Abstract
Genetic polymorphisms in drug metabolizing enzymes and drug transporters may affect irinotecan toxicity. Although genetic polymorphisms have been shown to influence the irinotecan toxicity, data are limited in Thai population. Thus, the aim of this study was to assess the allele and genotype frequencies and the relationship between CYP3A4/5, DPYD, UGT1A1, ABCB1, and ABCC2 genetic variations and irinotecan-induced toxicity in Thai colorectal cancer patients. One hundred and thirty-two patients were genotyped, and the effect of genetic variations on irinotecan-induced toxicity was assessed in 66 patients who received irinotecan-based chemotherapy. Allele frequencies of ABCB1 c.1236C > T, ABCB1 c.3435C > T, ABCC2 c.3972C > T, ABCG2 c.421C > A, CYP3A4*1B, CYP3A4*18, CYP3A5*3, DPYD*5, UGT1A1*28, and UGT1A1*6 were 0.67, 0.43, 0.23, 0.27, 0.01, 0.02, 0.64, 0.19, 0.16, and 0.09, respectively. DPYD*2A and DPYD c.1774C > T variants were not detected in our study population. The ABCC2 c.3972C > T was significantly associated with grade 1–4 neutropenia (P < 0.012) at the first cycle. Patients carrying both UGT1A1*28 and *6 were significantly associated with severe neutropenia at the first (P < 0.001) and second (P = 0.017) cycles. In addition, patients carrying UG1A1*28 and *6 had significantly lower absolute neutrophil count (ANC) nadir at first (P < 0.001) and second (P = 0.001) cycles. This finding suggests that UGT1A1*28, *6, and ABCC2 c.3972C > T might be an important predictor for irinotecan-induced severe neutropenia.
Similar content being viewed by others
Introduction
Irinotecan (CPT-11), a topoisomerase I inhibitor, is commonly used for the treatment of colorectal, gastric, and lung cancer. Although irinotecan is efficient, it causes severe neutropenia and diarrhea in 20–35% of the patients1,2. Irinotecan is metabolized by carboxylesterases (CESs) to form the primary pharmacologically active metabolite SN-38. SN-38, a topoisomerase I inhibitor, binds to and stabilizes the topoisomerase I-DNA complex preventing the re-ligation of DNA during replication and transcription, and subsequently results in double-stranded DNA breaks and apoptosis3,4. The SN-38 glucuronide (SN-38G), an inactive metabolite, is converted by uridine diphospho-glucuronosyltransferases (UGTs) in the liver and eliminated into bile by drug efflux transporters ABCB1, ABCC2, and ABCG25,6.
Several studies have described the complex pharmacogenetics of irinotecan7,8,9. UGT1A1 polymorphisms in promoter and coding regions are associated with reduced enzyme activity and accumulation of SN-38G which increases the toxicity of irinotecan. Previous studies reported that patients carrying UGT1A1*28 and *6 variants resulted in increased SN-38 activity leading to diarrhea and severe neutropenia10,11. ABCB1 c.1236 C allele was significantly associated with grade 3/4 toxicities in metastatic colorectal cancer patients12, and ABCB1 c.1236 T/T genotype was also associated with significantly increased exposure to irinotecan and its active metabolite SN-38 compared to those with heterozygous and wild-type13. ABCB1 c.3435C > T altered expression levels and transport efficiency in vitro and in vivo14,15. ABCC2 c.3972T/T genotype was related with higher areas under the plasma concentration–time curve (AUC) of irinotecan, (7-ethyl-10-[4-N-(5-aminopentanoic acid)-1-piperidino] carbonyloxycamptothecin) (APC) and SN-38G16. ABCG2 c.421C > A variant results in lower protein expression and higher drug exposure17. Clinically, the combination of irinotecan and 5-fluorouracil is frequently used to treat colorectal cancer patients, and DPYD polymorphisms are related to 5-fluorouracil-induced severe neutropenia and diarrhea18.
Although there have been reports of the relationship between genetic polymorphisms and irinotecan induced-toxicity, there are few reports in Thai colorectal cancer patients. Therefore, the aim of this study was to extensively investigate the association between genetic polymorphisms in CYP3A4/5, DPYD, UGT1A1, ABCB1, ABCC2, and ABCG2 and irinotecan induced-toxicity in cohort of Thai colorectal cancer patient.
