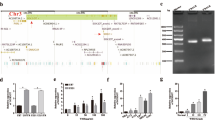
To verify the role of MUC1 in GBM cell growth, U373 and T98G cells were transduced with shLuc and shMUC1 lentivirus for 48 h and MUC1 repression was confirmed with RT-PCR analysis and western blot analysis (Fig. 2A).
MUC1 Knockdown inhibits the proliferation of glioblastoma cells. (A) GFP fluorescence images (magnification, × 100), RT-PCR and western blot (WB) analysis to confirm MUC1 depletion in U373 and T98G with shLuc or shMUC1 lentivirus infection. GADPH and ACTB were used as an internal control in RT-PCR and western blot, respectively. The blots were cropped and full length blots are presented in Supplementary Fig. S1. (B) Cell proliferation assay for 4 days and (C) Colony-formation assays with T98G and U373 cells transduced with shLuc or shMUC1 lentivirus. After 14 days, the cultured cells were fixed with methanol and stained simultaneously with staining solution (0.5% crystal violet in acetic acid/methanol 1:7) and colonies were counted manually. (B,C) were representative results from at least 3 independent experiments and were plotted with average of 3 independent experiments. Error bars, SEM. Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
Cell viability test using MTT assay showed that MUC1 knockdown decreased cell proliferation in U373 and T98G cells (Fig. 2B). Furthermore, Colony-formation assays also demonstrated reduced the size of the single colonies and the number of colonies in MUC1 knockdown U373 and T98G cells (Fig. 2C). These results indicate that MUC1 has a crucial role in GBM cell proliferation.
MUC1 knockdown attenuates cell cycle progression at G1 phase
To explore the underlying mechanism of decreased cell viability by MUC1 knockdown, cell cycle analysis was performed. Flow cytometry showed that the cell cycle was stagnated in the G0/G1 phase after MUC1 knockdown (Fig. 3A,B). Among the G1 cell cycle regulators, CDKN1B proteins were upregulated and phosphorylation of retinoblastoma 1 (RB1) was decreased upon MUC1 knockdown (Fig. 3C). Control cells and MUC1 knockdown cells were stained by annexin V and 7-AAD to see whether the G1 arrest is related with apoptosis. However, no statistical difference in apoptotic population was noted between control cells and MUC1 knockdown cells (See Supplementary Fig. S4).
MUC1 knockdown attenuates cell cycle progression at G1 phase. (A), (B) Cell cycle distribution of T98G and U373 with shLuc or shMUC1 lentivirus infection. In MUC1 knockdown cell, cell cycle arrest in G0/G1 phase and reduction in S and G2/M phases were observed. Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001. (C) Western blot analysis of Rb and CDKN1B in control and MUC1 knockdown cells. The expression of GAPDH was used as an internal control. Target protein expression levels are were quantified by free image analyzer software. The blots were cropped and full length blots are presented in Supplementary Fig. S2.
Gene expression profiling reveals MUC1-related pathways other than cell cycle
To investigate the role of MUC1 in molecular mechanism underlying the tumorigenesis of GBM cells, we have profiled transcriptomes of both naive- and shMUC1-treated GBM cells (U373, T98G, and A172) by RNA-seq. Using differentially expressed gene analysis dataset, we performed gene set enrichment analysis (GSEA). After filtering based on significant nominal p values (p < 0.05) and NES score of ≥ 2 or ≤ -2, we found gene sets that were down-regulated by MUC1 knockdown were enriched in processes related to the EMT pathway, cell cycle-related pathway and telomere-related pathway (Fig. 4A,B). On the other hand, gene sets that were up-regulated by MUC1 knockdown were enriched in processes related to TGF-β signaling in the EMT pathway (Fig. 4A,B). Pathway network analysis using GSEA result showed telomere-related and cell cycle-related pathway gene sets associated with MUC1 knockdown were indirectly interconnected with each other (Fig. 4C). In MUC1 knockdown GBM cells, significantly down-regulated genes that regulate the G1 phase of cell cycle, telomere maintenance and EMT pathway are shown in the heatmap and hierarchical clustering analysis (Fig. 4D). Taken together, these results suggested the MUC1 promotes glioma tumorigenesis through cell cycle regulation, telomere maintenance mechanism and EMT.
