Abstract
Selection pressures exerted on Staphylococcus aureus by host factors during infection may lead to the emergence of regulatory phenotypes better adapted to the infection site. Traits convenient for persistence may be fixed by mutation thus turning these mutants into microevolution endpoints. The feasibility that stable, non-encapsulated S. aureus mutants can regain expression of key virulence factors for survival in the bloodstream was investigated. S. aureus agr mutant HU-14 (IS256 insertion in agrC) from a patient with chronic osteomyelitis was passed through the bloodstream using a bacteriemia mouse model and derivative P3.1 was obtained. Although IS256 remained inserted in agrC, P3.1 regained production of capsular polysaccharide type 5 (CP5) and staphyloxanthin. Furthermore, P3.1 expressed higher levels of asp23/SigB when compared with parental strain HU-14. Strain P3.1 displayed decreased osteoclastogenesis capacity, thus indicating decreased adaptability to bone compared with strain HU-14 and exhibited a trend to be more virulent than parental strain HU-14. Strain P3.1 exhibited the loss of one IS256 copy, which was originally located in the HU-14 noncoding region between dnaG (DNA primase) and rpoD (sigA). This loss may be associated with the observed phenotype change but the mechanism remains unknown. In conclusion, S. aureus organisms that escape the infected bone may recover the expression of key virulence factors through a rapid microevolution pathway involving SigB regulation of key virulence factors.
Similar content being viewed by others
Introduction
Staphylococcus aureus is a transient, sometimes permanent member of the human microbiota in the nares and skin of a significant number of healthy individuals. As predisposing conditions emerge in the host, this opportunistic species may cause infections with different severity, ranging from mild skin and soft tissue infection to severe disseminated disease1,2. Treatment of S. aureus infections is hampered by widespread dissemination of methicillin-resistant S. aureus (MRSA)3 and by the frequent emergence of S. aureus with low level resistance to vancomycin4. Whereas MRSA was primarily considered a nosocomial pathogen5, it is now unanimously accepted that MRSA also affects individuals of the general community with no previous exposure to health care settings6,7. Up to 1.5–2% of patients receiving an orthopedic prosthetic device becomes infected and a significant number of these infections are caused by S. aureus. Most osteomyelitis caused by S. aureus become refractory to antibiotic treatment soon after bacteria settles on the prosthetic device surface and in bone tissue1. S. aureus has the ability to swiftly adapt to the conditions encountered at the infection niche by adjusting its metabolism and/or regulating the expression of virulence factors required for successful establishment at the incipiently colonized tissue. Once S. aureus has adapted to the microenvironment certain traits may be fixed by mutations, which occur as osteomyelitis becomes chronic8,9. Indeed, selection pressure exerted by a vast number of yet undefined host factors would permit the emergence of bacterial variants more suitable to evade immune defense mechanisms and cause infection refractory to antibiotic treatment in the absence of genes that code antibiotic resistance.
Staphylococcus aureus possesses a vast repertoire of virulence and immune evasion factors that facilitates its dual life-style as either a commensal or a pathogen1,10. More important, S. aureus displays a complex regulatory network, composed of a number (as yet not fully known) of genes which allow the crosstalk between regulators11 thus permitting this species to rapidly switch on and off virulence factors to adapt to and survive in changing microenvironments. One of the strategies to investigate which factors may be up- or down-regulated during adaptation is to assess whether any of these factors is fixed by mutation during chronic infection. Among these factors, loss of short sequence–repeats in the protein A Xr region, small colony variant (SCV) emergence and loss of capsular polysaccharide (CP) expression can be mentioned12,13,14,15. Furthermore, we were able to demonstrate that the loss of CP expression due to a mutation in the agr occurs during chronic osteomyelitis9. This finding suggested that loss of RNAIII expression may yet be another endpoint in microevolution since agr regulates the expression of a vast number of virulence factors.
CP5 and CP8 are produced by a 75 to 80% of S. aureus isolates from humans and play a significant role in the pathogenesis of staphylococcal infections16. Isolates of S. aureus that fail to produce CP5 or CP8 and that produce non-mucoid colonies on solid media are defined as non-typeable (NT) regardless the mechanism responsible for the lack of CP expression17. CP5 and CP8 are virulence factors that permit S. aureus to avoid phagocytosis and facilitate bloodstream dissemination18,19. Once established in the bone of the patient with chronic osteomyelitis it appears advantageous for S. aureus to loose CP expression15 to remain undisturbed within the infected bone. To support the hypothesis that loss of CP expression may initially be due to regulation, in a very recent study it was demonstrated that S. aureus can express CP in vivo, even though production in vitro cannot be demonstrated20. These findings showed that S. aureus can switch on or off CP expression according to the in vivo microenvironment surrounding bacteria. In the present study, we investigated whether S. aureus can regain the ability to produce CP, even in the presence of a mutated non-functional Agr, when it is transferred from infected bone to the bloodstream.
Results
Mouse passages of strain HU-14
Strain HU-14 lacks CP5 expression due to an IS256 insertion in agrC. Strain HU-14 was passed through blood using the bacteremia mouse model and the emergence of colonies with positive reaction to anti-CP5 immune serum was monitored by colony immunoblot assay. After passage #3 (Fig. 1a) a colony (#1) reacting to anti-CP5 was pinpointed (Figs. 1b and 1c), which was recorded as strain P3.1. Other suspicious colonies from the same passage were tested for CP expression but only P3.1 was confirmed as CP5 positive (Fig. 1d). The P3.1 colony was amplified and subcultured several times to assess phenotype stability and after 7 passages on TSA the CP5 phenotype remained stable.
(a) Mouse experiment flowchart. Mice were injected by the i.p. route with 1 × 108 CFU of S. aureus HU-14. After 24 h mice were sacrificed and blood obtained by cardiac puncture. Blood was plated quantitatively and after an overnight culture all colonies from a plate were harvested and suspended in PBS, and the suspension adjusted to a density of 1 × 109 per ml for further injection to another group of mice. Suspension aliquots were plated for CFU count and colony immunoblots were performed on the grown TSB plates to detect the emergence of any colony producing CP5. (b) Colony immunoblot of strain HU-14 after three passages through mice. The arrows indicate strains Reynolds CP5, Reynolds CP8 and Reynolds NT. These three control strains were added by impregnation of the blot membrane. (c) Magnification of the photograph shown in (b). The arrow indicate colony #1, later identified as P3.1. (d) P3.1 was confirmed as CP5. Positive control was strain Reynolds CP5 (lower left strike). Negative control was Reynolds NT (lower right strike).
