Abstract
Toll-like receptors (TLRs) recognize pathogen-associated molecular patterns and activate innate and adaptive immune responses. Single nucleotide polymorphisms (SNPs) within the TLR genes may influence host–pathogen interactions and can have an impact on the progression of infectious diseases. The present study aimed to investigate the genotype distribution of TLR2 (2029C/T, rs121917864; 2258G/A, rs5743708), TLR4 (896A/G, rs4986790), and TLR9 (− 1237T/C, rs5743836; − 1486T/C, rs187084; 1174G/A, rs352139; and 2848C/T, rs352140) polymorphisms in 149 children and adolescents with infectious mononucleosis (IM) and 140 healthy individuals. The potential association of TLR SNPs with the clinical manifestations of EBV infection was also studied. The presence of TLR2, TLR4, and TLR9 SNPs was identified by polymerase chain reaction–restriction fragment length polymorphism (PCR–RFLP). EBV DNA loads were detected by quantitative real-time PCR assay. The TLR4 896 GG and the TLR9 1174 GA genotypes were associated with an increased risk of EBV-related IM in examined patients (p = 0.014 and p = 0.001, respectively). The heterozygous genotype of the TLR4 896A/G SNP was associated with an increased risk of elevated liver enzyme levels and leukocytosis (p < 0.05). Our preliminary study revealed that the TLR4 896A/G and the TLR9 1174G/A polymorphisms seem to be related to the course of acute EBV infection in children and adolescents.
Similar content being viewed by others
Introduction
Epstein–Barr virus (EBV) is an oncogenic, human γ-herpesvirus and one of the most ubiquitous pathogens, infecting more than 90% of the adult population worldwide1,2. Primary EBV infection is usually asymptomatic and occurs during infancy or childhood. In developing countries, however, primary EBV infection is often delayed until adolescence and early adulthood and it occurs as infectious mononucleosis (IM). It is a self-limiting disease characterized by fatigue, fever, headache, lymphadenopathy, exudative pharyngitis, enlarged spleen and liver, in laboratory findings with elevated activity of the liver enzyme in serum and the occurrence of atypical-lymphocytes in blood smear1,3,4. After primary infection, EBV becomes latent within the B lymphocytes and epithelial cells5,6. The virus establishes lifelong persistent infection in normal immunocompetent hosts as well as it is able to be reactivated for spreading to new hosts. Reactivated from latency EBV may be associated with the development of a wide spectrum of malignancies such as Burkitt’s lymphoma (BL), Hodgkin and non-Hodgkin lymphomas (NHL), posttransplant lymphoproliferative disorder (PTLD), as well as epithelial tumors such as nasopharyngeal and gastric carcinomas7,8. An association between infectious mononucleosis and an increased risk of classical Hodgkin lymphoma in young adults was observed9,10.
Toll-like receptors (TLRs) are a family of transmembrane, evolutionarily conserved receptors that recognize molecular patterns unique to pathogens and activate host innate immune response11. The innate immune response to EBV is initiated after the recognition of its major envelope glycoprotein gp350/220 by TLR2 or activation of complement receptors CR2/CD21 and/or CR1/CD35 on B cells12,13,14,15. In addition, EBV-encoded non-structural protein dUTPase, expressed during the lytic cycle of viral replication, activates the nuclear factor кB (NF-кB) via adaptor myeloid differentiated 88 (MyD88)-dependent signaling cascade and induces expression of proinflammatory cytokines in macrophages16. In plasmacytoid dendritic cells (pDCs) and primary monocytes, infection of EBV activates TLR7 and/or TLR9 signaling pathway leading to interferon (IFN)-α and IFN-γ production17,18,19. Although many studies on TLRs in EBV-associated malignancies have been carried out in the past years, little is known about the role of TLRs in the pathogenesis of IM in humans. Rare single TLRs mutations have been associated with increased susceptibility to herpesvirus infections20,21,22. Understanding the genetic basis of susceptibility to EBV infections is important to the development of antiviral strategies. In the present study, we have assessed the distribution of the seven single nucleotide polymorphisms (SNPs) in the TLR2 (2029C/T, 2258G/A), the TLR4 (896A/G), and the TLR9 (− 1237T/C, − 1486T/C, 1174G/A, and 2848C/T) genes in children and adolescents with acute EBV infection.
