Abstract
Staphylococcus aureus has evolved into diverse lineages, known as clonal complexes (CCs), which exhibit differences in the coding sequences of core virulence factors. Whether these alterations affect functionality is poorly understood. Here, we studied the highly polymorphic pore-forming toxin LukAB. We discovered that the LukAB toxin variants produced by S. aureus CC30 and CC45 kill human phagocytes regardless of whether CD11b, the previously established LukAB receptor, is present, and instead target the human hydrogen voltage-gated channel 1 (HVCN1). Biochemical studies identified the domain within human HVCN1 that drives LukAB species specificity, enabling the generation of humanized HVCN1 mice with enhanced susceptibility to CC30 LukAB and to bloodstream infection caused by CC30 S. aureus strains. Together, this work advances our understanding of an important S. aureus toxin and underscores the importance of considering genetic variation in characterizing virulence factors and understanding the tug of war between pathogens and the host.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data generated during this study are available upon request with no restrictions. Source data are provided with this paper.
Code availability
Any code generated during this study is available upon request with no restrictions.
References
Tong, S. Y., Davis, J. S., Eichenberger, E., Holland, T. L. & Fowler, V. G. Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661 (2015).
Alonzo, F. III & Torres, V. J. The bicomponent pore-forming leucocidins of Staphylococcus aureus. Microbiol. Mol. Biol. Rev. 78, 199–230 (2014).
Spaan, A. N., van Strijp, J. A. G. & Torres, V. J. Leukocidins: staphylococcal bi-component pore-forming toxins find their receptors. Nat. Rev. Microbiol. 15, 435–447 (2017).
Ventura, C. L. et al. Identification of a novel Staphylococcus aureus two-component leukotoxin using cell surface proteomics. PLoS ONE 5, e11634 (2010).
Dumont, A. L. et al. Characterization of a new cytotoxin that contributes to Staphylococcus aureus pathogenesis. Mol. Microbiol. 79, 814–825 (2011).
Melehani, J. H., James, D. B., DuMont, A. L., Torres, V. J. & Duncan, J. A. Staphylococcus aureus leukocidin A/B (LukAB) kills human monocytes via host NLRP3 and ASC when extracellular, but not intracellular. PLoS Pathog. 11, e1004970 (2015).
Berends, E. T. M. et al. Staphylococcus aureus impairs the function of and kills human dendritic cells via the LukAB toxin. mBio https://doi.org/10.1128/mBio.01918-18 (2019).
DuMont, A. L. et al. Staphylococcus aureus LukAB cytotoxin kills human neutrophils by targeting the CD11b subunit of the integrin Mac-1. Proc. Natl Acad. Sci. USA 110, 10794–10799 (2013).
Badarau, A., Trstenjak, N. & Nagy, E. Structure and function of the two-component cytotoxins of Staphylococcus aureus—learnings for designing novel therapeutics. Adv. Exp. Med. Biol. 966, 15–35 (2017).
Planet, P. J. Life after USA300: the rise and fall of a superbug. J. Infect. Dis. 215, S71–S77 (2017).
Carrel, M., Perencevich, E. N. & David, M. Z. USA300 methicillin-resistant Staphylococcus aureus, United States, 2000–2013. Emerg. Infect. Dis. 21, 1973–1980 (2015).
Malachowa, N. et al. Staphylococcus aureus leukotoxin GH promotes inflammation. J. Infect. Dis. 206, 1185–1193 (2012).
DuMont, A. L. et al. Staphylococcus aureus elaborates leukocidin AB to mediate escape from within human neutrophils. Infect. Immun. 81, 1830–1841 (2013).
Malachowa, N., Kobayashi, S. D., Freedman, B., Dorward, D. W. & DeLeo, F. R. Staphylococcus aureus leukotoxin GH promotes formation of neutrophil extracellular traps. J. Immunol. 191, 6022–6029 (2013).
DuMont, A. L. et al. Identification of a crucial residue required for Staphylococcus aureus LukAB cytotoxicity and receptor recognition. Infect. Immun. 82, 1268–1276 (2014).