Methods
Eligible patients
A total of 132 metastatic colorectal cancer patients who received chemotherapy were recruited in this retro- and prospective study between August 2012 and June 2016 from the Division of Cancer, Department of Medicine, Faculty of Medicine, Ramathibodi Hospital, Mahidol University, Thailand. The clinical eligibility criteria that was used to enroll patients were as follows: histologically or cytologically confirmed metastatic colorectal cancer; age at least 18 years; Eastern Cooperative Oncology Group (ECOG) performance status 0–2; life expectancy > 3 months; neutrophil count ≥ 1.5 × 109/L; platelet count ≥ 8 × 1010/L; serum creatinine ≤ 1.25 upper limit normal (ULN); total bilirubin ≤ 1.25 ULN; alanine aminotransferase and aspartate aminotransferase ≤ 2.5 ULN. All patients had peripheral blood samples taken and complete clinical information (Supplementary Table 1). Sixty-six patients who were treated with irinotecan based-chemotherapy were analyzed for toxicity assessment. The flow chart for patient screening is shown in Fig. 1.
Flow chart for patient screening. A total of 132 metastatic colorectal cancer patients were genotyped for genetic polymorphisms and 66 patients who did not treated with irinotecan-based chemotherapy were excluded. Of the 66 patients treated with irinotecan-based chemotherapy were included in this analysis.
This study was approved by the Ethics Review Committee on Human Research of the Faculty of Medicine Ramathibodi Hospital, Mahidol University, Thailand (MURA2015/299) and conducted in accordance with the Declaration of Helsinki. The study protocol was clearly explained to all patients and informed consent was given before the study.
Genotyping analysis
Peripheral blood was collected into an ethylenediaminetetraacetic acid (EDTA) tube, and genomic DNA was extracted using the MagNA Pure Compact System (Roche, Mannheim, Germany). DNA concentration was measured with a Thermo Scientific™ NanoDrop™ spectrophotometer and concentrations adjusted as recommended for each genotyping platforms. A total of 10 SNVs of CYP3A4*1B (c.-392A > G, Assay ID: C_1837671_50), CYP3A4*18 (c.878 T > C, C_27859823_20), CYP3A5*3 (c.6986A > G, C_26201809_30), DPYD*2A (IVS14 + 1G > A, C_30633851_20), DPYD*5 (c.1627A > G, C_1823316_20), DPYD c.1774C > T (C_90454263_10), ABCB1 c.3435C > T (C_7586657_20), ABCB1 c.1236C > T (C_758662_10), ABCC2 c.3972C > T (C_11214910_20), and ABCG2 c.421C > A (C_15854163_70) were genotyped by TaqMan® Genotyping Assays (Applied Biosystems™, Carlsbad, CA, USA) according to the manufacturer’s instructions. An additional 2 variants, UGT1A1*28 (A(TA)7TAA) and *6 (c.211G > A), were genotyped by pyrosequencing (Qiagen, Japan) analysis according to a previously described method19.
Drug administration
FOLFIRI regimen: Irinotecan 180 mg/m2, 90 min intravenous infusion on day 1; leucovorin (LV) 200 mg/m2 intravenous infusion on day 1; fluorouracil 400 mg/m2 intravenous bolus on day 1; fluorouracil 600 mg/m2 intravenous over the course of 46 h of continuous infusion; repeated every 2 weeks (28 patients).
FOLFIRI regimen plus cetuximab regimen: Cetuximab 400 mg/m2 intravenous infusion on day 1; Irinotecan 180 mg/m2, 90 min intravenous infusion on day 1; leucovorin (LV) 200 mg/m2 intravenous infusion on day 1; fluorouracil 400 mg/m2 intravenous bolus on day 1; fluorouracil 600 mg/m2 intravenous over the course of 46 h of continuous infusion; repeated every 2 weeks (seven patients).
FOLFIRI regimen plus bevacizumab regimen: Bevacizumab 5–10 mg/kg intravenous infusion once every 2 weeks; Irinotecan 180 mg/m2, 90 min intravenous infusion on day 1; leucovorin (LV) 200 mg/m2 intravenous infusion on day 1; fluorouracil 400 mg/m2 intravenous bolus on day 1; fluorouracil 600 mg/m2 intravenous over the course of 46 h of continuous infusion; repeated every 2 weeks (one patient).