Isolate P3.1 characterization
Phenotypic characterization revealed that strains HU-14 and P3.1 have almost identical mean generation times (MGT) (40 min and 39 min, respectively). Biofilm production was significantly higher (p < 0.0001, Student t test) in the P3.1 derivative [ODB/ODG = 0.60 ± 0.02 (n = 42)] when compared with the parental HU-14 strain [ODB/ODG = 0.35 ± 0.01 (n = 42)], as assessed by the crystal violet test. Total hemolytic activity was negative for both strains as well as for alpha- and beta-hemolysins. Proteolytic activity was negative for both strains too. Both strains were vancomycin susceptible. Transcriptional evaluation of the Agr system in HU-14 and P3.1 revealed that the expression of RNAIII and agrA was still abolished in P3.1 as ascertained through qRT-PCR (Fig. 2a). Amplification of the P3.1 agrC gene produced an amplicon of identical size (2,793 bp) to that observed in HU-14, which suggested that IS256 remained positioned in the agrC gene of isolate P3.1 (Fig. 2b). Sequence analysis revealed that the insertion site of IS256 in agrC was identical in both strains and positioned after base 218. In synthesis, S. aureus regained CP expression in the absence of a functional Agr system (lack of RNAIII and agrA expression). Another noticeable phenotypic feature was that P3.1 produced the classical yellowish-golden colonies that give the name to this bacterial species whereas its parental HU-14 strain produced whitish colonies on TSA (Fig. 3a). Methanolic extracts from HU-14 exhibited a significantly lower absorbance at 450 nm when compared with those of P3.1, which suggested that P3.1 produced increased levels of staphyloxanthin (Fig. 3b).
(a) Quantitative real time PCR of RNAIII and agrA transcripts from strains HU-14 and derivative P3.1. S. aureus strain 6,850 was used as reference. Changes in gene expression are shown as normalized mean fold change 2-ΔCt. Data were normalized to 16S expression. The data represent the mean of duplicate measurements from 3 independent experiments. (b) Electrophoretic run of agrC PCR amplicons. HU-14 and P3.1 rendered amplicons of identical size, larger than that obtained from control strain HU-8 with a conserved agrC. Lane (-) contains no DNA. The original gel image is shown in Suppl. Figure 3. The result obtained shows that the IS256 insertion remains in place within agrC in derivative P.3.1.
(a) Strain HU-14 and P3.1 colonies on TSA showing the different pigmentation level. Panel b: pigments were extracted S. aureus strains with methanol and the optical density ratios at 450 nm/600 nm were measured. Sample size in each column varied from 4 to 18. The P3.1 methanolic extract produced a significantly higher absorbance when compared with the parental HU-14 strain. (*) p = 0.0121, Student t test for unpaired samples. S. aureus RN6911 does not produce staphyloxanthin and SH1000 is a heavy staphyloxanthin producer strain.
Whole genome sequence analysis
Analysis using Illumina short read sequences confirmed that HU-14 and P3.1 were isogenic, with only one single nucleotide polymorphism (SNP) in P3.1. It consisted of replacement of C by T in position 589 of the icaA gene, leading to a change from alanine to threonine in position 197. Phenotypical analysis of the icaADBC product, namely the polysaccharide intercellular adhesin (PIA) revealed negligible PIA production with no significant differences between HU-14 and P3.1 (Fig. 4). In addition to agrC, which was mutated by the IS256 insertion in both strains, further short read sequence analysis determined that no genomic modification of any other known major regulator occurred which may explain the P3.1 capsulated phenotype. Furthermore, no SNP was found in the intergenic regions which may affect the expression of any regulator.
PIA in biofilms produced by S. aureus HU-14 and P3.1. Strains SA113 and MBD034 were included as positive standard and negative control, respectively. Each bar represents the arithmetic mean ± SEM from 4 to 6 wells from 3 separate experiments. PIA production values are the OD at 595 nm of crystal violet (ODB) relative to the final culture density (ODG) after 24 h incubation. There was no significant difference between HU-14 and P3.1 (ordinary one-way ANOVA, Tukey´s test, Brown-Forsythe post-test).
Pacific Biosciences sequencing was performed to assess whether there was any structural difference between the HU-14 and P3.1 genomes. Hybrid Unicycler assembly of PacBio and Illumina reads resulted in two genome fragments. The smaller fragment in the HU-14 and P3.1 hybrid Unicycler assemblies was a circular sequence that shared 99.98% and 99.80% identity respectively with plasmid pCM05 (PLSDB database). Annotation with Prokka using the pCM05 plasmid from the NCBI GenBank Database (NC_013332.1) as a reference and SnapGene side-by-side analysis with the pCM05 GenBank file confirmed that the circular sequence was indeed pCM05 in both assemblies, and contained all the same genes and restriction sites. The plasmid sequences were also aligned using ProgressiveMaude, but these did not show any rearrangements.
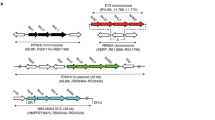
The size of the larger fragment (2,972,382 bp and 2,971,047 bp in HU-14 and P3.1, respectively) was consistent with that of the S. aureus chromosome. Whole genome alignment of the chromosome sequences was performed using progressiveMauve, showing no chromosomal rearrangements between the two. However, annotation with Prokka revealed that P3.1 has lost one IS256 copy originally located in the non-coding region downstream of DNA primase (dnaG, LJDFIFNA_00432) and upstream of RNA polymerase sigma factor RpoD (rpoD, LJDFIFNA_00434) (sigA) (Suppl Fig. 1). IS256 excision in P3.1 was clean and left a conserved region between dnaG and rpoD identical to those of N315, USA300 and other ST5 isolates from our collection.
Interestingly, when both hybrid assembly and only short read analysis was performed the IS256 was definitively missing in P3.1 but such excision was not pinpointed by performing long read analysis only. Finally, SNP-calling between the two closed hybrid genomes was carried out using Snippy, with the Prokka-annotated HU-14 closed reference genome and the P3.1 Illumina reads, and vice-versa. This revealed the same single SNP as the same analysis using just the Illumina reads: an inconsequential change from alanine in HU-14 to threonine in P3.1, as described above.