Badarau, A. et al. Structure–function analysis of heterodimer formation, oligomerization, and receptor binding of the Staphylococcus aureus bi-component toxin LukGH. J. Biol. Chem. 290, 142–156 (2015).
Badarau, A. et al. Context matters: the importance of dimerization-induced conformation of the LukGH leukocidin of Staphylococcus aureus for the generation of neutralizing antibodies. mAbs 8, 1347–1360 (2016).
Boguslawski, K. M. et al. Exploiting species specificity to understand the tropism of a human-specific toxin. Sci. Adv. 6, eaax7515 (2020).
Bal, A. M. et al. Genomic insights into the emergence and spread of international clones of healthcare-, community- and livestock-associated meticillin-resistant Staphylococcus aureus: blurring of the traditional definitions. J. Glob. Antimicrob. Resist. 6, 95–101 (2016).
Diekema, D. J. et al. Continued emergence of USA300 methicillin-resistant Staphylococcus aureus in the United States: results from a nationwide surveillance study. Infect. Control Hosp. Epidemiol. 35, 285–292 (2014).
Lakhundi, S. & Zhang, K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. https://doi.org/10.1128/CMR.00020-18 (2018).
Planet, P. J. et al. Architecture of a species: phylogenomics of Staphylococcus aureus. Trends Microbiol. 25, 153–166 (2017).
Hughes, A. L. & Friedman, R. Nucleotide substitution and recombination at orthologous loci in Staphylococcus aureus. J. Bacteriol. 187, 2698–2704 (2005).
Springer, T. A., Thompson, W. S., Miller, L. J., Schmalstieg, F. C. & Anderson, D. C. Inherited deficiency of the Mac-1, LFA-1, p150,95 glycoprotein family and its molecular basis. J. Exp. Med. 160, 1901–1918 (1984).
McKillop, W. M., Barrett, J. W., Pasternak, S. H., Chan, B. M. & Dekaban, G. A. The extracellular domain of CD11d regulates its cell surface expression. J. Leukoc. Biol. 86, 851–862 (2009).
Shalem, O. et al. Genome-scale CRISPR–Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
DeCoursey, T. E., Morgan, D. & Cherny, V. V. The voltage dependence of NADPH oxidase reveals why phagocytes need proton channels. Nature 422, 531–534 (2003).
Morgan, D. et al. Voltage-gated proton channels maintain pH in human neutrophils during phagocytosis. Proc. Natl Acad. Sci. USA 106, 18022–18027 (2009).
Ramsey, I. S., Ruchti, E., Kaczmarek, J. S. & Clapham, D. E. Hv1 proton channels are required for high-level NADPH oxidase-dependent superoxide production during the phagocyte respiratory burst. Proc. Natl Acad. Sci. USA 106, 7642–7647 (2009).
DeCoursey, T. E. Voltage-gated proton channels find their dream job managing the respiratory burst in phagocytes. Physiology 25, 27–40 (2010).
Spaan, A. N. et al. The staphylococcal toxins γ-haemolysin AB and CB differentially target phagocytes by employing specific chemokine receptors. Nat. Commun. 5, 5438 (2014).
Okochi, Y., Sasaki, M., Iwasaki, H. & Okamura, Y. Voltage-gated proton channel is expressed on phagosomes. Biochem. Biophys. Res. Commun. 382, 274–279 (2009).
El Chemaly, A., Nunes, P., Jimaja, W., Castelbou, C. & Demaurex, N. Hv1 proton channels differentially regulate the pH of neutrophil and macrophage phagosomes by sustaining the production of phagosomal ROS that inhibit the delivery of vacuolar ATPases. J. Leukoc. Biol. 95, 827–839 (2014).
Uhlen, M. et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science https://doi.org/10.1126/science.aax9198 (2019).
Schilling, T., Gratopp, A., DeCoursey, T. E. & Eder, C. Voltage-activated proton currents in human lymphocytes. J. Physiol. 545, 93–105 (2002).