Modified FOLFIRI regimen: Irinotecan 180 mg/m2, 90 min intravenous infusion on day 1; leucovorin (LV) 400 mg/m2 intravenous infusion on day 1; fluorouracil 400 mg/m2 intravenous bolus on day 1; fluorouracil 1,200 mg/m2 intravenous over the course of 46 h of continuous infusion; repeated every 2 weeks (18 patients).
Single irinotecan regimen: Irinotecan 100 mg/m2, 90 min intravenous infusion on day 1 (eight patients).
Irinotecan plus cetuximab or irinotecan plus capecitabine regimen: Irinotecan 100 mg/m2, 90 min intravenous infusion on day 1; cetuximab 100–130 mg/m2 intravenous infusion on day 1 or irinotecan 100 mg/m2, 90 min intravenous infusion on day 1; capecitabine 1,000 mg/m2 (four patients).
Toxicity criteria
Toxicity was assessed at first and second cycles of treatment according to National Cancer Institute Common Toxicity Criteria for Adverse Events, version 5.0. Grade 3–4 toxicity was considered as severe toxicity.
Statistical analysis
Deviation from Hardy–Weinberg equilibrium was assessed using Fisher’s exact and chi-square test. Allele and genotype frequencies were determined by direct counting. Comparisons of allele and genotype frequencies and grades of toxicity were performed using the χ2 test. Mann–Whitney U test was performed according to difference of genetic groups and nonparametric data [absolute neutrophil count (ANC) nadir and ANC ratio]. Logistic regression analysis was performed to assess univariate and multivariate relationships genetic polymorphisms, and other parameters. All statistics were calculated using SPSS version 18 (SPSS Inc., Chicago, IL, USA) and differences were significant when P values were < 0.05.
Results
Clinical characteristics and genotyping data
A total of 132 metastatic colorectal cancer patients were genotyped for CYP3A4*1B, CYP3A4*18, CYP3A5*3, DPYD*2A, DPYD*5, DPYD c.1774C > T, UGT1A1*28, UGT1A1*6, ABCB1 c.1236C > T, ABCB1 c.3435C > T, ABCC2 c.3972C > T, and ABCG2 c.421C > A. The genotype and allele frequencies are shown in Table 1. The most prevalent alleles were ABCB1 c.1236C > T (0.67), CYP3A5*3 (0.64), and ABCB1 c.3435C > T (0.43), respectively. DPYD*2A and DPYD c.1774C > T were not detected in our samples.
Sixty-six patients with metastatic colorectal cancer receiving an irinotecan-based regimen were enrolled for association analysis. Their clinical characteristics are summarized in Table 2. The average age of the 66 patients was 62 years (range 25–74) with 42 (63.6%) male and 24 (36.4%) female. Most patients showed an ECOG performance status of zero. The most common site of disease was the rectum. The liver was the dominant site for metastases. There were no statistically significant differences between clinical characteristics and hematological toxicity including neutropenia, leucopenia, thrombocytopenia, and anemia (data not shown).
Association between genetic polymorphisms and irinotecan-induced neutropenia
The association analysis is summarized in Table 3. At the first cycle of treatment, ABCC2 c.3972C > T was significantly associated with all grades neutropenia [grade 1–4 neutropenia; odds ratio (OR) 3, 95% confidence intervals (CI) 1.3–7; P < 0.012]. In addition, UGT1A1*6 was significantly associated with grade 1–4 and severe neutropenia (grade 3–4) (OR 20.3, 95% CI 4.3–95.6; P < 0.001, and OR 4, 95% CI 1.2–13; P < 0.026, respectively). Although there were no significant differences between grade 3–4 neutropenia and patients with UGT1A1*28, the incidence of severe neutropenia in patients with hetero- and homozygous *28 was higher than patients with homozygous wild type, (OR 2.7, 95% CI 0.8–8.8; P = 0.087).
At the second cycle, an association was observed between UGT1A1*28 and grade 1–4 neutropenia (OR 3.1, 95% CI 1.2–7.97; P = 0.017). Similarly, UGT1A1*6 was significantly associated with grade 1–4 (OR 20.3, 95% CI 4.3–95.6; P < 0.001) and severe neutropenia (OR 12.5, 95% CI 3.4–45.7; P < 0.001).