Regulation of CP5 expression
It is known that regulation of CP5(8) expression in S. aureus is complex and involves a number of redundant mechanisms that act in concert. Evaluation of main S. aureus regulators sae and sarA at the transcriptional level by qRT-PCR revealed that no functional modification occurred (data not shown). However, transcription of asp23, a gene that is solely regulated by SigB, was significantly increased in P3.1 when compared with HU-14 (Fig. 5). Since staphyloxanthin production by S. aureus is positively regulated by SigB, the increased pigmentation displayed by P3.1 supports the hypothesis that S. aureus P3.1 reacquired the capacity to produce CP5 through a mechanism related to recovery of SigB functionality, in the absence of an operative Agr system.
Quantitative real time PCR of asp23 and sigB transcripts from strains HU-14 and derivative P3.1. S. aureus strain 6850 was used as reference. Changes in gene expression are shown as normalized mean fold change 2-ΔCt. Data were normalized to 16S expression. The data represent the median of duplicate measurements from 3 independent experiments. (*) Significant difference with p = 0.0070 (Mann–Whitney test).
Virulence study
The virulence of strain HU-14 and derivative P3.1 was tested in a mouse model of intraperitoneal injection. Mortality 5 days after challenge with P3.1 was higher than that in the group challenged with HU-14 but was not significantly different (Suppl Fig. 2a). Body weight was recorded over 5 days after challenge and the results showed that mice challenged with the P3.1 derivative lost more weight than those injected with the parental strain HU-14, but the differences were again not significant (Suppl Fig. 2b). Interestingly, mice challenged with P3.1 exhibited more prominent signs of distress, such as hunched posture and spiky fur, when compared with those challenged with HU-14.
Osteoclastogenesis and cytokine production studies
The differential capacity of strains HU-14 and P3.1 to trigger osteoclastogenesis was investigated in vitro. Osteoclast precursors were stimulated with S. aureus in the presence of M-CSF. Forty-eight hours after stimulation cells were fixed and multinucleated TRAP ( +) cells were counted. Rapid differentiation into osteoclasts was induced by HU-14 with formation of multinucleated cells (3 or more nuclei) (Fig. 6a,b). In contrast, significantly decreased numbers of osteoclasts were observed when the osteoclast precursors were stimulated with P3.1 (Fig. 6a,b). S. aureus HU-14 and its P3.1 derivative were tested for their capacity to induce the production of TNF-α, a proinflammatory cytokine involved in osteoclastogenesis, by osteoclast precursors in culture. The P3.1 derivative exhibited a decreased capacity to trigger TNFα secretion (Fig. 6c).
(a,b) RAW 264.7 cells were stimulated with the S. aureus HU-14 isolate or its derivative P3.1 (Heat-killed bacteria, 107 CFU/ml) for 48 h. Media alone was used as negative control. (a) Photographs at 20 × magnification. (b) The number of mature osteoclasts was quantified by light microscopy using TRAP staining. TRAP + cells with 3 or more nuclei were considered mature osteoclasts. The mean and standard error of cumulative data from 3 independent experiments (6 wells) is shown. Levels of significance were obtained by ordinary one-way ANOVA (Tukey´s multi comparisons test, Brown-Forsythe post-test). (c) TNF-α production was quantified by ELISA in the culture media. The mean and standard error of cumulative data from 3 independent experiments (6 wells) is shown. Levels of significance were obtained by ordinary one-way ANOVA (Tukey’s multi comparisons test, Brown-Forsythe post-test).
Discussion
Staphylococcus aureus has the potential to adapt to diverse niches with defined microenvironments21. In a recent study we showed that a stable non-encapsulated derivative emerged during chronic osteomyelitis and displaced a more virulent CP8 parental strain which had been isolated several months before from the same infection site9 . The S. aureus derivative, which bore a frameshift mutation in agrC that caused loss of both RNAIII transcription and CP8 expression, was considered a microevolution endpoint adapted to bone. Lack of CP expression benefits the interaction between surface S. aureus adhesins and eukaryotic cell receptors thus promoting internalization of bacteria into the eukaryotic cell milieu8. Intracellular location, reduced capacity to cause inflammation13,22, increased biofilm production and SCV formation23 may explain why not only S. aureus pulmonary infection of cystic fibrosis patients24 but also S. aureus osteomyelitis become refractory to antibiotic treatment in spite of appropriate antibiotic treatment. From our bacterial collection we selected non-encapsulated S. aureus isolate HU-14 with a non-functional agr due to an IS256 insertion in agrC. Although HU-14 may be considered a microevolution endpoint, in the present study we show that the passage of this isolate through blood in an experimental mouse model induced the reversion from a NT to a CP5 phenotype. Interestingly, it was not a true genetic reversion since IS256 remained inserted at the same location in the agrC of the CP5 isogenic derivative. Therefore, loss of the agr functionality due to a stable mutation (in this case caused by an IS256 insertion) may only be considered a microevolution endpoint at the precise niche where selection initially took place.
A variable number of relevant S. aureus isolates are non-reactive to antibodies anti-CP5 and anti-CP8 as phenotypically ascertained in vitro in the clinical bacteriology laboratory. Diverse mechanisms responsible for the loss CP5(8) expression have been reported, one of which is related to dysfunction of the main global regulator Agr17. There is ample evidence that agr-deficient strains may not only be selected during infection25 but also colonize healthy hosts26. In a study on 195 S. aureus isolates collected from different sources it was found that 49% of the isolates carried an inactive agr locus, as shown by the lack of a RNAIII transcript27. In the same study, interestingly, 55% of the strains with a non-functional agr produced capsule (CP5 or CP8). Such finding permits to speculate that the type of S. aureus derivative obtained after passage through the bloodstream described in the present report, which produce CP5 in the absence of a functional agr, may not be a rare event in clinical practice after all. CP5(8) is a strong candidate for construction of a multicomponent vaccine to prevent S. aureus severe infections28. Indeed, the most promising preparations contain CP conjugated to a toxoid plus other relevant antigens16,29,30,31. The finding that prevalent clone USA300 in the USA is non-encapsulated due to mutations in the capsular genes rather than in a capsule regulator32 appeared to cast doubt on the inclusion of CP as vaccine component. But on the other hand, many S. aureus clinical isolates carry a conserved cap cluster and, furthermore, it was recently demonstrated that clinical strains that do not produce CP in vitro are able to produce CP in vivo20. Our present study demonstrates the feasibility that S. aureus may recover in a stable manner CP5 production, which facilitates phagocytosis evasion once bacteria reach the bloodstream, in spite of a non-functional Agr. The results of our study suggest that the inclusion of CP in a multicomponent vaccine preparation should not be dismissed.
