Abstract
Maritime ecosystems in Vietnam such as mangroves and mud flats are characterized by high biodiversity. However, elements of its biodiversity remain unclear and highly threatened. In this context, the assessment of rare species is a starting point to develop effective strategies for the conservation of entire ecosystems. In this paper, we report upon cryptic amphibolid gastropods in Vietnamese mangrove forests from the Mekong Delta. The snail fauna in the mangrove forests was previously known from published literature and three museum specimens as three amphibolid species, ‘Amphibola’ burmana, A. quadrasi, Salinator fragilis and ‘S.’ quadrasi. We investigated the identities of such snails using molecular and morphological methods. The amphibolids found in this survey were identified to belong to the genus Naranjia, new for Vietnam fauna. In addition, our phylogenetic analyses suggested that the Vietnamese amphibolids were the same species as Naranjia sp. reported from Thailand, and the amphibolids have both genetic and morphological polymorphisms within the population. These findings add to the great biodiversity of Vietnamese mangrove forests and mudflats.
Similar content being viewed by others
Introduction
The country of Vietnam straddles some 13 degrees of latitude with a corresponding long coastline with biodiversity hotspot which are highly threatened1,2,3,4,5,6. In particular, mangrove forests which mainly occur in the Mekong Delta along the southern coastline, and also around the Red River Delta in the north, are being destroyed rapidly in spite of their importance to economics, coastal stability, and habitat for organisms7,8,9. Within the Mekong Delta, mangrove forests are particularly developed in Ca Mau and Bac Lieu provinces9. Besides these areas, a mangrove belt still covers part of the shoreline in Kien Giang Province also in the Mekong Delta, belonging to Kien Giang Biosphere Reserve, recognized by UNESCO in 200610,11,12. Though molluscs are known as one of the most diverse, abundant and important components of the mangrove ecosystem, the estuarine malacofauna of Vietnam, especially in the Mekong Delta is still poorly studied13,14,15. In the delta, several molluscan species occurring in the Ca Mau mangroves were reported by N. Thach in his general monographs on Vietnam molluscan fauna16,17,18, and three other additional species were recorded from the Ba Tai tidal mangrove forest in Kien Luong District of the Kien Giang Province19, 20.
Our first malacological survey in 2017 encompassing the mangrove forests, tidal mudflats, and estuaries along the coastline of Kien Giang Province revealed the presence of more than 60 gastropod and bivalve species in the Ba Tai tidal mangrove forest (An Binh Commune, Kien Luong District), which is recognized as a molluscan diversity hot spot21. The forest growing between the limestone Ba Tai hills and brackish water Ba Tai Lake belongs to the Kien Luong Protected Area which encompasses 200 km of low coastline and supports one of the last significant areas of the natural mangroves and mud flats in Kien Giang Province12. In order to further understand the molluscan components of the Mekong Delta, the second field survey was undertaken in the same mangrove forest by an international research team in May 2018. As a result, some rare and new records for estuarine Vietnamese molluscan taxa including those Amphibolidae were recorded22. These findings are of great importance in the absence of comprehensive biodiversity data on coastal ecosystems in Vietnam. The assessment of rare species is useful in the development of effective conservation strategies for entire ecosystems23,24,25.
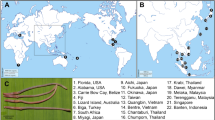
Amphibolid snails are an air-breathing taxon that inhabit mangrove forests, mudflats and salt marshes26. This family is poorly studied in Vietnam. In fact, no recent Vietnamese molluscan monographs have recorded this family16,17,18, 27. Thus, Vietnamese amphibolids have not been examined in morphological studies involving the taxonomic revision of the superfamily Amphiboloidea28, 29. However, there is a single record30 in the literature and the three GBIF online records as well as one lot in the malacology collection in the Academy of Natural Science, Philadelphia (ANSP)31,32,33,34 (Fig. 1; three species, ‘Amphibola’ burmana, A. quadrasi, Salinator fragilis and ‘S.’ quadrasi). Note that the ANSP record may refer to the same material as those reported upon by Davis (1974) because the locality name, collector, collection date and station name were all the same. In this study, we report details of the occurrence of these rare mangrove amphibolid species and examine for the first time the molecular sequences of these rare mangal amphibolid snails in Vietnam.