Trstenjak, N. et al. Molecular mechanism of leukocidin GH-integrin CD11b/CD18 recognition and species specificity. Proc. Natl Acad. Sci. USA 117, 317–327 (2020).
Alonzo, F. III et al. Staphylococcus aureus leucocidin ED contributes to systemic infection by targeting neutrophils and promoting bacterial growth in vivo. Mol. Microbiol. 83, 423–435 (2012).
Alonzo, F. III et al. CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature 493, 51–55 (2013).
Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013).
Pelzek, A. J. et al. Human memory B cells targeting Staphylococcus aureus exotoxins are prevalent with skin and soft tissue infection. mBio https://doi.org/10.1128/mBio.02125-17 (2018).
Chambers, H. F. & Deleo, F. R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7, 629–641 (2009).
Nienaber, J. J. et al. Methicillin-susceptible Staphylococcus aureus endocarditis isolates are associated with clonal complex 30 genotype and a distinct repertoire of enterotoxins and adhesins. J. Infect. Dis. 204, 704–713 (2011).
Fowler, V. G. Jr. et al. Potential associations between hematogenous complications and bacterial genotype in Staphylococcus aureus infection. J. Infect. Dis. 196, 738–747 (2007).
Holden, M. T. et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl Acad. Sci. USA 101, 9786–9791 (2004).
DeLeo, F. R. et al. Molecular differentiation of historic phage-type 80/81 and contemporary epidemic Staphylococcus aureus. Proc. Natl Acad. Sci. USA 108, 18091–18096 (2011).
Tromp, A. T. et al. Human CD45 is an F-component-specific receptor for the staphylococcal toxin Panton–Valentine leukocidin. Nat. Microbiol. 3, 708–717 (2018).
Monk, I. R., Tree, J. J., Howden, B. P., Stinear, T. P. & Foster, T. J. Complete bypass of restriction systems for major Staphylococcus aureus lineages. mBio 6, e00308-15 (2015).
Harper, L. et al. Staphylococcus aureus responds to the central metabolite pyruvate to regulate virulence. mBio https://doi.org/10.1128/mBio.02272-17 (2018).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Lanave, C., Preparata, G., Saccone, C. & Serio, G. A new method for calculating evolutionary substitution rates. J. Mol. Evol. 20, 86–93 (1984).
Yang, Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J. Mol. Evol. 39, 306–314 (1994).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Nelson, C. W., Moncla, L. H. & Hughes, A. L. SNPGenie: estimating evolutionary parameters to detect natural selection using pooled next-generation sequencing data. Bioinformatics 31, 3709–3711 (2015).
Nei, M & Kumar, S. Molecular Evolution and Phylogenetics (Oxford Univ. Press, 2000).
Kosakovsky Pond, S. L. & Frost, S. D. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22, 1208–1222 (2005).
Murrell, B. et al. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8, e1002764 (2012).
Almagro Armenteros, J. J. et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423 (2019).
Osicka, R. et al. Bordetella adenylate cyclase toxin is a unique ligand of the integrin complement receptor 3. eLife 4, e10766 (2015).
Campeau, E. et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS ONE 4, e6529 (2009).
Schoggins, J. W. et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 (2011).
Virreira Winter, S., Zychlinsky, A. & Bardoel, B. W. Genome-wide CRISPR screen reveals novel host factors required for Staphylococcus aureus α-hemolysin-mediated toxicity. Sci. Rep. 6, 24242 (2016).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Radke, E. E. et al. Hierarchy of human IgG recognition within the Staphylococcus aureus immunome. Sci. Rep. 8, 13296 (2018).
Reyes-Robles, T., Lubkin, A., Alonzo, F. III, Lacy, D. B. & Torres, V. J. Exploiting dominant-negative toxins to combat Staphylococcus aureus pathogenesis. EMBO Rep. 17, 780 (2016).
Monk, I. R., Shah, I. M., Xu, M., Tan, M. W. & Foster, T. J. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio https://doi.org/10.1128/mBio.00277-11 (2012).