The combination of UGT1A1*28 and *6 showed a significant increased risk for all grades of neutropenia (P < 0.001) and severe neutropenia (P = 0.002) at first cycle. Similarly in the second cycle, patients with hetero- and homozygous variant had a high incidence of all grades of neutropenia (P < 0.001) and severe neutropenia (P = 0.016).
A multivariate logistic regression analysis was performed to analyze the influence of CYP3A4*1B, CYP3A4*18, CYP3A5*3, DPYD*2A, DPYD*5, DPYD c.1774C > T, UGT1A1*28, UGT1A1*6, ABCB1 c.1236C > T, ABCB1 c.3435C > T, ABCC2 c.3972C > T, and ABCG2 c.421C > A on neutropenia (all grades and severe neutropenia) at first and second cycles. The result showed that ABCC2 c.3972C > T was significantly associated with grade 1–4 neutropenia (P = 0.015). In the second cycle, we found patients with UGT1A1*28 were at significant increased risk for grade 1–4 neutropenia compared with wild type patients (P = 0.011). Moreover, patients with UGT1A1*6 were at significantly increased risk for grades 1–4 and severe neutropenia compared with wild type patients (P = 0.002, P = 0.001, respectively), as shown in Table 4.
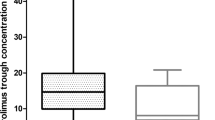
The association of genetic polymorphisms and absolute neutrophil count (ANC) nadir was also assessed at the first and second cycle. Regarding UGT1A1*6, the ANC nadir of G/A was significantly lower than A/A in both first (1,600:2,560.7/mm3, P = 0.004) and second (1,201.8:2,379.8/mm3, P < 0.001) cycles. Hetero- and homozygous UGT1A1*28 or *6 carriers showed decreased ANC nadir compared to wild type carriers at first (1,595.9:2,894.1/mm3, P < 0.001) and second 1,528.4:2,793/mm3, P = 0.001) cycles (Fig. 2).
Using ANC ratio (ANC nadir to ANC baseline), patients carrying a variant of UGT1A1 had lower ANC ratio at first (0.41:0.79; P < 0.001) and second (0.45:0.79; P = 0.001) cycles, (Fig. 3). ANC ratio in patients with UGT1A1 c.211 G/A was significantly lower than G/G at the first (0.4:0.6, P = 0.020) and second (0.3:0.7, P = 0.010) cycles, and UGT1A1*28 was significantly associated with decreased ANC ratio (variant: wild type; 0.47:0.72, P = 0.047) at the first cycle.
Discussion
In this study, the association between irinotecan-induced toxicity and pharmacogenetics of drug metabolizing enzymes and drug transporters was investigated. Our results showed that combined analysis of UGT1A1*28 and *6 polymorphisms and ABCC2 c.3972C > T were closely related with neutropenia toxicity in Thai colorectal cancer patients.
The UGT1A1*28 allele is the most important risk factor for severe neutropenia or diarrhea. In 2005, the U.S. Food and Drug Administration (FDA) informed that patients with homozygous UGT1A1*28 are at increased risk of severe neutropenia following initiation of irinotecan treatment20. Several studies have investigated the relationship of UGT1A1*28 and severe neutropenia and diarrhea during irinotecan treatment21,22,23. Wang et al.24 reported a significantly high risk for grade 3–4 leukopenia and neutropenia in patients carrying heterozygous UGT1A1*28 compared to homozygous wild type patients. Similarly, Rouits et al.25 reported that patients carrying the homozygous or heterozygous UGT1A1*28 had significantly higher risk of neutropenia than those with UGT1A1*1. In this study, although there were no significant differences between grade 3–4 neutropenia and patients with UGT1A1*28, the incidence of severe neutropenia in patients with hetero- and homozygous *28 was higher than patients with homozygous wild type at the first cycle (OR 2.7, 95% CI 0.8–8.8; P = 0.087). At the second cycle, patient carried UGT1A1*28 was a significantly higher risk of neutropenia than patients with homozygous wild type (OR 3.1, 95% CI 1.2–7.97; P = 0.017). A multivariate analysis was performed to analyze the influence of UGT1A1*28. This result shown that patients with UGT1A1*28 was at significant increased risk for grade 1–4 neutropenia compared with wild type patients (P = 0.011) in second cycle. Using ANC ratio, patient carried heterozygous and homozygous UGT1A1*28 had lower ANC nadir and ANC ratio than wild type at first cycle (P = 0.047). These results shown an increased risk of neutropenia in patient with UGT1A1*28 genotype.