Results
Clinical outcome assessment
At the time of this study 149 participants with acute IM, including 93 children (median age: 4.3 years; range 3 months–9.0 years) and 56 adolescents (median age: 14.2 years; range 10.0 years–17.5 years), and 140 healthy volunteers were examined. In all patients with IM, EBV infection was confirmed by the serologic and/or virologic tests. In brief, EBV-IgM was positive for 109 individuals, viral capsid antigen (VCA)-IgG for 46, EBV-IgG for 71, and EBNA-IgG was positive for 30 participants. Almost all patients (38/40) in whom specific IgM was undetected were children < 7 years of age. The results are presented separately for the two patient groups (children and adolescents) in Table 1. Higher frequency of EBV-specific IgM in adolescents than children (p = 0.008) was found, whereas children presented higher levels of the EBNA IgG antibodies (p = 0.037). The EBV DNA was detected in whole blood samples from 69 (74.2%) children and 35 (62.5%) adolescents (mean 2.13 × 104 copies/ml ± 1.23 × 105 copies/ml and mean 1.20 × 104 copies/ml ± 4.74 × 104 copies/ml, respectively). In all patients with IM, the presence of the EBV DNA in mouth saliva and/or throat swabs was found (mouth saliva: mean 8.77 × 105 copies/ml ± 1.75 × 106 copies/ml; throat swabs: mean 5.55 × 104 copies/ml ± 2.23 × 105 copies/ml). Patients were classified as having clinically confirmed IM when the following signs or symptoms were observed: atypical-lymphocytes, pharyngitis, lymphadenopathy, fever, fatigue, and enlarged spleen and/or liver. Clinical manifestations of acute IM were found in all examined children and youth patients. However, as expected from other studies, specific signs and symptoms of IM (elevated liver enzyme levels, atypical-lymphocytes, and thrombocytopenia) were detected more frequently in adolescents than in children (Table 1). It was found that elevated liver enzyme levels were more common in adolescents than children (82.1% vs. 61.3%, p = 0.010). There were also more adolescents than children with atypical-lymphocytes (66.1% vs. 48.4%, p = 0.042) and thrombocytopenia (42.9% vs. 19.4%, p = 0.003). In contrast, incidents of amoxicillin rash were observed more frequently among children than adolescents (25.8% vs. 10.7%, p = 0.034).
The heterozygous genotype of the TLR9 1174G/A SNP occurs more frequently in patients with IM
The TLR2 2029C/T, 2258G/A, TLR4 896A/G, and TLR9 − 1237T/C, − 1486T/C, 1174G/A, and 2848C/T SNPs were genotyped in 149 patients with IM and 140 healthy volunteers (Table 2). For the TLR2 2029C/T SNP and the TLR4 896A/G SNP, the homozygous recessive genotypes were detected more frequently in children and adolescents with IM than in healthy subjects (4.0% vs. 0.0%, p = 0.030 and 6.7% vs. 0.7%, p = 0.011, respectively; Table 2). Consequently, the wild-type C alleles of these SNPs were detected more frequently in uninfected individuals compared with EBV-infected cases (p = 0.039 and p = 0.004, respectively; Table 2). The heterozygous genotype of the TLR9 1174G/A polymorphism was more common in patients with IM than in healthy individuals (47.0% vs. 30.0%, p = 0.004), while no difference in the frequency of the alleles was observed (p > 0.05). Mutation present in at least one allele of the TLR9 2848C/T SNP occurred more frequently in patients with IM than in healthy subjects (55.4% vs. 38.9%, p = 0.0001; Table 2). No significant differences for the TLR2 2258G/A and the TLR9 − 1237T/C and − 1486C/T SNPs were found. Moreover, no sex differences in the TLR SNPs frequency among patients with IM were observed. The expected genotype frequencies for TLR9 − 1486T/C and 2848C/T SNPs were not in Hardy–Weinberg Equilibrium (HWE; p < 0.001 for both) and were excluded from further analysis.
The TLR4 896 GG and the TLR9 1174 GA genotypes are associated with increased risk of IM
The TLR4 896 GG genotype was associated with a tenfold increased risk of IM (OR 10.00; 95% CI 1.26–79.14; p = 0.004, in the recessive model; Table 3). This SNP showed a higher risk of IM even after Bonferroni correction for multiple testing (pB = 0.01). A higher risk of IM in patients with the TLR9 1174 GA genotype was also found (OR 1.90; 95% CI 1.17–3.11; p = 0.001, in the codominant model), although a lower incidence of IM in patients with AA genotype was observed (OR 0.18; 95% CI 0.04–0.82; p = 0.010, in the recessive model). In addition, IM patients with the heterozygous genotype of the TLR2 2029C/T had a slightly increased risk of the disease compared to healthy individuals (OR 1.39; 95% CI 0.84–2.29; p = 0.008, in the codominant model).