Staphylococcus aureus possesses an intricate regulatory system that governs the production of soluble virulence factors and exoenzymes. In addition to Agr, main regulators include a number of two-component systems, such as SaeRS, transcriptional factors33, such as SarA and its homologues34 and factor σB (sigB)35. Bioinformatic analysis of strains HU-14 and P3.1 sequences revealed that the sequences of the aforementioned regulators were conserved, with no differences between the parental strain and the derivative. Since it was recently demonstrated that CP production is up regulated by SigB we focused our attention on this regulator36. The SigB regulon is known to respond to different stress signals and accordingly regulates stress responses33. One of the marker genes upregulated by SigB is asp2337. An increase of SigB activity in P3.1 was suspected by noteworthy pigmentation of P3.1 colonies. Indeed, another known SigB-regulated trait is production of staphyloxanthin38. Further assessment revealed that P3.1 produced pigments whereas the parental strain HU-14 did not, as determined spectrophotometrically. In the present report, the significant increase in asp23 expression revealed that the increase in SigB functionality was associated with the recovery of CP5 production. It is hypothesized that adaptation of S. aureus through a long-term process in the host may have led to reduced SigB expression and perhaps loss of the Agr function seen in HU-14. Experimental passage of this isolate through the bloodstream induced reacquisition of CP expression with parallel increase of SigB expression, as ascertained by increased asp23 expression. In favor of this hypothesis, Marbach and coworkers39 has found that S. aureus infecting the bovine udder may not require sigB expression once it is established in the mammary gland of the cow with chronic mastitis. Interestingly, as we hypothesized before, loss of SigB expression in the infected cow was eventually fixed by mutation8. In this case the mutated gene was rsbU39.
Whole genome sequence analysis from reads obtained by Illumina sequencing did not reveal any genetic lesion directly or indirectly responsible for the observed SigB increased expression. Therefore, the presence of structural changes in P3.1 was then investigated by long-read sequencing. Hybrid (PacBio and Illumina) sequence analysis of the HU-14 and P3.1 chromosomes as well as of plasmid pCM05 suggested a structural change in P3.1 compared with HU-14, which consisted of an IS256 excision located between dnaG (DNA primase) and rpoD (sigA). This preliminary observation was not surprising since mobile genetic elements are key drivers of evolution in S. aureus40. Giulieri and coworkers41 have indeed demonstrated that insertion of IS256 elements enhances genetic diversity during infection thus representing an effective driver of within-host microevolution. Furthermore, insertion of IS256 in agrC was found to promote increased fitness of S. aureus as a compensatory mechanism for the biological cost resulting from acquisition of a high number of multiresistant traits by hospital isolates42. In this regard, it is speculated that reacquisition of SigB activity and CP5 as well as staphyloxanthin production was perhaps achieved by restoration of SigA activity since IS256 was excised without affecting the original conserved intergenic non-coding region between dnaG (DNA primase) and rpoD (sigA). In support of this hypothesis, it has been shown that the sigB operon is transcribed from at least two differentially controlled promoters, one of which is a putative σA-dependent promoter termed sigBP143. Therefore, sigB expression is driven by SigA, which leads to the transcription of the whole operon, as well as through autoregulation by SigB itself, leading to transcription of rsbV and rsbW33. Further research is required to better understand the complex mechanisms that permit concomitant reacquisition of CP5 and pigment production by S. aureus in the presence of an inactive agr and a functional sigB operon, which is known to be driven by SigA.
Altman and coworkers have shown that within-host loss of agr function was associated with increased genetic divergence between distinct S. aureus subclones44. In synthesis, whereas agr dysfunction is adaptive for survival in the infected bone, it may be counter adaptive outside such niche. As observed before, agr mutations do not tend persist in natural S. aureus populations45. In the present report we show that S. aureus can regain CP production when subjected to selection pressure in vivo through a mechanism that does not require RNAIII expression. Indeed, in a previous study it has been shown that genetic changes occurring in an agr-defective infecting S. aureus strain resulted in increased virulence in a murine model of bloodstream infection thus bypassing the agr mutation44. The S. aureus derivative obtained after passage through the bloodstream displayed a significant reduction in the capacity to trigger TNFα production and became less adapted to bone as ascertained by its decreased capacity to induce osteoclastogenesis. The S. aureus strain adapted to blood not only conserved but increased its capacity to produce biofilm. Recovery of at least two important traits such as CP5 and pigment production permit bacteria to evade innate immune mechanisms of defense in the blood46,47,48,49 and readapt to a new environment elsewhere in the host with the potential to cause methastatic infection. The fact that S. aureus can adapt to blood by regaining the capacity to produce CP5 and pigments does not necessarily mean that virulence should increase. P3.1 exhibited a trend to be more virulent, as ascertained in the mouse model of intraperitoneal challenge, but the differences were not significant. This was not a surprising fact since the Agr remained non-functional and many other Agr-regulated virulence factors are not being expressed, such as the hemolysins. Therefore, as much as S. aureus is a multifactorial pathogen, it is speculated that its adaptation to a given niche in the host is most probably multifactorial too. In the present study, modification of virulence factor expression due to increased SigB activity was enough to reduce adaptation to bone but not to increase virulence in a significant fashion.
Conclusion
Here we demonstrated that S. aureus adapted to infected bone can regain the ability to produce CP, even in the presence of a mutated, non-functional Agr when transferred to the bloodstream. S. aureus organisms that escape the infected bone can potentially become a threat because they can recover in a stable manner the expression of traits that permit survival in blood, such as CP and staphyloxanthin. Therefore, a supposedly harmless endpoint-of-microevolution may turn back into a dangerous infective derivative. Given that CP is one of the factors identified in this microevolution process, our findings support the idea that S. aureus CP should not be dismissed as a multifactorial vaccine candidate component.