The prevalence of UGT1A1*6 in Asian is higher than Caucasian population. The UGT1A1*6 polymorphism is the most common allele that is correlated with reduced SN-38 glucuronidation activity and drug toxicity26. Han et al.27 demonstrated that UGT1A1*6/*6 was significantly associated with higher SN-38 AUC and may increase the risk for toxicities. Onoue et al.28 performed a prospective study of 135 Japanese cancer patients treated with irinotecan, and found that severe neutropenia was highly correlated with homozygous UGT1A1*6 in a multiple logistic regression analysis. Similarly, UGT1A1*6 was significantly associated with grade 1–4 and severe neutropenia at the first and second cycles in this study. In contrast, there were significant differences between grade 1–4 neutropenia and patients with UGT1A1*28 at the second cycle. However, UGT1A1 genotype was associated with an increased risk of grade 1–4 and severe neutropenia at the first and second cycle. Similar to the study by Yang et al.29, UGT1A1*28 and *6 were significantly associated with higher incidence of grade 3–4 neutropenia. A meta-analysis by Han et al.30 found that Asian cancer patients with UGT1A1*28 and *6 are at increased risk of irinotecan-induced neutropenia. Moreover, we also found an association between UGT1A1*28 or/and UGT1A1*6 and ANC nadir. This revealed that patients carrying variant of UGT1A1 genotype had a significantly lower ANC nadir in the first and second cycle. Moriya et al.31 reported that ANC nadir in patients carrying UGT1A1*6/*28, *6/*6 were significantly lower compared with those with *1/*1.
ABCC2 protein is expressed in liver, kidney, and small intestine, and also plays a primary role in biliary excretion of irinotecan and its metabolites32,33. Interestingly, our result suggested that ABCC2 c.3972C > T is associated with grade 1–4 neutropenia at the first cycle. Multivariate analysis indicated that ABCC2 c.3972C > T is a risk factor for the occurrence of grade 1–4 neutropenia at the first cycle in patients who receive irinotecan-based chemotherapy. Innocenti et al.16 reported that ABCC2 c.3972T/T genotype correlates with higher AUC of irinotecan, APC, and SN-38G. This result suggests that ABCC2 c.3972C > T is associated with decreased hepatobiliary excretion of irinotecan and its metabolites.
ABCB1 c.3435C > T is associated with significantly lower AUC SN-38G levels, and homozygous ABCB1 c.3435T/T may be related to higher P-glycoprotein (MDR1) activity34. However, ABCB1 c.3435C > T was not associated with irinotecan induced severe neutropenia and diarrhea in Chinese cancer patients who received irinotecan chemotherapy35. Cote et al.36 reported that no statistically significant difference was found in ABCB1 c.3435C > T polymorphism and occurrence of severe hematologic toxicity or severe neutropenia.
The ABCB1 c.1236C > T has been reported to be associated with increased AUC of irinotecan and SN-38 in Caucasian cancer patients13, and ABCB1 c.1236T/T had significantly higher plasma irinotecan and SN-38 concentrations than C/C or C/T. However, Han et al.34 reported that no significant effect of ABCB1 c.1236C > T on irinotecan or its metabolites concentrations. Han et al.27 reported that no significant association between ABCB1 c.1236C > T and severe neutropenia and diarrhea was observed. In vitro studies have shown that ABCG2, an efflux drug transporter, had a higher affinity with SN-38 and SN-38G37, and ABCG2 c.421C > A is related with reduced expression of ABCG2 protein and transporter activity. However, de Jong et al.38 reported that no significant changes in irinotecan pharmacokinetics relative to the ABCG2 c.421C > A in Caucasian cancer patients. ABCG2 variants had no effect on SN-38 exposure or ANC nadir in 78 irinotecan-treated patients39.
Irinotecan is converted by CYP3A4/5 to APC metabolite in the liver, and correlation between these genes and irinotecan induced-toxicity found in this study may be due to low allele frequency of CYP3A4*1B and *18 in sampled population40. Similarly, Han et al.34 did not find any significant association between CYP3A5*3 and toxicity.