Fuller, J. R. et al. Identification of a lactate-quinone oxidoreductase in Staphylococcus aureus that is essential for virulence. Front. Cell Infect. Microbiol. 1, 19 (2011).
Berends, E. T. et al. Distinct localization of the complement C5b–9 complex on Gram-positive bacteria. Cell Microbiol. 15, 1955–1968 (2013).
Acknowledgements
We are thankful to members of the Torres and Planet laboratories for insightful discussions and comments on the manuscript, and to H. Behrens, B. Dunn, M. Abrams and R. Plessel for help with reagents, and Z. Chen for help with data analysis. We thank T. Lhakhang (NYU Grossman School of Medicine’s Applied Bioinformatics Laboratories) for help with the bioinformatics analysis of the CRISPR–Cas9 screen, and the NYU Grossman School of Medicine’s Genome Technology Center for help with next-generation sequencing. Last, we acknowledge E. Campeau, P. Kaufman, C. Rice, N. Alto, J. Schoggins, K. Mostov, T. Foster, R. Osicka and F. Zhang for providing reagents. This work was supported in part by the National Institutes of Health–National Institute of Allergy and Infectious Diseases award numbers R01 AI099394, R01 AI105129, R01 AI121244 and contract HHSN272201400019C (to V.J.T.), R01 AI137336 and R01 AI140754 (to B.S. and V.J.T.), T32 AI007180 (to D.B.A.J., E.E.Z. and K.T.), and F32 AI122486 (to D.B.A.J.). S.S.P. was supported by the Jan Vilcek/David Goldfarb Fellowship Endowment Funds and the Bernard B. Levine Program for Postdoctoral Research Fellowships in Immunology. K.M.B. was supported by the Vilcek MSTP Scholars award and T32 GM007308, and C.W.N. was supported by a Gerstner Scholars Fellowship from the Gerstner Family Foundation at the American Museum of Natural History, and a Postdoctoral Research Fellowship from Academia Sinica. P.J.P. and A.M.M. were supported by R01 AI137526. The Genome Technology Center and the flow cytometry technologies are partially supported by the Cancer Center Support Grant P30 CA016087 from the National Institutes of Health–National Cancer Institute at the Laura and Isaac Perlmutter Cancer Center. V.J.T. is a Burroughs Wellcome Fund Investigator in the pathogenesis of infectious diseases.
Author information
Authors and Affiliations
Contributions
S.S.P., D.B.A.J., K.M.B. and V.J.T. designed the study. S.S.P., D.B.A.J., K.M.B. and J.K.I. performed experiments. C.W.N., A.N., A.M.M. and P.J.P. performed the bioinformatics analyses, K.T. generated the monoclonal antibody. E.E.Z. performed isolation and sorting of the primary human leukocytes, and R.C. purified recombinant toxins. B.S. provided bacterial strains and insight into the clinical relevance of CC30 strains. S.S.P., K.M.B., S.Y.K. and S.B.K. generated the hHVCN1 mice. N.V. and M.B.P. bred the mice. R.A.P., A.M. and M.D. provided guidance and assisted with microscopy and image analyses. D.C.E. and G.B. provided expertise with purification of recombinant HCVN1–Strep as well as with Octet measurements. S.S.P. and V.J.T. wrote the manuscript, and all authors commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
V.J.T. is an inventor on patents and patent applications filed by New York University, which are currently under commercial license to Janssen Biotech Inc. (patent nos. US10669329; US10202440; US10087243; MY-175062-A; BR112013032774-0; 60 2014 068 192.1; 40000719B; 1190640B9; 10-2050267; 2012273125; 2012273123; 10,781,246; 10,301,378; 9,783,597; 9,657,103; 9,644,023; 9,481,723; 9,480,726; 9,091,689; 8,846,609; 6758363; 6452765; 6,170,913; 6,093,760; 3441474; 3403669; 3011012; 2,720,754; 2720714; 2,635,462; 2,613,135; 2,609,650; 730359; 710,439; 619,942; 619,938; 357,938; 343,589; 340,446; and 229922). Janssen Biotech Inc. provides research funding and other payments associated with the licensing agreement. All other authors declare no conflicts of interest.