MUC1 knockdown is associated with the cell cycle and telomere maintenance pathway. (A) Gene set enrichment analysis (GSEA) of control and MUC1 knockdown glioblastoma (GBM) cell lines (T98G, U373, A172). TGF- β receptor signaling in epithelial mesenchymal transition (EMT) positively enriched in MUC1 knockdown cell whereas gene sets associated with cell cycle and telomere maintenance pathway are negatively enriched. “N” indicates gene counts in enriched gene set. The asterisks indicate FDR q-values (*< 0.25; **< 0.1; ***< 0.05; ****< 0.005; ***** < 0.0005). Pathways with p-value < 0.05 and NES score of > 2 or < -2 are shown. (B) Examples of GSEA charts revealing the role of MUC1 in GBM. Enrichment plot showing negative enrichment of cell cycle and telomere maintenance pathways and positive enrichment of TGF- β signaling in EMT pathway between control and MUC1 knockdown cell lines. (C) Enrichment map was drawn using Cytoscape with the GSEA result (C5 gene set from MSigDb), FDR cut off value 0.01, Edge cut off 0.5. Cell cycle pathways and telomere related pathways are interconnected together by common genes and all are negatively enriched in the MUC1 knockdown cell lines. (D) Heatmap comparison of cell cycle, telomere maintenance and EMT related gene lists in control vs MUC1 knockdown GBM cell lines.
MUC1 knockdown induces changes in telomere maintenance mechanism
The perpetual maintenance of telomere length is an essential characteristic of tumorigenesis, which mechanism achieved either by activation of telomerase or by alternative lengthening of telomeres (ALT). ALT is characterized by the presence of ALT-associated promyelocytic leukemia bodies (APB), extrachromosomal telomeric circular DNA (c-circle), heterogeneous telomere length, and increased telomeric recombination. Here we used C-circle assay and telomere length measurement to detect the ALT activity25.
In order to validate the association of MUC1 and telomere maintenance pathway, we tested hTERT expression and telomerase activity in GBM cells after MUC1 knockdown, and we could observe significant reduction of hTERT expression as well as telomerase activity (Fig. 5A,B). We also performed the c-circle assay to elucidate whether MUC1 knockdown affects the switch of telomere maintenance mechanism. MUC1 knockdown induced a significantly increased c-circle formation (Fig. 5C). Additionally, telomere restriction fragment (TRF) analysis revealed telomere lengths were slightly increased in the MUC1 knockdown GBM cells although there was no significant difference (Fig. 5D). Data are representative results from at least 3 independent experiments. Taken together, these results suggested that MUC1 depletion contributes to the switching of telomere maintenance mechanism from classic telomerase activation to ALT in GBM cells.
MUC1 knockdown induces changes in telomere maintenance mechanism. (A) qRT-PCR results showing significantly lower TERT mRNA expression in MUC1 knockdown cells. Data representative results from at least 3 independent experiments and were plotted with average of 3 independent experiments. Error bars, SEM. Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001. (B) MUC1 knockdown cells showing significantly reduce telomerase activity. Telomerase activities were analyzed using telomerase PCR ELISA kit and was quantified as ΔA (A450-A690) and then calculated as RTA (%). Data representative results from at least 3 independent experiments and were plotted with average of 3 independent experiments. Error bars, SEM. Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001. (C) Blot showing positive formation of C-circles in MUC1 knockdown glioblastoma (GBM) cells. Genomic DNA isolated from control (CTR) and MUC1 knockdown T98G and U373 cells used for the C-circle assay. U2OS and WI-38 VA13 cells were used as the positive control and 293 T cell was used as the negative control. The blots were cropped and full length blots are presented in Supplementary Fig. S3. (D) Telomere restriction fragment (TRF) analysis using T98G and U373 cells infected with shLuc or shMUC1 lentivirus infection. Mean telomere restriction fragments (TRF) detected by southern blot analysis. Data are representative results from at least 3 independent experiments.