Locality map of Vietnamese Amphibolidae. Red circles indicate localities (see Fig. 2) where amphibolids were collected in this study. Black circles indicate localities where amphibolids were recorded based on published literature and museum collections. The reference numbers above the scientific names indicate the catalogue number of each specimen. This map was created from GADM database, version 3.445using QGIS version 2.1846.
Material and methods
Vietnamese amphibolids were first discovered at two sites in the Kien Luong District, Kien Giang Province on May 3rd 201822. One site was at a mangrove forest near Ha Tien Town (17 specimens collected; Fig. 1: site 1, Fig. 2a), and the other was in the Ba Tai mangroves (one specimen collected; Fig. 1: site 2, Fig. 2b). Molluscs were hand-picked from mud flats and open mangrove forests with Rhizophora and Avicennia trees. Three individuals from site 1 (ScNM 486, 487 and 488; Fig. 2c–e) were preserved in 99.5% ethanol for molecular analysis and other 14 individuals from site 1 (Federal Scientific Center of the East Asia Terrestrial Biodiversity [FSCEATB] 96v2018-10/1-14; Fig. 2g) were stored in 95% ethanol for morphological study after collecting (for details, see Supplementary Information 1). Total DNA was isolated from each of three individuals by DNeasy Blood & Tissue kit (Qiagen Inc., Hilden, Germany). Then, two mitochondrial (cytochrome oxidase subunit 1 [CO1] and 16S ribosomal DNA [16S]) and one nuclear marker (28S ribosomal DNA [28S]) were amplified according to the methods of Golding26. However, an annealing temperature of strictly 63°C was used for 28S instead of 52°C to 63°C26. Each amplified gene fragment was sequenced after purification via ExoSAP-IT (Affymetrix Inc., California, USA). Sequencing was performed by Eurofins Genomics (Tokyo, Japan). All sequences were deposited in the GenBank database (LC575065-LC575072). For phylogenetic analysis, each sequence was firstly aligned using the default settings of MUSCLE v3.8.3135 with sequences of 26 amphibolid materials and one outgroup material of Golding (2012; dataset B)26. Then, to eliminate the uncertainty of the 16S and 28S alignments, trimAl 1.236 was used to select regions of the aligned sequences for analyses (output files: Supplementary Information 2–3). Secondly, to select an appropriate evolutionary model and partitioning schemes of each region, we used PartitionFinder 237 with Akaike's information criterion with correction for small sample size (AICc). Based on selected evolutionary models and partitioning schemes by PartitionFinder 2 (Supplementary Information 4), phylogenetic trees were obtained by Bayesian inference (BI) and maximum likelihood (ML) methods. BI analysis was conducted using MrBayes5D Version 3.1.2.2012.12.1338, which is an extended version of MrBayes 3.1.239, and ML analysis was performed using IQ-TREE version 1.6.740. In MrBayes5D, we used four simultaneous chains for one million generations, and trees were sampled every 100 generations. We discarded the first 1001 trees as burn-in after examining convergence and effective sample size (ESS; Over 200) using Tracer v. 1.641; the remaining samples were used to estimate phylogeny. IQ-TREE was run under the evolutionary models and partitioning schemes selected by PartitionFinder 2 with –spp option (the same set of branch lengths across partitions with allowing each partition to evolve under its rate). Bootstrap analysis was executed using IQ-TREE’s ultra fast bootstrap option (-bb)42 with 5000 replicates. In addition to the analysis using the combined three regions, we analysed mitochondrial and nuclear regions respectively using the same methods (for detail parameters: Supplementary Information 5–6). Furthermore, we calculated K2P-corrected pairwise distances between CO1 sequences of Naranjia species to compare with Golding26 using MEGA 6.0643. Finally, to examine the past records in GBIF, we investigated museum specimens with the help of the curators.