Additional information
Peer review information Nature Microbiology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Nonsynonymous (dN) and synonymous divergence (dS) values and ratios for lukA and lukB between clades.
a-b, Mean between-clade dN and dS values are shown below the diagonal, after applying a Jukes-Cantor correction for multiple substitutions at the same sites. Error bars show the standard error of the mean (SEM), estimated using 10,000 bootstrap replicates (codon unit). Corresponding mean between-clade dN/dS ratios are shown above the diagonal, with significance evaluated using a Z-test of the null hypothesis that dN = dS (10,000 bootstrap replicates, codon unit). All P-values are <1.82×10-6 (see corresponding Figure Source Data) such that asterisks *** indicate a significance level of 0.0001 after Bonferroni correction. Mean between-clade estimates were derived by comparing each member of one clade to each member of a second clade for all pairs of clades. For lukA, the numbers of taxa (n) and independent aligned codon positions (L) for each clade are n = 49 and L = 1013 (clade 1/5/8); 6 and 1013 (30/45); 2 and 1008 (10/395); 8 and 1012 (398); 3 and 860 (S. schweitzeri, labeled as S. sch.); and 6 and 1010 (S. argenteus, labeled as S. arg.). For lukB, the numbers of taxa (n) and independent aligned codon positions (L) for each clade are n = 51 and L = 1004 (clade 1/5/8); 8 and 1004 (30/45); 3 and 1004 (10/395); 3 and 1004 (398); 5 and 1004 (S. schweitzeri); and 6 and 966 (S. argenteus). The number of aligned codon positions (L) are given after eliminating positions with ≤2 defined (non-gap, non-ambiguous) codons in either clade, separately for each between-clade comparison.
Extended Data Fig. 2 Comparison between amino acid sequences of selected mature LukA and LukB.
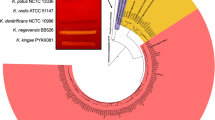
a,c, Phylogenetic tree based on amino acid sequences of mature LukA (a) and LukB (c) produced by S. aureus belonging to the indicated clonal complexes (CCs). The branch length in the tree is proportional to the number of amino acid substitutions per 100 residues. b,d, Percent identity and divergence of mature LukA (b) and LukB (d) proteins produced by S. aureus belonging to the indicated CCs.
Extended Data Fig. 3 Biolayer interferometry binding curves of CC30 and CC8 LukAB variants binding to CD11b I-domain.
a,b, Binding curves of CC30 (a) and CC8 (b) LukAB toxins to CD11b I-domain. The association and dissociation kinetics of LukAB with the I-domain coated sensor are represented in blue. Toxin concentrations are 400 nM, 200 nM, and 100 nM for CC30 LukAB, or 125 nM, 62.5 nM, and 31.3 nM for CC8 LukAB. Red curves show the best global fit using a 1:1 binding model.
Extended Data Fig. 4 HVCN1 expression in human white blood cells.
Consensus human blood cell type expression of HVCN1 derived from RNA-seq data from internally generated Human Protein Atlas (HPA) data1. Transcript expression values are presented as Normalized eXpression (NX), resulting from the internal normalization pipeline for 18 blood cell types and total peripheral blood mononuclear cells (PBMC). Data is available at v20.proteinatlas.org/ENSG00000122986-HVCN1/blood, Human Protein Atlas available from www.proteinatlas.org34.