Discussion
In this study, we explored a functional role of MUC1 in tumorigenesis of GBM based on results from both in vitro experiments and genomic data of human samples. We observed that MUC1 knockdown attenuated cell proliferation and impaired the cell cycle progression to S-phase. In fact, the role of MUC1 as a cell cycle regulator and poor prognostic marker has been studied in various human cancer including breast cancer, pancreatic cancer and lung cancer via various mechanism including β-catenin, NF-κB, platelet-derived growth factor (PDGF), epidermal growth factor receptor (EGFR), and MAPK and PI3K/Akt pathways11,15,26,27. However, the functional role of MUC1 in GBM has not been clearly clarified so far. As mentioned above, the inhibition of cell proliferation and cell cycle arrest induced by MUC1 knockdown, as detected by in vitro experiments in this study, suggest an oncogenic role of MUC1 in GBM. More specifically, our work shows that the cell cycle was stagnated in the G0/G1 phase after MUC1 knockdown involving CDKN1B up-regulation and decreased phosphorylation of RB1. The CDKN1B gene encodes for the p27Kip1 protein, which firstly described as an inhibitor of G1 cycle progression by binding a broad range of cyclin-CDK (cyclin-dependent kinase) complexes28. Moreover, RB1 plays a central role in G1-S checkpoint control and phosphorylation of RB1 is the most common mechanism of inactivation of this gene29. We found that MUC1 Knockdown leads to increased CDKN1B expression and decreased phosphorylation of RB1 in GBM cell lines. Taken together, these results suggested the effects of MUC1 in cell cycle regulation, especially at the G1 phase in GBM.
Transcriptome profiling revealed large numbers of gene sets that are up- or down-regulated by MUC1 knockdown. GSEA for the biological function of the genes up-regulated by MUC1 knockdown identifies functions related to TGF-β signaling in EMT pathway. Previous study shows that TGF-β induces G1 growth arrest and the accumulation of unphosphorylated Rb30. These reports support our finding of role of MUC1 as a G1 phase regulator of cell cycle as shown in Fig. 3. The mechanism how MUC1 gene silencing induced TGF-β signaling require further study. On the other hand, GSEA of genes down-regulated by MUC1 knockdown identifies cell cycle-related pathway, EMT pathway, and telomere-related pathway. This result supports the experimental result showing cell cycle arrest after decreased MUC1 expression. Telomere-related pathway, another suppressed pathway after MUC1 knockdown, is one of the core elements in tumorigenesis. It has been proposed that up to 90% of the human cancers utilize telomerase activation for the maintenance of their telomere length, while the other 10% of tumors utilize ALT31. As has been reported in other cancers including gliomas, there is a correlation between telomerase activity with the grade of malignancy and prognosis in glial tumors32,33,34. However, others suggested that telomerase-associated parameters might have limited value as independent prognostic markers in a younger GBM patient34. Recent study by Hu et al. showed a possibility of switching phenomenon of telomerase maintenance mechanism in cancer cells by inducing telomeric DNA damages and knockdown of ATRX/DAXX complex35. Interestingly, we could observe the similar switching phenomenon from telomerase-positive to ALT-positive cells after MUC1 knockdown. The identification of the role of MUC1 in playing as a regulator for telomere maintenance mechanism is a novel discovery. A further study is needed to develop the detailed understanding of this switching mechanism of telomerase maintenance associated by MUC1.
We investigated the role of MUC1, a gene which was one of the widely studied in cancer except for GBM, focusing on its anti-cancer mechanism by inhibition. Our research suggested the role of MUC1 as a regulator of cell cycle and telomere maintenance mechanism in GBM. It is expected that abrogating MUC1 can be considered as one of the therapeutic strategies for GBM.
Materials and methods
Cell culture
Human glioblastoma cell lines T98G, U373 and A172 were from Korean Cell Line Bank (Seoul, Republic of Korea) and the human embryonic kidney cell line 293 T was from American Type Culture Collection (ATCC). Cells were maintained in DMEM supplemented with 10% fetal bovine serum (J R Scientific), 100 U/ml penicillin and 100 μg /ml streptomycin sulfate (Welgene).
Tumor specimens
We used prospectively collected samples of 30 histologically verified GBM patients who had undergone surgical resection. This study was performed under the approval of the Institutional Review Board of Seoul National University Hospital (IRB approval No., H-1608-139-787), and all experiments were performed in accordance with relevant guidelines and regulations. Written informed consent was obtained from all patients for the usage of samples.