Polymorphisms in TLR genes influence the risk of the IM symptoms
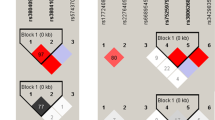
The TLR2 2029 CC genotype was associated with elevated liver enzyme levels (OR 3.025; 95% CI 1.321–6.927; p = 0.009) and leukocytosis (OR 2.574; 95% CI 1.129–5.866; p = 0.024) (Table 4). In patients with this SNP, the liver enzyme levels ranged as follows: serum aspartate aminotransferase (AST), 18–467 IU/l; alanine aminotransferase (ALT), 11–907 IU/l; and gamma-glutamyl transpeptidase (GGT), 12–196 IU/l. In contrast, the heterozygous genotype of the TLR2 2029C/T SNP was detected in 50.0% (15/30) patients with rash and was associated with at least threefold increased risk of incidence of this finding (OR 3.435; 95% CI 1.363–8.660; p = 0.009). The carriers of the heterozygous variant of the TLR4 896 A/G SNP had at least tenfold higher risk of elevated ALT and AST levels (OR 10.000; 95% CI 1.122–89.113; p = 0.039 and OR 13.846; 95% CI 1.544–124.135; p = 0.019, respectively) (Table 4). This genotype was also associated with an almost sevenfold increased risk of thrombocytopenia (OR 6.909; 95% CI 1.325–36.035; p = 0.022), although this genotype was detected in a small number of patients (14.5%). For the TLR9 1174G/A SNP, the GA genotype was associated with higher risk of elevated liver enzyme levels and leukocytosis (OR 3.055; 95% CI 1.333–7.000; p = 0.008 and OR 2.412; 95% CI 1.143–5.088; p = 0.021, respectively), whereas the risk of these symptoms were decreased in patients with wild-type genotype (OR 0.370; 95% CI 0.164–0.836; p = 0.017 and OR 0.447; 95% CI 0.213–0.939; p = 0.034, respectively). Pie charts show the percentage distribution of the mononucleosis symptoms in patients with the TLR2 2029C/T and the TLR9 1174G/A SNPs (Fig. 1). Furthermore, coexistent mutations in the TLR4 (896 A/G SNP) and the TLR9 (117G/A SNP) genes were detected in 11/104 (10.6%) of IM patients and were associated with an almost a sevenfold higher risk of elevated AST level (OR 6.827; 95% CI 1.247–37.380; p = 0.027, unadjusted model; OR 8.750; 95% CI 1.331–57.528; p = 0.024, adjusted model). A tendency towards a higher risk of occurrence of atypical-lymphocytes in patients with mutation in both SNPs was also observed (OR 7.420; 95% CI 0.881–62.477; p = 0.065, unadjusted model; OR 8.272; 95% CI 0.887–77.130; p = 0.044, adjusted model). No other significant correlation between coexisting SNPs and IM symptoms or viral load were found (p > 0.05).
The wild-type genotype of the TLR2 2029C/T polymorphism is associated with the EBV replication
To further examine the association between the TLR polymorphisms and viral infection, we correlated the specific TLR SNPs with the EBV DNA levels in the peripheral blood of IM patients. As shown in Fig. 2, the median EBV DNA levels in blood were lower among individuals who had a wild-type genotype for the TLR2 2029C/T (mean 1.09 × 104 copies/ml ± 2.66 × 104 copies/ml) compared with those who were heterozygous or homozygous recessive (mean 3.40 × 104 copies/ml ± 1.58 × 105 copies/ml) for this polymorphism (p = 0.014). Age group-specific analyses confirmed that children carrying the wild-type genotype for the TLR2 2029C/T SNP had lower EBV DNAemia than those with heterozygous or homozygous recessive genotypes (mean 1.37 × 104 copies/ml ± 3.24 × 104 copies/ml vs. mean 3.21 × 104 copies/ml ± 1.9 × 105 copies/ml; p = 0.015). Such a correlation was observed also for adolescents (mean 7.81 × 103 copies/ml ± 1.59 × 104 copies/ml vs. mean 2.07 × 104 copies/ml ± 8.05 × 104 copies/ml; p = 0.042, respectively). No association was observed between the EBV DNAemia and any other TLR polymorphisms (p > 0.05) in both study groups.
TLR haplotypes in patients with IM
Haplotype analysis of the TLR2 2029C/T, 2258G/A, the TLR4 896A/G, and the TLR9 − 1237T/C, and 1174G/A SNPs showed that the most frequent haplotype was CGATG (53.5%), which was detected in 48.9% of patients with IM and 58.1% of healthy individuals. The CAATA haplotype was detected at a minor frequency in 1.5% of patients with IM and 2.0% of healthy volunteers, whereas haplotype TAATG was determined only in EBV-infected patients (3.8%). We found no evidence for linkage disequilibrium (LD) for the all examined TLR SNPs (p > 0.05, r2 < 0.2).
Discussion
TLRs have been demonstrated to play a crucial role in modulating innate recognition of viruses not only by serving as pathogen sensors but also by activating signaling pathways that result in the increased production of proinflammatory cytokines and type I IFNs. This preliminary study provides the first evidence that TLR polymorphisms seem to influence the outcome of the infectious mononucleosis in children and adolescents. Patients with the TLR4 896A/G and the TLR9 1174G/A polymorphisms had a higher risk of IM and significantly increased the frequency of specific manifestations when compared with subjects with the wild-type genotype.
No studies were examining the allele frequency distribution and the role of TLR SNPs in patients with EBV-related IM. We observed that the TLR4 896A/G SNP and an intronic polymorphism 1174G/A in exon 2 of the TLR9 gene were significantly associated with increased risk of EBV infection in our population. Polymorphisms in TLR4 gene, which is located in chromosome 9q33.1, has been implicated so far in the pathogenesis of several infectious diseases, including bacterial23,24,25, fungal26, parasitic27, and some viral infection, such as human immunodeficiency virus28,29, Kaposi sarcoma-associated herpesvirus30, and respiratory syncytial virus (RSV)31. It was previously described that missense mutations and single nucleotide polymorphisms in the TLR4 gene could alter the function of the receptor and impairs recognition of the pathogens30,32. The aspartic acid to glycine substitution at residue 299 polymorphism (896A/G) of the TLR4 was found to be associated with susceptibility to bacterial pneumonia, possibly through impaired first-line defense mechanisms33. Individuals carrying the TLR4 896A/G polymorphisms were significantly less responsive to bacterial peptides derived from Escherichia coli and Porphyromonas gingivalis compared with the wild-type subjects34,35. Functional studies have demonstrated that this SNP was also associated with impaired NF-κB and IFN regulatory factor 3 activation in response to lipopolysaccharide, F protein from RSV, and chlamydial Hsp6036. It was found that TLR9 contributes to the recognition of EBV and is expressed on B cells, a natural target of the virus infection18,37. The TLR9 1174 G/A SNP was linked to rapid disease progression in children with malaria and HIV-infected patients38,39 and was also associated with nonresponse to anti-TNF treatment among patients with inflammatory bowel disease40. In our previous studies, no significant differences in the frequencies of these polymorphisms were detected among CMV-infected and healthy infants within the same ethnic group20,41. However, a reduced risk of the CMV infection in infants with the 1174 GG genotype was noticed20. We suggest that both the TLR4 896A/G and the TLR9 1174 G/A polymorphisms may influence the immune response and impair NF-κB activation in EBV infection.