Materials and methods
Bacterial strains and cultures
Staphylococcus aureus clinical isolate HU-14 from our strain collection was originally recovered in 2005 from a 34-year old male patient with chronic osteomyelitis of the right femur with a prosthetic implant (“Hospital Ramón Carrillo”, Buenos Aires, Argentina). Species was confirmed by a species-specific PCR50. Strain HU-14 is ST5, CC5, SCCmec type 1, Agr type II and Spa t14951. All strains utilized in the study were kept frozen in trypticase soy broth (TSB) with 20% glycerol at − 80 °C and S. aureus was routinely cultured at 37 °C for 24 h on TSB unless otherwise indicated. In addition, S. aureus strain 6850 was used as a qRT-PCR positive control for Agr and SigB functionality testing. S. aureus strains RN6911 and SH1000 were utilized as negative and positive controls, respectively, for staphyloxanthin production. S. aureus strains Sa113 and MBD034 were utilized as positive and negative controls, respectively, for PIA production. Strain HU-8 (CC5, SCCmec type 1, Agr type II, Spa t149 and produces CP5), which bears a functional agrC, was also utilized as control.
Mouse models
CF1 outbred mice were bred and maintained in the vivarium of the “Instituto de Investigaciones en Microbiología y Parasitología Médica” (IMPaM-Universidad de Buenos Aires-CONICET, Buenos Aires, Argentina). Animal care was in accordance with the recommendations of the guidelines set forth by the 11th report of the BVAAWF/FRAME/RSPCA/UFAW Joint Working Group on Refinement52. The animal research protocols utilized in this study were approved by the “Comité Institucional para el Uso y Cuidado de los Animales de Laboratorio,” through resolutions No. 885/19 issued on June 5, 2019 and No 2780/18 issued on February 5, 2019 by the “Consejo Directivo de la Facultad de Medicina, Universidad de Buenos Aires,” Argentina. The experimental scheme of the mouse bacteriemia model is shown in Fig. 1. Passage of S. aureus strain HU-14 through blood was performed on groups of 4 male mice in each cycle. An inoculum of 1 × 108 CFU in 100 µl of physiologic saline solution (PSS) was injected in mice by the ip route. After 24 h mice were sacrificed by exposure to CO2 and blood was drawn by cardiac puncture and quantitatively plated on trypticase soy agar (TSA). Plates were incubated for 24 h at 37 °C and colonies were suspended PSS to ca. 1 × 109 CFU to prepare a 108 CFU inoculum to be re-injected in the next group of mice. This cycle was repeated seven times. To assess the differential virulence of the strains under investigation mortality curves were compared. Groups of 10 CF1 outbred mice were injected with 1 × 108 CFU of the strain to be tested suspended in 0.5 ml of 2% (w/v) Brewer´s yeast (Sigma Chemical Co.) in TSB broth. Mortality and body weight were assessed daily for 5 days. Surviving mice were euthanized by cervical dislocation. The Kaplan–Meier mortality curve was obtained using the Graph-Pad Prism software (GraphPad Software, Inc., La Jolla, USA; version 6.00).
Illumina whole genome sequencing and analysis
DNA was extracted directly from plated microbial cultures using the Nextera DNA Flex Microbial Colony Extraction protocol (Illumina, San Diego, CA, USA). Library preparation was performed according to the Nextera DNA Flex Library Prep Kit using Nextera DNA 24 CD Indexes (Illumina, San Diego, CA, USA). Paired-end whole genome sequencing was performed in a MiSeq instrument (Illumina, San Diego, CA, USA) using the MiSeq Reagent kit v2 150 bp. The resulting fastq files were analyzed with the FastQC High Throughput Sequence QC Report Version 0.11.7 (https://www.bioinformatics.babraham.ac.uk/projects/). Only isolates that fulfilled the quality parameters of the FastQC software (e.g. per base sequence quality, per tile sequence quality, per sequence GC content) were used for analysis. The FastQ files were assembled using SPAdes v3.10.153. The genome was annotated using Prokka v1.14.554 and variant calling was performed using Snippy v3.2 (https://github.com/tseemann/snippy). The sequences analyzed in this study have been deposited in the European Nucleotide Archive (ENA) under Bioproject PRJEB35732.
Pacific biosciences genome sequencing and analysis
High molecular weight (HMW) gDNA was extracted from HU-14 and P3.1. Bacteria were grown overnight on TSA plates, then gDNA was extracted using Qiagen’s MagAttract HMW DNA Kit (Qiagen, Manchester, UK) according to the manufacturer’s instructions for Gram Positive species, using 20 µl of lysostaphin (10 mg/ml) instead of lysozyme. The extracted HMW gDNA was sent to the Centre for Genomic Research at the University of Liverpool, UK, where long-read sequencing and base modification analysis were conducted on the Pacific Biosciences (Pacific Biosciences, Menlo Park, CA, USA) Sequel system. The raw subreads for each sample were filtered to 200 × coverage of the S. aureus genome using Filtlong v0.2.0 (available at https://github.com/rrwick/Filtlong) and then corrected using Canu v1.955. Hybrid de novo assemblies were produced for both samples by Unicycler v0.4.856, using the PacBio long reads and Illumina short reads. In hybrid mode, Unicycler uses SPAdes v3.14.053 to assemble the short reads, then bridges the SPAdes contigs and closes the genome using the long reads with minimap and miniasm (built-in Unicycler v0.4.8 versions)57, followed by long-read polishing with Racon v1.4.1158 and short-read polishing with Pilon v1.2259. The completeness of the closed genome sequences was assessed using BUSCO v4.0.260. The “bacteria” database was used to search the genomes for 120 universal bacterial genes, and the more specific “Bacillales” database was used to search for 450 genes common to all Bacillales species. Prokka v1.14.554 was used to annotate the closed genome sequences, using the protein sequences from the S. aureus strain N315 (NC_00275.2) as a reference. The annotated GenBank format files were then used to call SNPs between the two samples with Snippy v4.5.1 (https://github.com/tseemann/snippy); the HU-14 GenBank file was used as the reference with the P3.1 Illumina reads, and vice-versa. The plasmid sequence for each sample was compared to the PLSDB plasmid database v.2019_10_0761 (11), and annotated with Prokka (1.14.5)54 to confirm its identity. The closed chromosome and plasmid sequences from each sample were rotated to start with the same genes (dnaA for the chromosome, repA for the plasmid) and aligned using progressiveMauve (build version: Sep 16 2015). The output files were inspected visually for any rearrangements using the progressiveMauve GUI.