Gene knockdown
For gene silencing, shLuc and shMUC1 targeting CTTCGAAATGTCCGTTCGGTT of firefly luciferase gene and GACACAGTTCAATCAGTATA of human MUC1, respectively, were cloned into pLB lentiviral vector (generous gift from Dr. H. Y. Chung of Hanyang University). Lentivirus was produced by co-transfection of shLuc or shMUC1 with pMD2.G and psPAX2 into 293 T cells (ATCC) by calcium-phosphate method as described by the RNAi Consortium (2007). T98G and U373 cells were infected with lentiviral supernatant with 8 μg /ml protamine sulfate for 10 h. Gene knockdown was examined 48 h post infection36.
RNA sequencing and analysis
Total RNA was isolated from GBM tissues using RNeasy Lipid Tissue Mini Kit (Qiagen) and the compatible library was prepared using the TruSeq stranded total RNA LT sample prep kit (Illumina, San Diego, CA, USA) according to the instructions specified by the manufacturer. Sequencing was done using NovaSeq 6000 system (Illumina). RNA expression levels were estimated using HISAT2 (version 2.1.0). The reference genome sequence (hg19, Genome Reference Consortium GRCh37) and annotation data were downloaded from the UCSC website (https://genome.uscs.edu). The transcript counts in gene level, and the relative transcript abundances in FPKM (fragments per kilobase of exon per million fragments mapped) were calculated using StringTie (version 1.3.4d).
Gene set enrichment analysis (GSEA) and pathway network construction
GSEA was carried out using the standard GSEA tool for Windows version 4.0.3 (https://www.gsea-msigdb.org/gsea/index.jsp) with 1000 gene set permutations. Normalized count of the genes were inputted. Gene sets used for the analysis were Hallmark, canonical pathways and the Gene Ontology (GO). The upregulated and downregulated pathways were then filtered by normalized enrichment score ≥ 2 or ≤ -2, nominal P value < 0.05, FDR q value ≤ 0.25. The results obtained from the GSEA were then used to create a network map of the pathways. Enrichment map app from the Cytoscape program (version 3.8.0) was used to create the pathway network.37.
Cell proliferation assay
Cell proliferation assay was performed with EZ-Cytox (Daeillab Service) on cells initially plated at 1 × 103 cells/well in 96-well plates and cultured for indicated times. The absorbance was measured using a microplate reader (Molecular Devices) at a wavelength of 450 nm.
Colony-forming assay
The cells were seeded in 6-well plates at a density of 1000 cells/well incubated at 37 °C under an atmosphere of 5% CO2 for 14 days to grow colonies. After 14 days, the cell colonies were fixed and stained with 0.05% crystal violet–methanol–acetic acid solution. Plates with stained colonies were scanned and scored.
Cell cycle analysis
Cells were stained with propidium iodide and analyzed by flow cytometry as described38. The distribution of the cell cycle was determined by flow cytometry with FACSCanto II flow cytometer and FACSDiva software (BD Biosciences).
Annexin V apoptosis assay
Cells were stained with the Annexin V-APC and 7-aminoactinomycinD (7-AAD) (BD Biosciences) according to the manufacturer’s protocol. The apoptosis rate was then analyzed using a FACS Calibur flow cytometer (BD Biosciences) using Cell Quest Pro software (BD Biosciences).
RT-PCR and quantitative PCR (qPCR)
Total RNA was isolated from the tissues using RNeasy Lipid Tissue Mini Kit (Qiagen) and cDNA was cDNA was synthesized using RNA to cDNA EcoDry Premix Oligo dT(Takara) according to the manufacturer’s protocol. Real-time quantitative PCR (qPCR) was performed using PowerSYBR Green PCR Master Mix (Applied biosystems). Primers used for RT-PCR reactions were as follows: TERT (forward, 5′-GGAGCAAGTTGCAAAGCATTG-3′; reverse, 5′-TCCCACGACGTAGTCCATGTT-3′), MUC1 (forward, 5′- TACCGATCGTAGCCCCTATG-3′; reverse, 5′-CTCACCAGCCCAAACAGG-3′), GAPDH (forward, 5′-TGGTCACCAGGGCTGCTT-3′; reverse, 5′-AGCTTCCCGTTCTCAGCCTT-3′).