Several studies have implicated that TLR polymorphisms are involved in the development of lymphoid and non-lymphoid EBV-associated malignancies42,43,44,45,46,47,48,49,50. It has been previously reported that the − 16933T/A SNP in the TLR2 gene increased the risk of follicular lymphoma and decreased the risk of chronic lymphocytic leukemia43. Mutation present in both alleles of the TLR2 2029C/T and 2258G/A SNPs were associated with susceptibility to gastric carcinoma in China42. In the TLR4 gene, the 896A/G polymorphism influences the risk of mucosa-associated lymphoid tissue lymphoma and Hodgkin lymphoma42, but not with non-Hodgkin lymphoma45,46. The combined CT/TT genotype of the 1196C/T SNP and the GC genotype of the 11350G/C polymorphism in the 3′-untranslated region of the TLR4 gene may alter its expression, influence the expression of inflammatory cytokines and chemokines, and increased the risk of nasopharyngeal carcinoma (NPC)49,50. The occurrence and progression of NPC were also associated with the TLR9 − 1486T/C SNP48. The TLR9 − 1486 CC genotype increased the susceptibility of NPC in the Chinese population and patients with this genotype were inclined to advanced tumor stage and lymph node metastasis. No association between the TLR9 − 1237T/C and 2848C/T SNPs and the risk of NPC was found48. Nevertheless, carriers of the − 1237 C and 2848 A allele had been reported to be associated with an increased risk of Hodgkin’s lymphoma in the Caucasian population47. The − 1237T/C polymorphism was associated with non-Hodgkin lymphoma in Portuguese and Italian, but not in the United States cohort of patients44. Analysis in silico of the TLR9 promoter showed that the mutated − 1237T/C and − 1486T/C variants create a putative c-Rel/NF-kB transcription factor binding sites that influence transcriptional regulation of the gene51. The NF-kB binding site in the C allele at the position − 1237 enhanced the transcriptional activity of the TLR9 gene and affect activation of proinflammatory cytokines, chemokines, and the adaptive immune response52. In contrast, another study revealed that the TT allelic variant of − 1237T/C SNP had higher transcriptional activity53.
A few investigations indicated that the risk of developing infectious mononucleosis after primary infection with EBV correlates with the age of patients and adolescents would react more strongly against EBV infection than children54,55,56. It is also known that the frequency of IM symptoms depends on age, ethnicity and geographic location57,58,59. Most people are exposed to the virus as children, when the disease produces flu-like symptoms such as pharyngitis, whereas a characteristic triad of fever, pharyngitis, and lymphadenopathy is more frequently presented in adolescents and young adults. In this study, we observed that elevated liver enzyme levels, thrombocytopenia, as well as higher atypical-lymphocytes were more frequent in adolescents than children. This result supports the hypothesis that the prevalence of acute IM rises significantly with patient age, and is common in adolescents54,55. These findings are coincident with that described by Wang et al., who demonstrated that young patients had significantly increased levels of liver enzymes and atypical-lymphocytes, while no significant difference in the prevalence of fever between children and adolescents was observed58. The most significant laboratory abnormality found in the present study was atypical-lymphocytes, which occurred more frequently in adolescents than in children (66.1% vs. 48.5%). The low platelet count, in course of IM, was observed in 26.3% adult patients in France (age 16–53 years)59, less than 1% young adults in the United States60, 2.3–4.5% preschool children and youth patients in China58,61, and 7.3% Mexican children (age 0–17.5 years)62. The higher incidence of rash in children than in adolescents (25.8% vs. 10.7%) was observed in our study. Almost one-third population of Israeli patients up to 18 years had a rash during the IM, but the age of the patients was not associated with the development of this symptom57. Similarly, no statistically significant difference in patient age and the incidence of rash was found among preschool children (21.5%) and youth patients (13.9%) in Beijing, China58. Part of the explanation that would elucidate the more frequent risk of IM in adolescents and young adults than in children is that the percentage of CD8+ T cell counts increased and CD4+ T cell counts decreased with age increment61. Moreover, significantly higher levels of early-differentiated CD56dim NKG2A+ killer-cell immunoglobulin-like receptors (KIR)− NK cells in the peripheral blood of children than in adolescents and young adults may affect the course of EBV infection63,64. It was also found that polymorphisms in the HLA class I locus may predispose patients to the development of IM upon primary EBV infection65. McAulay et al. described that the nature of primary EBV infection and the level of viral persistence can be determined by the genetic variation in T cell responses65. It is also possible that adolescents receive a larger amount of the virus through deep kissing, while young children probably acquire EBV from parents or guardians, who during reactivation of infection transmit smaller infectious amounts of EBV, or from siblings or other children54,66,67. Furthermore, the heterophile antibody test is less sensitive and often unreliable in young children3,56,68. It is also known that EBV VCAs cause lifelong persistent IgG titers, while antibodies of the IgM type are produced only transiently but are not necessarily produced in all patients with primary infections69. However, to understand the differential impact age on susceptibility to primary EBV infection, further research in age-matched patient groups are needed.