The 5-FU-based regimen may cause neutropenia, however, over 80% of 5-FU is metabolized by dihydropyrimidine dehydrogenase (DPD) in the liver41. DPYD variants may be related to severe 5-FU-associated toxicities. DPYD*2A and c.1774 C > T variants were not found in this study. Even though, the variant allele of DPYD*5 had a frequency of 0.19, there was no association between DPYD*5 and hematological toxicities.
A retrospective study design and small sample size are limitations of this study. A prospective study involving larger numbers of patients should confirm our study hypothesis. Secondly, rare genetic variants and multiple genes play a role in the irinotecan pathway. Those variants were not considered in our study. Lastly, non-hematologic toxicity (especially severe diarrhea) was not assesses in our study.
In conclusion, combination of UGT1A1*28 and *6 and ABCC2 c.3972C > T genotype are associated with the occurrence of grade 1–4 and severe neutropenia in Thai patients with metastatic colorectal cancer who receive irinotecan-based chemotherapy. Our findings suggest that UGT1A1 genotype and ABCC2 c.3972C > T might be an important predictor for irinotecan induced-toxicity.
References
Innocenti, F. et al. Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan. J. Clin. Oncol.22, 1382–1388. https://doi.org/10.1200/jco.2004.07.173 (2004).
Ishida, H. et al. Regimen selection for first-line FOLFIRI and FOLFOX based on UGT1A1 genotype and physical background is feasible in Japanese patients with advanced colorectal cancer. Jpn. J. Clin. Oncol.41, 617–623. https://doi.org/10.1093/jjco/hyr010 (2011).
Fuchs, C. S. et al. Phase III comparison of two irinotecan dosing regimens in second-line therapy of metastatic colorectal cancer. J. Clin. Oncol.21, 807–814 (2003).
Bencharit, S. et al. Structural insights into CPT-11 activation by mammalian carboxylesterases. Nat. Struct. Biol.9, 337–342. https://doi.org/10.1038/nsb790 (2002).
Cresteil, T. et al. Regioselective metabolism of taxoids by human CYP3A4 and 2C8: Structure–activity relationship. Drug Metab. Dispos.30, 438–445 (2002).
Allen, J. D., Brinkhuis, R. F., van Deemter, L., Wijnholds, J. & Schinkel, A. H. Extensive contribution of the multidrug transporters P-glycoprotein and Mrp1 to basal drug resistance. Cancer Res.60, 5761–5766 (2000).
Nagar, S. & Blanchard, R. L. Pharmacogenetics of uridine diphosphoglucuronosyltransferase (UGT) 1A family members and its role in patient response to irinotecan. Drug Metab. Rev.38, 393–409. https://doi.org/10.1080/03602530600739835 (2006).
Smith, N. F., Figg, W. D. & Sparreboom, A. Pharmacogenetics of irinotecan metabolism and transport: An update. Toxicol. In Vitro20, 163–175. https://doi.org/10.1016/j.tiv.2005.06.045 (2006).
Ando, Y. & Hasegawa, Y. Clinical pharmacogenetics of irinotecan (CPT-11). Drug Metab. Rev.37, 565–574. https://doi.org/10.1080/03602530500316254 (2005).
Huang, S. H. et al. Concurrence of UGT1A polymorphism and end-stage renal disease leads to severe toxicities of irinotecan in a patient with metastatic colon cancer. Tumori97, 243–247. https://doi.org/10.1700/667.7793 (2011).
Takano, M. et al. Clinical significance of UDP-glucuronosyltransferase 1A1*6 for toxicities of combination chemotherapy with irinotecan and cisplatin in gynecologic cancers: A prospective multi-institutional study. Oncology76, 315–321. https://doi.org/10.1159/000209335 (2009).
Miura, M. et al. Influence of drug transporters and UGT polymorphisms on pharmacokinetics of phenolic glucuronide metabolite of mycophenolic acid in Japanese renal transplant recipients. Ther. Drug Monit.30, 559–564. https://doi.org/10.1097/FTD.0b013e3181838063 (2008).
Mathijssen, R. H. et al. Irinotecan pathway genotype analysis to predict pharmacokinetics. Clin. Cancer Res.9, 3246–3253 (2003).