Western blot analysis
Western blot analysis was performed as described39. Antibodies against CDKN1B (sc-528), RB1 (sc-50) were from Santa Cruz Biotechnology, GAPDH (2118), phosphorylated RB1 at S780 (p-S780, 9307A), MUC1 (4538) and ACTB (4967) were from Cell Signaling Technology, HRP-conjugated IgGs (111-035-003 and 115-035-003) were from Jackson Immune Research. Immunoblots were visualized with ChemiDoc XRS system (Bio-Rad). The densities of bands were measured using free image analyzer software (ImageJ V1.8x; National Institutes of Health, USA, https://rsb.info.nih.gov/ij/).
Telomere repeat amplification protocol (TRAP) assay with ELISA
The enzymatic activity of telomerase was measured using TeloTAGGG Telomerase PCR ELISA PLUS kit (Roche) according to the manufacturer’s protocol. GBM tissues and cells were homogenized in ice-cold lysis buffer using automill (Tokken). Briefly, after BCA protein quantification of the lysates, 10 µg of proteins were incubated in total volume of 50 µl reaction mixture at 25 °C for 30 min to allow the telomerase to add telomeric repeats to the end of the biotin-labeled primer. Consequently, PCR was conducted for 33 cycles of 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 90 s, followed by additional extension time of 10 min at 72 °C and holding at 4 °C. The telomerase activity was measured at 450 nm and the reference wavelength 690 nm. Relative telomerase activity (RTA) of each sample was calculated according to the instruction of TeloTAGGG Telomerase PCR-ELISA PLUS Kit.
C-circle assay
Detection of C-circles was performed as previously described40. Briefly, 30 ng DNA was combined with 10 µl 2X Φ29 Buffer, 7.5 U Φ29 DNA polymerase (NEB), 0.2 mg/ml BSA, 0.1% (v/v) Tween 20, 1 mM each dATP, dGTP and dTTP and incubated at 30 °C for 4 h and 8 h followed by 20 min at 70 °C. Amplification products were deposited on a Hybond N + nylon membrane (Bio-Rad) and developed using the TeloTAGGG Telomere Length Assay Kit (Roche). Chemiluminescent signals was visualized with ChemiDoc XRS system (Bio-Rad) and the intensity of the spots was quantified with ImageQuant TL software (Bio-Rad).
Telomere length fragmentation assay
Telomere length was determined by southern blot using TeloTAGGG Telomere Length Assay Kit (Roche) according to the manufacturer’s protocol. Briefly, 1 µg DNA was digested with Rsa I and Hinf I for O/N at 37 °C, then electrophoresed on 0.8% agarose gel at 50 V for 4 h then transferred to a nylon membrane by Southern blotting. The blotting membrane was blocked and hybridized to a digoxigenin (DIG)-labeled probe specific for telomeric repeats for O/N. Washed blot was incubated with anti-DIG-alkaline phosphatase (1:10,00 dilution) for 30 min and developed using substrate in TeloTAGGG Telomere Length Assay kit (Roche). After Chemiluminescent signals was visualized with ChemiDoc XRS system (Bio-Rad), terminal restriction fragment analysis was performed with Telo Tool version 1.3.
Statistical analysis
The results were analyzed by using IBM SPSS Statistics software (version 20.0; SPSS, Armonk, NY, USA). Data were expressed as the mean ± SE. Statistical significance was determined using the Student’s t-test. Kaplan–Meier curve analysis was used for analyzing patient survival time. P values < 0.05 were considered statistically significant.
References
DeAngelis, L. M. Brain tumors. N. Engl. J. Med. 344, 114–123. https://doi.org/10.1056/nejm200101113440207 (2001).
Stupp, R. et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 10, 459–466. https://doi.org/10.1016/s1470-2045(09)70025-7 (2009).
Buckner, J. C. Factors influencing survival in high-grade gliomas. Semin. Oncol. 30, 10–14. https://doi.org/10.1053/j.seminoncol.2003.11.031 (2003).
Aquilanti, E., Miller, J., Santagata, S., Cahill, D. P. & Brastianos, P. K. Updates in prognostic markers for gliomas. Neuro-Oncology 20, vii17–vii26. https://doi.org/10.1093/neuonc/noy158 (2018).
Eckel-Passow, J. E. et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N. Engl. J. Med. 372, 2499–2508. https://doi.org/10.1056/NEJMoa1407279 (2015).