The present study is first to examine the distribution and possible association between the TLR gene polymorphisms and the clinical and/or laboratory findings of infectious mononucleosis. Similar to other genetic association studies, this study has some potential limitations. First, the present study may be limited by a relatively small number of participants. However, we enrolled patients from a well-characterized population and all noticeable symptoms were verified by experienced clinicians. Moreover, serological documentation of EBV infection indicates the higher incidence of advanced-stage disease or the delayed recognition of IM in the examined children than adolescents. Further studies in a larger sample group are needed to confirm our findings and to evaluate the function of the disease-associated TLR polymorphisms.
Methods
Study population
A total of 289 blood samples was obtained from 149 patients with IM (median age: 8.2 years; range 3 months–17.5 years) and 140 unrelated seronegative and aviremic healthy volunteers, without clinical symptoms/signs of EBV infection. Whole mouth saliva and throat swabs samples from patients with IM were also collected for medical research. The subjects involved in the study were recruited from the Department of Pediatric Infectious Diseases, Wroclaw Medical University in Wroclaw (between April 2014 and August 2018) and Polish Mother’s Memorial Hospital Research Institute in Lodz (between September 2011 and April 2018). The presence of EBNA-IgG, EBV-IgM, and EBV-IgG or VCA-IgG was measured using an immunochemiluminescence assay (CLIA, DiaSorin, Saluggia, Italy) according to the manufacturer’s instructions. The presence of EBV DNA in the whole blood and/or the presence of IgM antibodies to VCA in the absence of measurable antibodies to EBNA-1 was considered the evidence for primary EBV infection as described previously3,70. AST, ALT, and GGT were determined by dry chemistry method using Johnson & Johnson Vitros 5.1 FS Chemistry Analyzer (Ortho Clinical Diagnostics, Raritan, NJ, USA) or by the kinetic method using Architect 2000 (Abbott Laboratories, Abbott Park, IL, USA). Mild elevations of ALT and AST were commonly discovered in individuals with mild or no symptoms and were defined as increased liver enzyme values higher than 2–5 times the upper limit of normal. Liver disease was diagnosed in patients with elevated liver enzyme levels greater than 5 times the upper limit of the reference range. Platelet counts were tested using optical and/or impedance methods using the Sysmex XE-2000 Analyzer (Sysmex Corp., Kobe, Japan). Thrombocytopenia was recognized when the platelet count fell below 150 × 109/l of blood, whereas leukocytosis was classified as elevated white blood cells count above the age-specific reference values. All the individuals with EBV infection were ethnically classified as European descents and were enrolled from the Central and South-West areas of Poland. Clinical data from EBV infected patients were summarized in Table 1. This study was approved by the appropriate Bioethics Committee of the Medical University of Lodz (RNN/120/09/KE) and the Ethics Committee of the Polish Mother’s Memorial Hospital Research Institute (7/2014). Written informed consent was provided from parents of all children who participated in the study. All experiments were performed in accordance with relevant guidelines and regulations.
Genotyping of TLRs polymorphisms
Total genomic DNA was extracted from the peripheral blood, whole mouth saliva, and throat swabs samples using the QIAamp DNA Blood Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instruction. Molecular typing of TLR2 (2029C/T, rs121917864; 2258G/A, rs5743708), TLR4 (896A/G, rs4986790) and TLR9 (− 1237T/C, rs5743836; − 1486T/C, rs187084; 1174G/A, rs352139; and 2848C/T, rs352140) SNPs were performed by polymerase chain reaction–restriction fragment length polymorphism (PCR–RFLP) as described elsewhere20,41. The results were confirmed by the sequencing of randomly selected samples of each TLR gene using the BigDye Terminator v3.1 Cycle Sequencing Kit and the 96-capillary 3730xl DNA Analyzer (Applied Biosystems).
Assessment of EBV replication
EBV DNA copy numbers in clinical samples were determined using a 7900HT Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) with a commercially available Epstein Barr Virus (Human Herpes virus 4) nonglycosylated membrane protein (BNRF1) gene Kit (Primerdesign Ltd., York House, UK) according to the manufacturer’s instruction. The PCR conditions were as follows: 2 min at 95 °C, followed by 50 cycles of 95 °C for 10 s and 60 °C for 60 s. A negative control without template DNA was included in every amplification run. The sensitivity of the assay has been determined to be 200 copies/ml of whole blood, mouth saliva, and throat swabs.