Phenotypic studies
CP5 production was evaluated by colony immunoblot on TSA plates as described previously62. Phenotypic expression of α- and β-hemolysin was performed by evaluating the production of the hemolysis in rabbit and goat blood agar (α- and β-hemolysin, respectively). Proteolytic activity was assessed in TSA supplemented with 10% whole milk. After 18 h incubation at 37 °C the hemolytic and proteolytic halos were evaluated, respectively. Ethanolic extracts of the carotenoid pigment from bacterial suspensions with equal optical density (OD) at 600 nm were quantified spectrophotometrically at 450 nm63. The quantitative assessment of biofilm formation was performed as previously described with modifications64. Briefly, S. aureus strains were grown for 18 h and diluted 1:100 in TSB supplemented with 0.25% of glucose (TSBg). After 24 h of static incubation at 37 °C in 96-well microtiter plates, bacterial growth in each well was measured by optical density reading at 595 nm (ODG) using a microplate reader (Multiskan EX). Then, the wells were washed two times with PBS, the biofilms were fixed with 100% methanol for 15 min, stained with 0.5% crystal violet for 20 min, and washed twice again gently under running tap water. The amount of biofilm biomass was measured after addition of 30% glacial acetic acid by OD reading at 595 nm (ODB). The biofilm biomass was expressed relative to the final cell density measured prior to crystal violet staining (biofilm ODB/ODG). Three independent experiments were performed in sixtuplicate measurements. PIA production was assessed by ELISA according to a protocol described elsewhere64. Briefly, the procedure was similar to that described above for biofilm except that, after blocking solution removal,100 μl per well was added of a 75 ng/ml wheat germ agglutinin (WGA)-HRP conjugate, a lectin that binds to PIA sugar residues.
Real-time quantitative reverse transcription PCR (qRT-PCR)
Bacterial RNA was extracted from S. aureus grown in TSB until post-exponential phase, using TRIzol Reagent (Invitrogen Life Technologies), according to the manufacturer’s protocol. RNA was subjected to DNAse treatment using a RQ1 RNAse free DNAse (Promega). cDNA synthesis was performed with an ImProm-II Reverse Transcriptase kit (Promega). qRT-PCR for RNAIII, agrA and asp23 expression were performed using the primers and conditions described in a previous publication9 and the SYBR Green PCR Master Mix (Applied Biosystems) equipment and kits. The 16S gene was used to normalize data. The (− ΔCt) value represents the difference in threshold cycle (Ct) between the target and control(16S) genes65.
Osteoclastogenesis assay and TNFα detection
To determine the differential capacity of strains HU-14 and P3.1 to trigger osteoclastogenesis RAW 264.7 cells were plated on coverslips (2,5 × 104 cells/well, 24 well plate) in the presence of M-CSF 30 ng/ml (MACS Miltenyi Biotec) and 107 CFU/ml or each S. aureus strain or media alone. At 48 h after stimulation cells were fixed in 4% paraformaldehyde and stained for Tartrate-Resistant Acid Phosphatase (TRAP) (Sigma-Aldrich, St. Louis, MO, USA). Multinucleated (more than three nuclei), TRAP-positive cells were defined as osteoclasts and counted66. TNFα concentrations were determined quantitatively in culture supernatants collected at 24 h after stimulation by ELISA using specific antibody pairs (Beckton-Dickinson) as described previously67.
Statistical considerations
Multiple data statistical comparison was performed by ordinary one-way ANOVA using the Tukey´s multi comparisons test and the Bartlett-Forsythe post-test. Statistical comparison of two data groups with normal distribution was performed with the Student t test for unpaired samples. Statistical comparison of two data groups with non-normal distribution was performed with the Mann–Whitney test. P < 0.05 was considered statistically significant. Graph-Pad Prism software (GraphPad Software, Inc., La Jolla, USA; version 6.00) was used for all statistical analyses.
Data availability
The sequences analyzed in this report have been deposited in the European Nucleotide Archive (ENA) under Bioproject PRJEB35732. Other datasets generated and/or analyzed during the current study are available from the corresponding author upon request.
References
Tong, S. Y. C. C., Davis, J. S., Eichenberger, E., Holland, T. L. & Fowler, V. G. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661 (2015).
Goetghebeur, M., Landry, P. A., Han, D. & Vicente, C. Methicillin-resistant Staphylococcus aureus: A public health issue with economic consequences. Can. J. Infect. Dis. Med. Microbiol. 18, 27–34 (2007).
Prestinaci, F., Pezzotti, P. & Pantosti, A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog. Glob. Health 109, 309–318 (2015).
Howden, B. P. et al. Different bacterial gene expression patterns and attenuated host immune responses are associated with the evolution of low-level vancomycin resistance during persistent methicillin-resistant Staphylococcus aureus bacteraemia. BMC Microbiol. 8, 39. https://doi.org/10.1186/1471-2180-8-39 (2008).
Kallen, A. J. et al. Health care-associated invasive MRSA infections, 2005–2008. JAMA 304, 641–648 (2010).
David, M. Z. & Daum, R. S. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 23, 616–687 (2010).
Fernandez, S. et al. High virulence of methicillin resistant Staphylococcus aureus ST30-SCCmecIVc-spat019, the dominant community-associated clone in Argentina. Int. J. Med. Microbiol. 307, 191–199 (2017).
Tuchscherr, L., Loffler, B., Buzzola, F. R. & Sordelli, D. O. Staphylococcus aureus adaptation to the host and persistence: role of loss of capsular polysaccharide expression. Future Microbiol. 5, 1823–1832 (2010).
Suligoy, C. M. et al. Mutation of Agr Is associated with the adaptation of Staphylococcus aureus to the host during chronic osteomyelitis. Front. Cell. Infect. Microbiol. 8, 18. https://doi.org/10.3389/fcimb.2018.00018 (2018).