(a) The environment of the locality; site 1, mangrove forest near Ha Tien Town. (b) The environment of the locality; site 2, Ba Tai mangroves. (c) Shell morphology of Naranjia sp. (Specimen No. ScNM487). This specimen was collected from site 1 (Fig. 1). Scale bar = 1 mm. Shell length (SL) is around 6.6 mm. Deposited in Tohoku University. (d) Shell morphology of Naranjia sp. (Specimen No. ScNM488). This specimen was collected from site 1 (Fig. 1). Scale bar = 1 mm. SL is around 7.2 mm. Deposited in Tohoku University. (e) Operculum of Naranjia sp. (Specimen No. ScNM488). Above: the front side, below: the back side. Scale bar = 1 mm. Deposited in Tohoku University. (f) Live specimens of Naranjia sp. from site 1. Molecular data of these specimens were not examined. (g) Head and organs of mantle cavity of Naranjia sp. from site 1. (Specimen No. 96v2018-10/4). Deposited in Federal Scientific Center of the East Asia Terrestrial Biodiversity (FSCEATB). Scale bar = 1 mm. al anal lobe, bv blood vessel, ct opposed ciliary tracts, e eye, he heart, hg hypobranchial gland, ki kidney, m mantle, p papillae on the distal surface of the spermovipositor, pn pneumostome, pr prostate gland, s snout, so spermovipositor, te tentacle.
Results and discussion
First, inferred phylogeny showed that Vietnamese amphibolids belonged to the genus Naranjia Golding, Ponder & Byrne, 2007 because of fully supported monophyly with two Naranjia sp. materials26 from Thailand (Fig. 3; Bayesian posterior probability = 1.00/Bootstrap value = 100). Furthermore, this monophyletic group was sister to a clade of N. cf. swatowensis (Yen, 1939) from Hong Kong, type species of the Naranjia28 and this monophyletic clade was relatively well-supported (0.98/94). This clade was in turn sister to another clade of Naranjia sp. from Malaysia and Singapore and was fully supported (1.00/100). Thus, Vietnamese amphibolids are here regarded as belonging to the genus Naranjia on the basis of molecular phylogeny, and this is the first record of the genus Naranjia in Vietnam.
Bayesian phylogenetic tree inferred from a combined dataset of two mtDNA and one ncDNA sequences (mtCO1, mt16S; nc28S; total 2209 bp). Numbers at the branch nodes represent Bayesian posterior probability and Ultrafast bootstrap value in IQtree. Red OTUs indicate Vietnamese amphibolids. Refer to Golding (2012)26 for detailed information of other amphibolids.
In addition to molecular phylogeny, we examined the morphology of the shell, operculum and soft tissue of Vietnamese amphibolids (Fig. 2c–g). The morphology of organs in the mantle cavity is described on the basis of dissection of specimens deposited in FSCEATB. We used terminology following Golding et al.28, 29. The morphological treats are as follows: shell globose, diameter to 8 mm, umbilicate, with short spire and large last whorl, nearly smooth; exterior pale-orange to brownish-orange, uniformly coloured or sometimes with one to two dark brown spiral bands; aperture semi-lunar; umbilicus of intermediate width (Fig. 2c,d). Operculum thin, corneous, semi-transparent pale yellow with eccentric, paucispiral nucleus, inner surface of nucleus slightly raised, smooth and bright (Fig. 2e). Foot short, anterior and posterior region slightly convex; head with short, bilobed snout, unpigmented distally and light yellow proximally, separated from foot by deep furrow; cephalic tentacles short, flattened-semicircular; eyes at outer base (Fig. 2f,g). Pneumostome with large white coloured pneumostomal lobe attached to anal lobe on right side of mantle edge; mantle roof with few blood vessels; opposed ciliary tracts not short; kidney on roof of mantle cavity, ovate-triangular, large; hypobranchial gland large triangular, white in ethanol-preserved specimens, located at right anterior corner of mantle cavity roof between kidney and opposed ciliary tracts; prostate gland large, oval, white, formed by a tightly packed tube, uniform for their entire length; large spermovipositor covered by soft finger-shaped papillae on its distal surface (Fig. 2g). These morphological traits were consistent with those of Naranjia cf. swatowensis28, the only Naranjia species documented in detail, although we could not examine the central nervous system and radula. In particular, our specimens had shells which were brownish-orange and the spermovipositor distally covered by finger-like papillae, which Golding (2008) indicated as the distinguishing traits of the genus Naranjia.