Statistical analysis
Data were statistically analyzed using the GraphPad Prism 5.00 (GraphPad Software, San Diego, CA, USA) and SPSS statistical software package for Windows 25.0 (SPSS, Chicago, IL, USA). Categorical data were analyzed by the Chi-square test and Fisher’s exact test, while the Mann–Whitney U test was used to estimate the association between TLR SNPs and the viral load. The association of polymorphisms and clinical characteristics of the patients was estimated by odds ratio (OR) with a 95% confidence interval (95% CI). A p-value of less than 0.05 was considered statistically significant. The HWE, LD, and haplotype analyses were performed using the SNPStats software (https://www.snpstats.net/start.htm). The Bonferroni correction of the significance level was applied for five multiple comparisons; the significance level for pB is 0.01 instead standard 0.05.
References
Stock, I. Infectious mononucleosis-a “childhood disease” of great medical concern. Med. Monatsschr. Pharm. 36, 364–368 (2013).
Macsween, K. F. & Crawford, D. H. Epstein–Barr virus-recent advances. Lancet Infect. Dis. 3, 131–140 (2003).
Balfour, H. H. Jr. et al. Behavioral, virologic, and immunologic factors associated with acquisition and severity of primary Epstein–Barr virus infection in university students. J. Infect. Dis. 207, 80–88 (2013).
Rea, T. D., Russo, J. E., Katon, W., Ashley, R. L. & Buchwald, D. S. Prospective study of the natural history of infectious mononucleosis caused by Epstein–Barr virus. J. Am. Board. Fam. Pract. 14, 234–242 (2001).
Hadinoto, V. et al. On the dynamics of acute EBV infection and the pathogenesis of infectious mononucleosis. Blood 111, 1420–1427 (2008).
Kurth, J. et al. EBV-infected B cells in infectious mononucleosis: Viral strategies for spreading in the B cell compartment and establishing latency. Immunity 13, 485–495 (2000).
Maeda, E. et al. Spectrum of Epstein–Barr virus-related diseases: A pictorial review. Jpn. J. Radiol. 27, 4–19 (2009).
Young, L. S. & Rickinson, A. B. Epstein–Barr virus: 40 years on. Nat. Rev. Cancer. 4, 757–768 (2004).
Hjalgrim, H. et al. Infectious mononucleosis, childhood social environment, and risk of Hodgkin lymphoma. Cancer Res. 67, 2382–2388 (2007).
Hjalgrim, H. et al. Characteristics of Hodgkin’s lymphoma after infectious mononucleosis. N. Engl. J. Med. 349, 1324–1332 (2003).
Kawai, T. & Akira, S. TLR signaling. Semin. Immunol. 9, 24–32 (2007).
Ogembo, J. G. et al. Human complement receptor type 1/CD35 is an Epstein–Barr virus receptor. Cell Rep. 3, 371–385 (2013).
Gaudreault, E., Fiola, S., Olivier, M. & Gosselin, J. Epstein–Barr virus induces MCP-1 secretion by human monocytes via TLR2. J. Virol. 81, 8016–8024 (2007).
Tanner, J., Weis, J., Fearon, D., Whang, Y. & Kieff, E. Epstein–Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell 50, 203–213 (1987).
Fingeroth, J. D. et al. Epstein–Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc. Natl. Acad. Sci. U. S. A. 81, 4510–4514 (1984).
Ariza, M. E., Glaser, R., Kaumaya, P. T., Jones, C. & Williams, M. V. The EBV-encoded dUTPase activates NF-kappa B through the TLR2 and MyD88-dependent signaling pathway. J. Immunol. 182, 851–859 (2009).
Valente, R. M. et al. Toll-like receptor 7 stimulates the expression of Epstein–Barr virus latent membrane protein 1. PLoS ONE 7, e43317 (2012).
Fiola, S., Gosselin, D., Takada, K. & Gosselin, J. TLR9 contributes to recognition of EBV by primary monocytes and plasmacytoid dendritic cells. J. Immunol. 185, 3620–3631 (2010).
Lim, W. H., Kireta, S., Russ, G. R. & Coates, P. T. Human plasmacytoid dendritic cells regulate immune responses to Epstein–Barr virus (EBV) infection and delay EBV-related mortality in humanized NOD-SCID mice. Blood 109, 1043–1050 (2007).
Paradowska, E. et al. TLR9 -1486T/C and 2848C/T SNPs are associated with human cytomegalovirus infection in infants. PLoS ONE 11, e0154100 (2016).
Taniguchi, R. et al. Polymorphisms in TLR-2 are associated with congenital cytomegalovirus (CMV) infection but not with congenital CMV disease. Int. J. Infect. Dis. 17, e1092-1097 (2013).
Bochud, P. Y., Magaret, A. S., Koelle, D. M., Aderem, A. & Wald, A. Polymorphism in TLR2 are associated with increased viral shedding and lesional rate in patients with genital herpes simplex virus Type 2 infection. J. Infect. Dis. 196, 505–509 (2007).
Rani, A., Nawaz, S. K., Arshad, M. & Irfan, S. Role of rs4986790 polymorphism of TLR4 gene in susceptibility towards malaria infection in the Pakistan population. Iran J. Public Health. 47, 735–741 (2018).
Loganathan, R. et al. Genetic variants of TLR4 and TLR9 are risk factors for chronic Helicobacter pylorii infection in South Indian Tamils. Hum. Immunol. 78, 216–220 (2017).