Siegsmund, M. et al. Association of the P-glycoprotein transporter MDR1(C3435T) polymorphism with the susceptibility to renal epithelial tumors. J. Am. Soc. Nephrol.13, 1847–1854 (2002).
Drescher, S. et al. MDR1 gene polymorphisms and disposition of the P-glycoprotein substrate fexofenadine. Br. J. Clin. Pharmacol.53, 526–534 (2002).
Innocenti, F. et al. Pharmacogenetic analysis of interindividual irinotecan (CPT-11) pharmacokinetic (PK) variability: Evidence for a functional variant of ABCC2. J. Clin. Oncol. 22, 2010. https://doi.org/10.1200/jco.2004.22.90140.2010 (2004).
Sparreboom, A. et al. Diflomotecan pharmacokinetics in relation to ABCG2 421C>A genotype. Clin. Pharmacol. Ther.76, 38–44. https://doi.org/10.1016/j.clpt.2004.03.003 (2004).
Gentile, G. et al. Genotype–phenotype correlations in 5-fluorouracil metabolism: A candidate DPYD haplotype to improve toxicity prediction. Pharmacogenom. J.16, 320–325. https://doi.org/10.1038/tpj.2015.56 (2016).
Sukasem, C. et al. Development of pyrosequencing method for detection of UGT1A1 polymorphisms in thai colorectal cancers. J. Clin. Lab. Anal.30, 84–89. https://doi.org/10.1002/jcla.21820 (2016).
Hoskins, J. M., Goldberg, R. M., Qu, P., Ibrahim, J. G. & McLeod, H. L. UGT1A1*28 genotype and irinotecan-induced neutropenia: Dose matters. J. Natl. Cancer Inst.99, 1290–1295. https://doi.org/10.1093/jnci/djm115 (2007).
Xu, J. M. et al. Severe irinotecan-induced toxicity in a patient with UGT1A1 28 and UGT1A1 6 polymorphisms. World J. Gastroenterol.19, 3899–3903. https://doi.org/10.3748/wjg.v19.i24.3899 (2013).
Shulman, K. et al. Clinical implications of UGT1A1*28 genotype testing in colorectal cancer patients. Cancer117, 3156–3162. https://doi.org/10.1002/cncr.25735 (2011).
Zhou, C. F. et al. UGT1A1 gene polymorphisms and the toxicities of FOLFIRI in Chinese Han patients with gastrointestinal cancer. Anticancer Agents Med. Chem.13, 235–241 (2013).
Wang, Y. et al. Distribution of uridine diphosphate glucuronosyltransferase 1A polymorphisms and their role in irinotecan-induced toxicity in patients with cancer. Oncol. Lett.14, 5743–5752. https://doi.org/10.3892/ol.2017.6933 (2017).
Rouits, E. et al. Relevance of different UGT1A1 polymorphisms in irinotecan-induced toxicity: A molecular and clinical study of 75 patients. Clin. Cancer Res.10, 5151–5159. https://doi.org/10.1158/1078-0432.ccr-03-0548 (2004).
Akiyama, Y. et al. Association of ABCC2 genotype with efficacy of first-line FOLFIRI in Japanese patients with advanced colorectal cancer. Drug Metab. Pharmacokinet.27, 325–335 (2012).
Han, J. Y., Lim, H. S., Park, Y. H., Lee, S. Y. & Lee, J. S. Integrated pharmacogenetic prediction of irinotecan pharmacokinetics and toxicity in patients with advanced non-small cell lung cancer. Lung Cancer (Amsterdam, Netherlands)63, 115–120. https://doi.org/10.1016/j.lungcan.2007.12.003 (2009).
Onoue, M. et al. UGT1A1*6 polymorphism is most predictive of severe neutropenia induced by irinotecan in Japanese cancer patients. Int. J. Clin. Oncol.14, 136–142. https://doi.org/10.1007/s10147-008-0821-z (2009).
Yang, C. et al. Relationship between UGT1A1*6/*28 polymorphisms and severe toxicities in Chinese patients with pancreatic or biliary tract cancer treated with irinotecan-containing regimens. Drug Des. Dev. Ther.9, 3677–3683. https://doi.org/10.2147/dddt.s86750 (2015).
Han, F. F. et al. Associations between UGT1A1*6 or UGT1A1*6/*28 polymorphisms and irinotecan-induced neutropenia in Asian cancer patients. Cancer Chemother. Pharmacol.73, 779–788. https://doi.org/10.1007/s00280-014-2405-0 (2014).