Vandenesch, F., Lina, G. & Henry, T. Staphylococcus aureus hemolysins, bi-component leukocidins, and cytolytic peptides: a redundant arsenal of membrane-damaging virulence factors? 2, 12. https://doi.org/10.3389/fcimb.2012.00012 (2012).
Tuchscherr, L. & Loffler, B. Staphylococcus aureus dynamically adapts global regulators and virulence factor expression in the course from acute to chronic infection. Curr. Genet. 62, 15–17 (2016).
Proctor, R. A. et al. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4, 295–305 (2006).
Garofalo, A. et al. The length of the Staphylococcus aureus protein a polymorphic region regulates inflammation: Impact on acute and chronic infection. J. Infect. Dis. 206, 81–90 (2012).
Das, S. et al. Natural mutations in a Staphylococcus aureus virulence regulator attenuate cytotoxicity but permit bacteremia and abscess formation. Proc. Natl. Acad. Sci. 113, E3101–E3110 (2016).
Lattar, S. M. et al. Capsule expression and genotypic differences among Staphylococcus aureus isolates from patients with chronic or acute osteomyelitis. Infect. Immun. 77, 1968–1975 (2009).
Riordan, K. O. & Lee, J. C. Staphylococcus aureus capsular polysaccharides. Clin. Microbiol. Rev. 17, 218–234 (2004).
Cocchiaro, J. L. et al. Molecular characterization of the capsule locus from non-typeable Staphylococcus aureus. Mol. Microbiol. 59, 948–960 (2006).
Nanra, J. S. et al. Capsular polysaccharides are an important immune evasion mechanism for Staphylococcus aureus. Hum. Vaccin. Immunother. 9, 480–487 (2013).
Thakker, M., Park, J. S., Carey, V. & Lee, J. C. Staphylococcus aureus serotype 5 capsular polysaccharide is antiphagocytic and enhances bacterial virulence in a murine bacteremia model. Infect. Immun. 66, 5183–5189 (1998).
Mohamed, N. et al. Molecular epidemiology and expression of capsular polysaccharides in Staphylococcus aureus clinical isolates in the United States. PLoS ONE 14, e0208356. https://doi.org/10.1371/journal.pone.0208356 (2019).
Dastgheyb, S. S. & Otto, M. Staphylococcal adaptation to diverse physiologic niches: an overview of transcriptomic and phenotypic changes in different biological environments. Future Microbiol. 10, 1981–1995 (2015).
Tan, X. et al. Chronic Staphylococcus aureus lung infection correlates with proteogenomic and metabolic adaptations leading to an increased intracellular persistence. Clin. Infect. Dis. 69, 1937–1945 (2019).
Tuchscherr, L. et al. Clinical S. aureus isolates vary in their virulence to promote adaptation to the host. Toxins (Basel) 11, 1. https://doi.org/10.3390/toxins11030135 (2019).
Kahl, B. C. et al. Population dynamics of persistent Staphylococcus aureus isolated from the airways of cystic fibrosis patients during a 6-year prospective study. J. Clin. Microbiol. 41, 4424–4427 (2003).
Traber, K. E. et al. agr function in clinical Staphylococcus aureus isolates. Microbiology 154, 2265–2274. https://doi.org/10.1099/mic.0.2007/011874-0 (2008).
Shopsin, B. et al. Prevalence of agr dysfunction among colonizing Staphylococcus aureus strains. J. Infect. Dis. 198, 1171–1174 (2008).
Fischer, J., Lee, J. C., Peters, G. & Kahl, B. C. Acapsular clinical Staphylococcus aureus isolates lack agr function. Clin. Microbiol. Infect. 20, O414–O417 (2014).
Proctor, R. A. Recent developments for Staphylococcus aureus vaccines: clinical and basic science challenges. Eur. Cell. Mater. 30, 315–326 (2015).
Anderson, A. S. et al. Development of a multicomponent Staphylococcus aureus vaccine designed to counter multiple bacterial virulence factors. Hum. Vaccin. Immunother. 8, 1585–1594 (2012).
Lattar, S. M. et al. Protein antigens increase the protective efficacy of a capsule-based vaccine against Staphylococcus aureus in a rat model of osteomyelitis. Infect. Immun. 82, 83–91 (2014).
Park, S., Gerber, S. & Lee, J. C. Antibodies to Staphylococcus aureus serotype 8 capsular polysaccharide react with and protect against serotype 5 and 8 Isolates. Infect. Immun. 82, 5049–5055 (2014).
Boyle-Vavra, S. et al. USA300 and USA500 clonal lineages of Staphylococcus aureus do not produce capsular polysaccharide due to conserved mutations in the cap5 locus. mBio 6, e02585-e2614. https://doi.org/10.1128/mBio.02585-14.1-10 (2015).
Jenul, C. & Horswill, A. R. Regulation of Staphylococcus aureus virulence. Microbiol. Spectr. 6, 1. https://doi.org/10.1128/microbiolspec.GPP3-0031-2018 (2018).
Ballal, A., Ray, B. & Manna, A. C. sarZ, a sarA family gene, is transcriptionally activated by MgrA and is involved in the regulation of genes encoding exoproteins in Staphylococcus aureus. J. Bacteriol. 191, 1656–1665 (2009).
Moisan, H. et al. Transcription of virulence factors in Staphylococcus aureus small-colony variants isolated from cystic fibrosis patients is influenced by SigB. J. Bacteriol. 188, 64–76 (2006).
Keinhörster, D. et al. Revisiting the regulation of the capsular polysaccharide biosynthesis gene cluster in Staphylococcus aureus. Mol. Microbiol. 112, 1083–1099 (2019).
Muller, M. et al. Deletion of membrane-associated Asp23 leads to upregulation of cell wall stress genes in Staphylococcus aureus. Mol. Microbiol. 93, 1259–1268 (2014).
Liu, H. et al. A novel SigB(Q225P) mutation in Staphylococcus aureus retains virulence but promotes biofilm formation. Emerg. Microbes Infect. 7, 72. https://doi.org/10.1038/s41426-018-0078-1 (2018).
Marbach, H. et al. Within-host evolution of bovine Staphylococcus aureus selects for a SigB-deficient pathotype characterized by reduced virulence but enhanced proteolytic activity and biofilm formation. Sci. Rep. 9, 13479. https://doi.org/10.1038/s41598-019-49981-6 (2019).