Sampath, V. et al. Toll-like receptor genetic variants are associated with Gram-negative infections in VLBW infants. J. Perinatol. 33, 772–777 (2013).
Carvalho, A. et al. Polymorphisms in toll-like receptor genes and susceptibility to pulmonary aspergillosis. J. Infect. Dis. 197, 618–621 (2008).
Ziakas, P. D., Prodromou, M. L., El Khoury, J., Zintzaras, E. & Mylonakis, E. The role of TLR4 896 A>G and 1196 C>T in susceptibility to infections: A review and meta-analysis of genetic association studies. PLoS ONE 8, e81047 (2013).
Vidyant, S., Chatterjee, A. & Dhole, T. N. A single-nucleotide polymorphism in TLR4 is linked with the risk of HIV-1 infection. Br. J. Biomed. Sci. 76, 59–63 (2019).
Papadopoulos, A. I. et al. Association of toll-like receptor 4 Asp299Gly and Thr399Ile polymorphisms with increased infection risk in patients with advanced HIV-1 infection. Clin. Infect. Dis. 51, 242–247 (2010).
Lagos, D. et al. Toll-like receptor 4 mediates innate immunity to Kaposi sarcoma herpesvirus. Cell Host Microbe. 4, 470–483 (2008).
Tal, G. et al. Association between common toll-like receptor 4 mutations and severe respiratory syncytial virus disease. J. Infect. Dis. 189, 2057–2063 (2004).
Arbour, N. C. et al. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat. Genet. 25, 187–191 (2000).
Tachado, S. D. et al. MyD88-dependent TLR4 signaling is selectively impaired in alveolar macrophages from asymptomatic HIV+ persons. Blood 115, 3606–3615 (2010).
Kinane, D. F. et al. Gingival epithelial cells heterozygous for toll-like receptor 4 polymorphisms Asp299Gly and Thr399Ile are hypo-responsive to Porphyromonas gingivalis. Genes Immun. 7, 190–200 (2006).
Miller, S. I., Ernst, R. K. & Bader, M. W. LPS, TLR4 and infectious disease diversity. Nat. Rev. Microbiol. 3, 36–46 (2005).
Figueroa, L. et al. The Asp299Gly polymorphism alters TLR4 signaling by interfering with recruitment of MyD88 and TRIF. J. Immunol. 188, 4506–4515 (2012).
Chijioke, O., Azzi, T., Nadal, D. & Münz, C. Innate immune responses against Epstein–Barr virus infection. J. Leukoc. Biol. 94, 1185–1190 (2013).
Omar, A. H. et al. Toll-like receptor 9 (TLR9) polymorphism associated with symptomatic malaria: A cohort study. Malar. J. 11, 168 (2012).
Bochud, P. Y. et al. Polymorphisms in toll-like receptor 9 influence the clinical course of HIV-1 infection. AIDS. 21, 441–446 (2007).
Bank, S. et al. Associations between functional polymorphisms in the NFκB signaling pathway and response to anti-TNF treatment in Danish patients with inflammatory bowel disease. Pharmacogenom. J. 14, 526–534 (2014).
Jabłońska, A. et al. Relationship between toll-like receptor 2 Arg677Trp and Arg753Gln and toll-like receptor 4 Asp299Gly polymorphisms and cytomegalovirus infection. Int. J. Infect. Dis. 25, 11–15 (2014).
Liu, S. et al. Toll-like receptor gene polymorphisms and susceptibility to Epstein–Barr virus-associated and -negative gastric carcinoma in Northern China. Saudi J. Gastroenterol. 21, 95–103 (2015).
Nieters, A., Beckmann, L., Deeg, E. & Becker, N. Gene polymorphisms in toll-like receptors, interleukin-10, and interleukin-10 receptor alpha and lymphoma risk. Genes Immun. 7, 615–624 (2006).
Carvalho, A. et al. The rs5743836 polymorphism in TLR9 confers a population-based increased risk of non-Hodgkin lymphoma. Genes Immun. 13, 197–201 (2012).
Purdue, M. P. et al. A pooled investigation of toll-like receptor gene variants and risk of non-Hodgkin lymphoma. Carcinogenesis 30, 275–281 (2009).
Forrest, M. S. et al. Polymorphisms in innate immunity genes and risk of non-Hodgkin lymphoma. Br. J. Haematol. 134, 180–183 (2006).
Mollaki, V. et al. Polymorphisms and haplotypes in TLR9 and MyD88 are associated with the development of Hodgkin’s lymphoma: A candidate-gene association study. J. Hum. Genet. 54, 655–659 (2009).
Dai, Q., Li, X. P., Chai, L., Long, H. A. & Yang, Z. H. Polymorphisms of toll-like receptor 9 are associated with nasopharyngeal carcinoma susceptibility. Tumour Biol. 35, 3247–3253 (2014).
Yang, Z. H., Dai, Q., Gu, Y. J., Guo, Q. X. & Gong, L. Cytokine and chemokine modification by toll-like receptor polymorphisms is associated with nasopharyngeal carcinoma. Cancer Sci. 103, 653–658 (2012).