Hattrup, C. L. & Gendler, S. J. Structure and function of the cell surface (tethered) mucins. Annu. Rev. Physiol. 70, 431–457. https://doi.org/10.1146/annurev.physiol.70.113006.100659 (2008).
Nath, S. & Mukherjee, P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol. Med. 20, 332–342. https://doi.org/10.1016/j.molmed.2014.02.007 (2014).
Gendler, S. J. MUC1, the renaissance molecule. J Mammary Gland Biol. Neoplasia 6, 339–353. https://doi.org/10.1023/a:1011379725811 (2001).
Hattrup, C. L. & Gendler, S. J. MUC1 alters oncogenic events and transcription in human breast cancer cells. Breast Cancer Res. 8, R37. https://doi.org/10.1186/bcr1515 (2006).
Li, Q. et al. Impact of Mucin1 knockdown on the phenotypic characteristics of the human hepatocellular carcinoma cell line SMMC-7721. Oncol. Rep. 31, 2811–2819. https://doi.org/10.3892/or.2014.3136 (2014).
Roy, L. D. et al. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene 30, 1449–1459. https://doi.org/10.1038/onc.2010.526 (2011).
Horm, T. M. & Schroeder, J. A. MUC1 and metastatic cancer: expression, function and therapeutic targeting. Cell Adhes. Migr. 7, 187–198. https://doi.org/10.4161/cam.23131 (2013).
Jing, X., Liang, H., Hao, C., Yang, X. & Cui, X. Overexpression of MUC1 predicts poor prognosis in patients with breast cancer. Oncol. Rep. 41, 801–810. https://doi.org/10.3892/or.2018.6887 (2019).
Pillai, K., Pourgholami, M. H., Chua, T. C. & Morris, D. L. MUC1 as a potential target in anticancer therapies. Am. J. Clin. Oncol. 38, 108–118. https://doi.org/10.1097/COC.0b013e31828f5a07 (2015).
Bernier, A. J. et al. Non-cysteine linked MUC1 cytoplasmic dimers are required for Src recruitment and ICAM-1 binding induced cell invasion. Mol. Cancer 10, 93. https://doi.org/10.1186/1476-4598-10-93 (2011).
Kalluri, R. & Weinberg, R. A. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428. https://doi.org/10.1172/jci39104 (2009).
Mohr, A. M. et al. MUC1 regulates expression of multiple microRNAs involved in pancreatic tumor progression, including the miR-200c/141 cluster. PLoS ONE 8, e73306. https://doi.org/10.1371/journal.pone.0073306 (2013).
Lebrun, J. J. The dual role of TGFβ in human cancer: from tumor suppression to cancer metastasis. ISRN Mol. Biol. 2012, 381428. https://doi.org/10.5402/2012/381428 (2012).
Siegel, P. M. & Massagué, J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat. Rev. Cancer 3, 807–821. https://doi.org/10.1038/nrc1208 (2003).
Lacerte, A. et al. Transforming growth factor-beta inhibits telomerase through SMAD3 and E2F transcription factors. Cell Signal 20, 50–59. https://doi.org/10.1016/j.cellsig.2007.08.012 (2008).
Xu, J., Lamouille, S. & Derynck, R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res 19, 156–172. https://doi.org/10.1038/cr.2009.5 (2009).
Seoane, J. & Gomis, R. R. TGF-β family signaling in tumor suppression and cancer progression. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a022277 (2017).
Grover, P. et al. SMAD4-independent activation of TGF-β signaling by MUC1 in a human pancreatic cancer cell line. Oncotarget 9, 6897–6910. https://doi.org/10.18632/oncotarget.23966 (2018).
Liu, Z. et al. LINC00909 promotes tumor progression in human glioma through regulation of miR-194/MUC1-C axis. Biomed. Pharmacother. 116, 108965. https://doi.org/10.1016/j.biopha.2019.108965 (2019).
Plantinga, M. J. et al. Telomerase suppresses formation of ALT-associated single-stranded telomeric C-circles. Mol. Cancer Res. 11, 557–567. https://doi.org/10.1158/1541-7786.Mcr-13-0013 (2013).
Sahraei, M. et al. MUC1 regulates PDGFA expression during pancreatic cancer progression. Oncogene 31, 4935–4945. https://doi.org/10.1038/onc.2011.651 (2012).