Malachowa, N. & DeLeo, F. R. Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 67, 3057–3071 (2010).
Giulieri, S. G. et al. Genomic exploration of sequential clinical isolates reveals a distinctive molecular signature of persistent Staphylococcus aureus bacteraemia. Genome Med. 10, 65. https://doi.org/10.1186/s13073-018-0574-x (2018).
Botelho, A. M. N. et al. Complete genome sequence of the MRSA isolate HC1335 from ST239 lineage displaying a truncated AgrC histidine kinase receptor. Genome Biol. Evol. 8, 3187–3192 (2016).
Senn, M. M. et al. Molecular analysis and organization of the sigmaB operon in Staphylococcus aureus. J. Bacteriol. 187, 8006–8019 (2005).
Altman, D. R. et al. Genome plasticity of agr-defective Staphylococcus aureus during clinical infection. Infect. Immun. 86, e00331-e418. https://doi.org/10.1128/IAI.00331-18 (2018).
Shopsin, B. et al. Mutations in agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 202, 1593–1599 (2010).
Clauditz, A., Resch, A., Wieland, K. P., Peschel, A. & Götz, F. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect. Immun. 74, 4950–4953 (2006).
Hall, J. W., Yang, J., Guo, H. & Ji, Y. The Staphylococcus aureus AirSR two-component system mediates reactive oxygen species resistance via transcriptional regulation of staphyloxanthin production. Infect. Immun. 85, e00838-e916. https://doi.org/10.1128/IAI.00838-16 (2017).
Liu, G. Y. et al. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J. Exp. Med. 202, 209–215 (2005).
Mishra, N. N. et al. Carotenoid-related alteration of cell membrane fluidity impacts Staphylococcus aureus susceptibility to host defense peptides. Antimicrob. Agents Chemother. 55, 526–531 (2011).
Martineau, F. et al. Species-specific and ubiquitous-DNA-based assays for rapid identification of Staphylococcus aureus. J. Clin. Microbiol. 36, 618–623 (1998).
Lattar, S. M. et al. Molecular fingerprinting of Staphylococcus aureus isolated from patients with osteomyelitis in Argentina and clonal distribution of the cap5(8) genes and of other selected virulence genes. Eur. J. Clin. Microbiol. Infect. Dis. 31, 2559–2566 (2012).
Hawkins, P. et al. A guide to defining and implementing protocols for the welfare assessment of laboratory animals: eleventh report of the BVAAWF/FRAME/RSPCA/UFAW Joint Working Group on Refinement. Lab. Anim. 45, 1. https://doi.org/10.1258/la.2010.010031 (2011).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Koren, S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595. https://doi.org/10.1371/journal.pcbi.1005595 (2017).
Li, H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32, 2103–2110 (2016).
Vaser, R., Sovic, I., Nagarajan, N. & Sikic, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746 (2017).
Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9, e112963. https://doi.org/10.1371/journal.pone.0112963 (2014).
Seppey, M., Manni, M. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness. Methods Mol. Biol. 1962, 227–245 (2019).
Galata, V., Fehlmann, T., Backes, C. & Keller, A. PLSDB: a resource of complete bacterial plasmids. Nucleic Acids Res. 47, D195–D202 (2019).
Lee, J. C., Liu, M.-J., Parsonnet, J. & Arbeit, R. D. Expression of type 8 capsular polysaccharide and production of toxic shock syndrome toxin 1 are associated among vaginal isolates of Staphylococcus aureus. J. Clin. Microbiol. 28, 2612–2615 (1990).
Morikawa, K. et al. Overexpression of sigma factor, sigma(B), urges Staphylococcus aureus to thicken the cell wall and to resist beta-lactams. Biochem. Biophys. Res. Commun. 288, 385–389 (2001).
Dotto, C. et al. The active component of aspirin, salicylic acid, promotes Staphylococcus aureus biofilm formation in a pia-dependent manner. 8, 4. https://doi.org/10.3389/fmicb.2017.00004 (2017).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Mendoza Bertelli, A. et al. Staphylococcus aureus protein A enhances osteoclastogenesis via TNFR1 and EGFR signaling. Biochim. Biophys. Acta 1862, 1975–1983 (2016).
Giai, C. et al. Staphylococcus aureus induces shedding of IL-1RII in monocytes and neutrophils. J. Innate Immun. 8, 284–298 (2016).
Acknowledgements
We are grateful to Lorena Medina for her valuable technical assistance. The authors thank Claudia Barberis for providing the clinical isolate and Graphic Designer Lucia C. Sordelli for her voluntary contribution of Fig. 1 illustrations. This study was partially supported by grants from CONICET, Buenos Aires, Argentina (PUE 0085-2016 to D.O.S. and F.R.B. and PIP 11220150100310 to F.R.B.); Secretaría de Ciencia y Técnica, Universidad de Buenos Aires (UBACyT), Argentina, #20020170100397BA to D.O.S., #20020150100126BA to F.R.B.; ANPCyT (PICT 2016-2678) to M.I.G.; Biotechnology and Biological Sciences Research Council (UK) #BB/K00638X/1 and institute strategic grant funding ISP2: BB/P013740/1 J.R.F.; Medical Research Council (UK) #MR/N02995X/1 to J.R.F and Wellcome Trust collaborative award #201531/Z/16/Z to J.R.F.
Author information
Authors and Affiliations
Contributions
F.R.B., C.M.S. and D.O.S. conceived and designed this study. C.M.S., R.D., A.K,G., M.I.G., S.W., M.N.L., N.R., G.Y., J.A., L.T. and F.R.B performed the experiments. D.O.S., F.R.B., M.I.G., L.T., B.L., N.R. and J.R.F. analyzed and interpreted the data. F.R.B., D.O.S., M.I.G., N.R. and C.M.S. wrote the manuscript. All authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Suligoy, C.M., Díaz, R.E., Gehrke, AK. et al. Acapsular Staphylococcus aureus with a non-functional agr regains capsule expression after passage through the bloodstream in a bacteremia mouse model. Sci Rep 10, 14108 (2020). https://doi.org/10.1038/s41598-020-70671-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-70671-1
This article is cited by
-
Bacterial capsules: Occurrence, mechanism, and function
npj Biofilms and Microbiomes (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.