Song, C., Chen, L. Z., Zhang, R. H., Yu, X. J. & Zeng, Y. X. Functional variant in the 3’-untranslated region of toll-like receptor 4 is associated with nasopharyngeal carcinoma risk. Cancer Biol. Ther. 5, 1285–1291 (2006).
Hamann, L. et al. Toll-like receptor (TLR)-9 promotor polymorphisms and atherosclerosis. Clin. Chim. Acta. 364, 303–307 (2006).
Ng, M. T. et al. Increase in NF-kappaB binding affinity of the variant C allele of the toll-like receptor 9-1237T/C polymorphism is associated with Helicobacter pylori-induced gastric disease. Infect. Immun. 78, 1345–1352 (2010).
Novak, N. et al. Putative association of a TLR9 promoter polymorphism with atopic eczema. Allergy 62, 766–772 (2007).
Balfour, H. H. Jr., Dunmire, S. K. & Hogquist, K. A. Infectious mononucleosis. Clin. Transl. Immunol. 4, e33 (2015).
Odumade, O. A., Hogquist, K. A. & Balfour, H. H. Jr. Progress and problems in understanding and managing primary Epstein–Barr virus infections. Clin. Microbiol. Rev. 24, 193–209 (2011).
Luzuriaga, K. & Sullivan, J. L. Infectious mononucleosis. N. Engl. J. Med. 362, 1993–2000 (2010).
Chovel-Sella, A. et al. Incidence of rash after amoxicillin treatment in children with infectious mononucleosis. Pediatrics 131, e1424–e1427 (2013).
Wang, Y., Li, J., Ren, Y. Y. & Zhao, H. The levels of liver enzymes and atypical lymphocytes are higher in youth patients with infectious mononucleosis than in preschool children. Clin. Mol. Hepatol. 19, 382–388 (2013).
Tattevin, P. et al. Increasing incidence of severe Epstein–Barr virus-related infectious mononucleosis: Surveillance study. J. Clin. Microbiol. 44, 1873–1874 (2006).
Dunmire, S. K., Hogquist, K. A. & Balfour, H. H. Infectious mononucleosis. Curr. Top. Microbiol. Immunol. 390, 211–240 (2015).
Gao, L. W., Xie, Z. D., Liu, Y. Y., Wang, Y. & Shen, K. L. Epidemiologic and clinical characteristics of infectious mononucleosis associated with Epstein–Barr virus infection in children in Beijing, China. World J. Pediatr. 7, 45–49 (2011).
González Saldaña, N., Monroy Colín, V. A., Piña Ruiz, G. & Juárez Olguín, H. Clinical and laboratory characteristics of infectious mononucleosis by Epstein–Barr virus in Mexican children. BMC Res. Notes. 5, 361 (2012).
Azzi, T. et al. Role for early-differentiated natural killer cells in infectious mononucleosis. Blood 124, 2533–2543 (2014).
Chijioke, O. et al. Human natural killer cells prevent infectious mononucleosis features by targeting lytic Epstein–Barr virus infection. Cell Rep. 5, 1489–1498 (2013).
McAulay, K. A. et al. HLA class I polymorphisms are associated with development of infectious mononucleosis upon primary EBV infection. J. Clin. Investig. 117, 3042–3048 (2007).
Condon, L. M. et al. Age-specific prevalence of Epstein–Barr virus infection among Minnesota children: Effects of race/ethnicity and family environment. Clin. Infect. Dis. 59, 501–508 (2014).
Crawford, D. H. et al. A cohort study among university students: Identification of risk factors for Epstein–Barr virus seroconversion and infectious mononucleosis. Clin. Infect. Dis. 43, 276–282 (2006).
Ebell, M. H. Epstein–Barr virus infectious mononucleosis. Am. Fam. Phys. 70, 1279–1287 (2004).
Lennette, E. H. Epstein–Barr virus (EBV). In Diagnostic Procedures for Viral, Rickettsial, and Chlamydial Infections 7th edn (eds Lennette, E. H. et al.) 299–312 (American Public Health Association, Washington, DC, 1995).
Chan, K. H., Ng, M. H., Seto, W. H. & Peiris, J. S. Epstein–Barr virus (EBV) DNA in sera of patients with primary EBV infection. J. Clin. Microbiol. 39, 4152–4154 (2001).
Acknowledgements
This work was supported by the Statutory Fund of IBM PAS. The authors are grateful to Prof. Teresa Woźniakowska-Gęsicka (Polish Mother’s Memorial Hospital Research Institute, Lodz) for providing samples from patients, Miss Sarah Woźniak for technical assistance, and all volunteers for their valuable contribution in the study.
Author information
Authors and Affiliations
Contributions
A.J. designed the study, performed experiments, analyzed and interpreted the data, and drafted the manuscript. M.S. performed experiments. L.S., M.W.L., M.K.S., T.G. provided samples for the study and collected the patients’ data. E.P. designed the study, analyzed and interpreted the data, drafted the manuscript, and supervised the work. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jabłońska, A., Studzińska, M., Szenborn, L. et al. TLR4 896A/G and TLR9 1174G/A polymorphisms are associated with the risk of infectious mononucleosis. Sci Rep 10, 13154 (2020). https://doi.org/10.1038/s41598-020-70129-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-70129-4
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.