Although statistical support was not high, Vietnamese Naranjia materials were divided into two lineages; one was closely related to Thai Naranjia sp., and another represented a potentially endemic lineage in Vietnam (Fig. 3). Although 28S sequences from two Thai Naranjia species were not included, our mitochondrial phylogeny also indicated similar relationships between the two Vietnamese lineages (Supplementary Information 5). However, K2P pairwise CO1 distances among the Vietnamese individuals were calculated to be 0.61% to 1.70%, and the CO1 distances between Vietnamese and Thai Naranjia sp. were 0.46 to 1.54% (Table 1). These values are close to the average intraspecies CO1 distances of amphibolids shown by Golding26. Accordingly, the Vietnamese Naranjia sp. and Thai Naranjia sp. seem to be the same species, and the presence of two lineages can imply a genetic polymorphism. In contrast, all of the CO1 distances between Vietnamese amphibolids and other Naranjia species were over 6.60% (Table 1). As Golding (2012) showed that CO1 distances of interspecies were 4.25 to 24.55%, the Vietnamese (and Thai) amphibolids can be considered to be genetically distinct from other Naranjia species including N. cf. swatowensis from Hong Kong. This suggests the existence of cryptic diversity in the Asian Naranjia species complex26. On the other hand, the shells of Vietnamese specimens display several different colors and band patterns (Fig. 2c,d,f; pale-orange to brownish-orange, no band: five, one band: seven, two bands: five in 17 individuals from site 1). In some Amphibolidae species, phenotypic polymorphism within species was reported28, 44. Thus, the Vietnamese amphibolids seem to show not only genetic but also morphological polymorphism such as shell color and band pattern.
In Vietnam, as mentioned above, there are only four records of amphibolids (Fig. 1), and all of these species (i.e., burmana, fragilis, quadrasi, as either Amphibola or Salinator) were likely to have been misidentified, as all recorded species are actually not distributed in Vietnam after recent taxonomic revision28. We examined museum materials of two amphibolids included in three GBIF records, and one shell of Amphibola quadrasi from ANSP collection (Supplementary Information 7). The shell of amphibolids does not have enough information to be identified to species and even genus28, and so we could not identify these specimens. Nevertheless, these specimens are included in Salinatorinae (Lactiforis, Naranjia or Salinator) based on shell size and morphology28. In addition, some specimens were polymorphic in the shell shape and size in spite of specimens from the same localities (Supplementary Information 7c–e), and it may contain two or more species, which merit further investigation.
Although further researches are needed to reveal the actual distribution of Vietnamese amphibolids, in any case, this paper is the first report of Vietnamese Amphibolidae. Vietnamese amphibolids appear to be rare, because the taxonomic revision of Amphiboloidea26, 28, catalogues of Vietnamese mollusks by Thach16,17,18, 27, and checklist of Vietnamese Khanh Hoa mangal molluscan fauna15 have not reported upon Amphiboloidea species. In contrast, our findings show that amphibolids are rare but distributed along the Vietnamese coastline sporadically and have genetic and phenotypic polymorphism from one locality. Therefore, immediate assessment of distribution and diversity of Amphiboloidea is required considering the rapid decline in the area of Vietnamese mangrove forests7, 8. Amphibolids may be an excellent indicator of the conservation status of cryptic biodiversity in Vietnamese mangrove forests.
Ethics declarations
All necessary permits for sampling and observational field studies have been obtained by the authors from the Institute of Tropical Biology (Vietnam Academy of Science and Technology) and local competent authorities.
Data availability
Sequences of Naranjia sp. were deposited in the GenBank (CO1:LC575065-LC575067/16S:LC575068-LC575070/28S: LC575071-LC575072). Additional data are available from supplementary materials.
References
IUCN. A Study on Aid to the Environment Sector in Vietnam. (Ministry of Planning and Investment and UNDP, 1999).
Myers, N. et al. Biodiversity hotspots for conservation priorities. Nature 403, 853–858 (2000).
Sterling, E. J. & Hurley, M. M. Conserving biodiversity in Vietnam: applying biogeography to conservation research. Proc. Calif. Acad. Sci. 56, Supplement I, No.9, 98–118 (2005).
Thuoc, P. & Long, N. Overview of the coastal fisheries of Vietnam. In Status and Management of Tropical Coastal Fisheries in Asia. (eds. Silvestre, G. & Pauly, D.) 96–106 (ICLARM, 1997).
Ng, P. K. L. & Tan, K. S. The state of marine biodiversity in the South China Sea. Raffles Bull. Zool. (Supplement No. 8), 3–7 (2000).