Moriya, H. et al. Association between the low-dose irinotecan regimen-induced occurrence of grade 4 neutropenia and genetic variants of UGT1A1 in patients with gynecological cancers. Oncol. Lett.7, 2035–2040. https://doi.org/10.3892/ol.2014.2046 (2014).
Fardel, O., Jigorel, E., Le Vee, M. & Payen, L. Physiological, pharmacological and clinical features of the multidrug resistance protein 2. Biomed. Pharmacother.59, 104–114. https://doi.org/10.1016/j.biopha.2005.01.005 (2005).
Suzuki, H. & Sugiyama, Y. Single nucleotide polymorphisms in multidrug resistance associated protein 2 (MRP2/ABCC2): Its impact on drug disposition. Adv. Drug Deliv. Rev.54, 1311–1331 (2002).
Han, J. Y. et al. Associations of ABCB1, ABCC2, and ABCG2 polymorphisms with irinotecan-pharmacokinetics and clinical outcome in patients with advanced non-small cell lung cancer. Cancer110, 138–147. https://doi.org/10.1002/cncr.22760 (2007).
Yan, L. et al. Effects of UGT1A1*6, UGT1A1*28, and ABCB1-3435C>T polymorphisms on irinotecan induced toxicity in Chinese cancer patients. Int. J. Clin. Pharmacol. Ther.54, 193–199. https://doi.org/10.5414/cp202442 (2016).
Cote, J. F. et al. UGT1A1 polymorphism can predict hematologic toxicity in patients treated with irinotecan. Clin. Cancer Res.13, 3269–3275. https://doi.org/10.1158/1078-0432.CCR-06-2290 (2007).
Nakatomi, K. et al. Transport of 7-ethyl-10-hydroxycamptothecin (SN-38) by breast cancer resistance protein ABCG2 in human lung cancer cells. Biochem. Biophys. Res. Commun.288, 827–832. https://doi.org/10.1006/bbrc.2001.5850 (2001).
de Jong, F. A. et al. ABCG2 pharmacogenetics: Ethnic differences in allele frequency and assessment of influence on irinotecan disposition. Clin. Cancer Res.10, 5889–5894. https://doi.org/10.1158/1078-0432.ccr-04-0144 (2004).
Li, M. et al. ABC transporter polymorphisms are associated with irinotecan pharmacokinetics and neutropenia. Pharmacogenom. J.18, 35–42. https://doi.org/10.1038/tpj.2016.75 (2018).
Haaz, M. C., Rivory, L., Riche, C., Vernillet, L. & Robert, J. Metabolism of irinotecan (CPT-11) by human hepatic microsomes: Participation of cytochrome P-450 3A and drug interactions. Cancer Res.58, 468–472 (1998).
Falvella, F. S. et al. Undetected toxicity risk in pharmacogenetic testing for dihydropyrimidine dehydrogenase. Int. J. Mol. Sci.16, 8884–8895. https://doi.org/10.3390/ijms16048884 (2015).
Acknowledgements
The authors thank (a) Suwannee Sirilerttrakul and Somthawin Lukerak for colorectal cancer collections; (b) Department of Pathology, Faculty of Medicine Ramathibodi Hospital, Mahidol University for the support of facilities; (c) Yupin Wisetpanit for colorectal cancer samples bank; and (d) This study was supported by the grants of the Ramathibodi Hospital Cancer Center, Bangkok, Thailand.
Author information
Authors and Affiliations
Contributions
All authors helped to perform the research; C.A. contribution included sample collection, data analysis and manuscript writing; P.C., E.S., T.R. contributed sample collection, drafting conception and design; S.S. contributed sample collection; M.C., A.P. contributed drafting conception and design; C.S. contributed conception and study design, data analysis, writing and revising the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Atasilp, C., Chansriwong, P., Sirachainan, E. et al. Effect of drug metabolizing enzymes and transporters in Thai colorectal cancer patients treated with irinotecan-based chemotherapy. Sci Rep 10, 13486 (2020). https://doi.org/10.1038/s41598-020-70351-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-70351-0
This article is cited by
-
Irinotecan-Induced Toxicity: A Pharmacogenetic Study Beyond UGT1A1
Clinical Pharmacokinetics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.