Extended Data Fig. 5 Generation and evaluation of the HVCN1 humanized mouse model.
a, Schematic representation of murine Hvcn1 locus and DNA template used to humanize exon 4. b, Genotyping strategy using genomic DNA isolated from wild type (WT), heterozygous (het), and homozygous (homo) hHVCN1 mice using primers VJT2065 and VJT2069. Images are representative of multiple independent experiments as routinely performed for hHVCN1 mouse genotyping. c-g, CFUs in the kidneys (c), livers (d), hearts (e), spleens (f), and lungs (g) collected from WT and hHVCN1 mice infected intravenously with 1 × 107 CFU of lukAB-deficient USA300 strain LAC. Data from 11 WT and 10 hHVCN1 mice are represented as mean values ±SEM. Statistical significance was determined by t-test (two-tailed), numbers above bars indicate P values. H-K, CFUs in the livers (h), hearts (i), spleens (j), and lungs (k) collected from WT and hHVCN1 mice infected intravenously with 5-10x107 CFU CC30 S. aureus MUZ211 (CFU obtained from 11 WT and 24 hHVCN1 mice) and 62300D1 (CFU obtained from 11 WT and 10 hHVCN1 mice). Data for each isolate are from mice infected over three independent experiments and is represented as mean values ±SEM. Statistical significance was determined by t-test (two-tailed), numbers above bars indicate P values.
Extended Data Fig. 6 Flow cytometry gating (part 1).
a, Flow cytometry gating scheme utilized to measure surface CD11b levels in scramble shRNA (top) and ITGAM shRNA (bottom) expressing THP1 cell (Fig. 2a) using APC-conjugated anti-CD11b antibody. b, Flow cytometry gating scheme utilized to measure binding of biotinylated LukAB (CC30 LukAB is shown as an example) to CHO cells expressing Fluc (top) or HVCN1 (bottom) using PerCP/Cy5.5-conjugated streptavidin staining (Fig. 5b,c). c, Flow cytometry gating scheme utilized to measure membrane damage in B cells following treatment with PBS control (top) and LukAB (CC30 LukAB is shown as an example, bottom) using Fixable Viability Dye eFluor™ 450 (Fig. 5e). d-e, Flow cytometry gating scheme utilized to measure membrane damage in CD4-positive (d) and CD8-positive (e) T cells following treatment with PBS control (top) and LukAB (CC30 LukAB is shown as an example, bottom) using Fixable Viability Dye eFluor™ 450 (Fig. 5e).
Extended Data Fig. 7 Flow cytometry gating (part 2).
a, Flow cytometry gating scheme utilized to measure membrane damage in PECs after treatment with PBS control (top) and leukocidins (LukED is shown as an example, bottom) using Fixable Viability Dye eFluor™ 450 (Fig. 6a). b, Flow cytometry gating scheme utilized to measure membrane damage in Lenti-X 293 T cells expressing C-terminal GFP-tagged wildtype HVCN1 and chimeric proteins (human HVCN1 is shown as an example) following treatment with PBS control (top) and CC30 LukAB (bottom) using Fixable Viability Dye eFluor™ 450 (Fig. 6d).
Supplementary information
Supplementary Information
Supplementary Tables 4 and 5 and supplementary references.
Supplementary Tables
Table 1: Related to Fig. 1. S. aureus sequences used in the phylogeny analyses. Table 2: Related to Fig. 1. Amino acid sequences of selected mature LukA and LukB produced by S. aureus belonging to the selected CCs. Table 3: Related to Fig. 3. sgRNAs enriched following CC30 LukAB selection.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 1
Unprocessed gel and western blot for Fig. 1c.
Source Data Fig. 3
Unprocessed gel Fig. 3d and western blot for Fig. 3e.
Source Data Fig. 4
Unprocessed gel Fig. 4a and western blot for Fig. 4b.
Source Data Fig. 5
Unprocessed gels and western blots for Fig. 5d.
Source Data Fig. 6
Unprocessed western blots for Fig. 6b.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed gel for Extended Data Fig. 5b.
Source Data Extended Data Fig. 5
Statistical source data.
Rights and permissions
About this article
Cite this article
Perelman, S.S., James, D.B.A., Boguslawski, K.M. et al. Genetic variation of staphylococcal LukAB toxin determines receptor tropism. Nat Microbiol 6, 731–745 (2021). https://doi.org/10.1038/s41564-021-00890-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-021-00890-3
This article is cited by
-
Staphylococcus aureus host interactions and adaptation
Nature Reviews Microbiology (2023)