Burke, L., Selig, E. & Spalding, M. Reefs at Risk in Southeast Asia. (Resource for the Future, 2002).
Benthem, W., van Lavieren, L. P. & Verheugt, W. J. M. Mangrove rehabilitation in the costal Mekong Delta, Vietnam. In An International Perspective on Wetland Rehabilitation. (ed. Streever, W.) 29–36 (Springer, 1999).
McNally, R., McEwin, A. & Holland, T. The Potential for Mangrove Carbon Projects in Vietnam. (Netherlands Development Organization [SNV], 2011).
Veettil, B. K. et al. Mangroves of Vietnam: historical development, current state of research and future threats. Estuar. Coast. Shelf Sci. 218, 212–236 (2019).
Duke, N. C. Mangroves of the Kien Giang Biosphere Reserve Viet Nam. (Deutsche Gesellschaft fur International Zusammenarbeit GmbH, 2012).
Cuong, C. V., Russell, M., Brown, S. & Dart, P. Using shoreline video assessment for coastal planning and restoration in the context of climate change in Kien Giang, Vietnam. Ocean Sci. J. 50, 413–432 (2015).
Nguyen, T. P., Luom, T. T. & Parnell, K. E. Mangrove allocation for coastal protection and livelihood improvement in Kien Giang Province, Vietnam: constraints and recommendations. Land Use Policy 63, 401–407 (2017).
Tue, N. T. et al. Food sources of macro-invertebrates in an important mangrove ecosystem of Vietnam determined by dual stable isotope signatures. J. Sea Res. 72, 14–21 (2012).
Thanh, N. V. et al. The Zoobenthos of the Can Gio Mangrove Ecosystem (Publishing House for Science and Technology, 2013).
Zvonareva, S. & Kantor, Y. Checklist of gastropod molluscs in mangroves of Khanh Hoa province, Vietnam. Zootaxa 4162, 401–437 (2016).
Thach, N. N. Shells of Vietnam (Conchbooks, 2005).
Thach, N. N. Recently Collected Shells of Vietnam (L’Informatore Piceno, 2007).
Thach, N. N. New Shells of Southeast Asia. Sea Shells & Land Snails (48HrBooks Company, 2017).
Raven, H. & Vermeulen, J. J. Notes on molluscs from NW Borneo and Singapore. 2. A synopsis of the Ellobiidae (Gastropoda, Pulmonata). Vita Malacol. 4, 29–62 (2007).
Lutaenko, K. A., Prozorova, L. A., Ngo, X. Q. & Bogatov, V. V. First reliable record of Mytilopsis sallei (Récluz, 1849) (Bivalvia: Dreissenidae) in Vietnam. Korean J. Malacol. 35, 355–360 (2019).
Prozorova, L. A. et al. Mangrove mollusk fauna of the Kien Giang Province in the Mekong River delta (South Vietnam). In The 1st International Conference on North East Asia Biodiversity, Vol. 1, 67 (2018a).
Prozorova, L. A. et al. New for the Mekong Delta and Vietnam fauna mollusk families. In The 1st International Conference on North East Asia Biodiversity Vol. 1, 65–66 (2018b).
Prendergast, J. R. et al. Rare species, the coincidence of diversity hotspots and conservation strategies. Nature 365, 335–337 (1993).
Raphael, M. G. & Molina, R. Conservation of Rare or Little-known Species: Biological, Social, and Economic Considerations (Island Press, Washington, D.C., 2013).
Mouillot, D. et al. Rare species support vulnerable functions in high-diversity ecosystems. PLoS Biol. 11, e1001569 (2013).
Golding, R. E. Molecular phylogenetic analysis of mudflat snails (Gastropoda: Euthyneura: Amphiboloidea) supports an Australasian centre of origin. Mol. Phylogenet. Evol. 63, 72–81 (2012).
Thach, N. N. Vietnamese New Mollusks. Seashells-Land Snails-Cephalopods, with 59 New Species. (Thach, N. N., 2016).
Golding, R. E., Ponder, W. F. & Byrne, M. Taxonomy and anatomy of Amphiboloidea (Gastropoda: Heterobranchia: Archaeopulmonata). Zootaxa 1476, 1–50 (2007).