Bitler, B. G., Goverdhan, A. & Schroeder, J. A. MUC1 regulates nuclear localization and function of the epidermal growth factor receptor. J. Cell Sci. 123, 1716–1723. https://doi.org/10.1242/jcs.062661 (2010).
Polyak, K. et al. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 8, 9–22. https://doi.org/10.1101/gad.8.1.9 (1994).
Macdonald, J. I. & Dick, F. A. Posttranslational modifications of the retinoblastoma tumor suppressor protein as determinants of function. Genes Cancer 3, 619–633. https://doi.org/10.1177/1947601912473305 (2012).
Laiho, M., DeCaprio, J. A., Ludlow, J. W., Livingston, D. M. & Massagué, J. Growth inhibition by TGF-beta linked to suppression of retinoblastoma protein phosphorylation. Cell 62, 175–185. https://doi.org/10.1016/0092-8674(90)90251-9 (1990).
Cesare, A. J. & Reddel, R. R. Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 11, 319–330. https://doi.org/10.1038/nrg2763 (2010).
Boldrini, L. et al. Telomerase activity and hTERT mRNA expression in glial tumors. Int. J. Oncol. 28, 1555–1560. https://doi.org/10.3892/ijo.28.6.1555 (2006).
Maes, L. et al. Relation between telomerase activity, hTERT and telomere length for intracranial tumours. Oncol. Rep. 18, 1571–1576. https://doi.org/10.3892/or.18.6.1571 (2007).
Lötsch, D. et al. Prognostic significance of telomerase-associated parameters in glioblastoma: effect of patient age. Neuro-oncology 15, 423–432. https://doi.org/10.1093/neuonc/nos329 (2013).
Hu, Y. et al. Switch telomerase to ALT mechanism by inducing telomeric DNA damages and dysfunction of ATRX and DAXX. Sci. Rep. 6, 32280. https://doi.org/10.1038/srep32280 (2016).
Jo, S. et al. Inhibition of PCGF2 enhances granulocytic differentiation of acute promyelocytic leukemia cell line HL-60 via induction of HOXA7. Biochem. Biophys. Res. Commun. 416, 86–91. https://doi.org/10.1016/j.bbrc.2011.10.152 (2011).
Isserlin, R., Merico, D., Voisin, V. & Bader, G. D. Enrichment Map: a Cytoscape app to visualize and explore OMICs pathway enrichment results. F1000Research 3, 141. https://doi.org/10.12688/f1000research.4536.1 (2014).
Chung, H. et al. The antitumor effect of LJ-529, a novel agonist to A3 adenosine receptor, in both estrogen receptor-positive and estrogen receptor-negative human breast cancers. Mol. Cancer Ther. 5, 685–692. https://doi.org/10.1158/1535-7163.Mct-05-0245 (2006).
Chung, H. et al. Anticancer effects of wogonin in both estrogen receptor-positive and -negative human breast cancer cell lines in vitro and in nude mice xenografts. Int. J. Cancer 122, 816–822. https://doi.org/10.1002/ijc.23182 (2008).
Henson, J. D. et al. The C-Circle Assay for alternative-lengthening-of-telomeres activity. Methods 114, 74–84. https://doi.org/10.1016/j.ymeth.2016.08.016 (2017).
Acknowledgements
This study was supported by Seoul National University Hospital Research Fund (0320200070) and the Bio & Medical Technology Development Program (NRF-2018M3A9H3021707) through the National Research Foundation of Korea (NRF) funded by the Ministry of Science & ICT of Republic of Korea.
Author information
Authors and Affiliations
Contributions
S.K., Y.S. wrote the manuscript. S.K., Y.S., C.-K.P. contributed to the conception and design of the project. S.K., T.C., H.J.Y., C.E.L., K.-M.K., and H.K. collected tissue samples and performed experimental works. H.J.K., S.-J.P., and K.K. supported experimental work design. All authors participated in drafting and revising the article for important intellectual content and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, S., Seo, Y., Chowdhury, T. et al. Inhibition of MUC1 exerts cell-cycle arrest and telomerase suppression in glioblastoma cells. Sci Rep 10, 18238 (2020). https://doi.org/10.1038/s41598-020-75457-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-75457-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.