Golding, R. E., Byrne, M. & Ponder, W. F. Novel copulatory structures and reproductive functions in Amphiboloidea (Gastropoda, Heterobranchia, Pulmonata). Invertebr. Biol. 127, 168–180 (2008).
Davis, G. M. Mollusks as indicators of the effects of herbicides on mangroves in South Vietnam. In The Effects of Herbicides in South Vietnam: Part B, Working papers. (ed. National Academy of Sciences, National Research Council) 1–29 (National Academy of Sciences, National Research Council, 1974).
Academy of Natural Sciences. MAL. Occurrence dataset. GBIF. https://doi.org/10.15468/xp1dhx (2019a).
Academy of Natural Sciences. ANSP Malacology Collection. The Academy of Natural Sciences, Philadelphia. https://clade.ansp.org/malacology/collections/index.html (2019b).
Akiba, M. & Sasaki, T. Mollusca specimens of Ryukyu University Museum (Fujukan). Version 1.1. National Museum of Nature and Science, Japan. GBIF. https://doi.org/10.15468/qgmdhb (2019).
Creuwels, J. Naturalis Biodiversity Center (NL) - Mollusca. Naturalis Biodiversity Center. GBIF. https://doi.org/10.15468/yefvnk (2019).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Lanfear, R. et al. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773 (2017).
Tanabe, A. S. Phylogears version 2.2.2012.02.13. 2012. Life is fifthdimension. https://www.fifthdimension.jp/ (2012).
Ronquist, F. & Huelsenbeck, J. P. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Rambaut, A., Drummond, A. J. & Suchard, M. Tracer v1.6. Molecular evolution, phylogenetics and epidemiology. https://tree.bio.ed.ac.uk/software/tracer/ (2013).
Minh, B. Q., Nguyen, M. A. T. & von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195 (2013).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Kuroda, T. Two families new to the Molluscan fauna of Japan. Venus 1, 10–15 (1928).
GADM. GADM database, version 3.4. GDAM maps and data. https://gadm.org/index.html (2018).
QGIS development team. QGIS geographic information system version 2.18, open source geospatial foundation project. QGIS. https://qgis.osgeo.org (2018).
Acknowledgements
We are grateful to Acad. V. Bogatov, Dr. E. Chernova (Far Eastern Branch Russian Academy of Science, Vladivostok) and Mr. T.T. Thai (Vietnam Academy of Science and Technology, Ho Chi Minh City) for supporting field surveys. We also thank Dr. T. Sasaki and Dr. M. Shimabukuro (University Museum Fujukan, University of the Ryukyus, Okinawa), Dr. P. Callomon and Dr. K. Seizova (Academy of Natural Science, Philadelphia), Dr. B. van der Bijl (Naturalis Biodiversity Center, Leiden) for help in the examination of museum specimens, as well as Dr. M. Byrne (University of Sydney, Sydney) for providing the references. Finally, we are grateful to two anonymous reviewers and the Editorial Board Member for valuable comments.
Funding
This study was funded by the Far East Branch, Russian Academy of Sciences Grant Number 17-007 and Vietnam Academy of Science and Technology Grant Number VAST.HTQT.NGA.01/17-18 and Grant-in-Aid for Japan Society for the Promotion of Science (JSPS) Research Fellow Grant Number 16J04692, 19J00392, JSPS KAKENHI Grant Number 18H02506 and 20K15866.
Author information
Authors and Affiliations
Contributions
L.P. and Q.N.X. arranged field survey, and T.S., L.P., and Q.N.X. conducted the field survey. L.P. discovered materials. L.P. and T.S. examined samples, and L.P. described the morphology of gastropods. T.S. conducted all analyses. T.S. wrote the manuscript with major contributions by L.P. and S.C. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Saito, T., Prozorova, L., Quang, N.X. et al. First molecular examination of Vietnamese mudflat snails in the genus Naranjia Golding, Ponder & Byrne, 2007 (Gastropoda: Amphibolidae). Sci Rep 10, 18714 (2020). https://doi.org/10.1038/s41598-020-75200-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-75200-8
This article is cited by
-
Stramonita Genus Exhibits a New Uncovered Species: A Cryptic Species Collected from Accra, Ghana (Eastern Atlantic Ocean)
Thalassas: An International Journal of Marine Sciences (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.