Abstract
The scientific community is exploiting the use of silver nanoparticles (AgNPs) in nanomedicine and other AgNPs combination like with biomaterials to reduce microbial contamination. In the field of nanomedicine and biomaterials, AgNPs are used as an antimicrobial agent. One of the most effective approaches for the production of AgNPs is green synthesis. Lysiloma acapulcensis (L. acapulcensis) is a perennial tree used in traditional medicine in Mexico. This tree contains abundant antimicrobial compounds. In the context of antimicrobial activity, the use of L. acapulcensis extracts can reduce silver to AgNPs and enhance its antimicrobial activity. In this work, we demonstrate such antimicrobial activity effect employing green synthesized AgNPs with L. acapulcensis. The FTIR and LC–MS results showed the presence of chemical groups that could act as either (i) reducing agents stabilizing the AgNPs or (ii) antimicrobial capping agents enhancing antimicrobial properties of AgNPs. The synthesized AgNPs with L. acapulcensis were crystalline with a spherical and quasi-spherical shape with diameters from 1.2 to 62 nm with an average size diameter of 5 nm. The disk diffusion method shows the magnitude of the susceptibility over four pathogenic microorganisms of clinical interest. The antimicrobial potency obtained was as follows: E. coli ≥ S. aureus ≥ P. aeruginosa > C. albicans. The results showed that green synthesized (biogenic) AgNPs possess higher antimicrobial potency than chemically produced AgNPs. The obtained results confirm a more significant antimicrobial effect of the biogenic AgNPs maintaining low-cytotoxicity than the AgNPs produced chemically.
Similar content being viewed by others
Introduction
The field of material sciences encourages to obtain materials of various types of nanoscale shapes and architectures1.NPs with a size range of 1–100 nm, and different shapes provide unique chemical2, physical3 and optical properties4,5. NPs can be synthesized with physical, chemical and biological methods6. These methods might have unique advantages and disadvantages depending on the end application7,8,9,10,11. For example, physical methods might have some disadvantages when applied in microbiology. The methods can be time-consuming and constrain to specific requirements like high temperature or pressure, which might result unattractive owing to equipment and associated cost12,13. A key advantage of chemical methods is the accessibility to get the NPs in suspension. After synthesis and purification, the NPs can immediately be accessible for functionality testing14. However, in some cases, the synthesis procedure might result expensive owing to the material type used (e.g., borohydride, 2-mercaptoethanol, thioglycerol and citrate). For chemical synthesis methods applied in microbiology, the most critical point is the toxic effect of the NPs or by-products generated, especially when released to the environment15. On the contrary, biological methods (e.g., plant extract) utilize fewer toxic reactants and additives. The reaction can occur at room temperature without harsh or stringent reaction constraints. Plant extract can then provide low or no cytotoxicity when combined with NPs. Therefore, biological methods using plant extracts could be cataloged safe, eco-friendly and low-cost, representing a viable alternative when applied in microbiology.
Microorganisms (yeast, fungi, bacteria, viruses and actinomycete) and plant-mediated synthesis are different primary sources for NPs synthesis by biological methods16. Synthesized NPs using plant extracts are more advantageous than intracellular synthesis using microorganisms since it does not need complex and specialized processes such as isolation, culture maintenance and multiple purification steps. Due to the above, it has become a major focus leading researchers to develop green methods using different parts of the plant, e.g., from leaf17,18,19, peel20 flower21, fruit22 and root23. Many compounds present in the plant extract (e.g., p1olyphenols, ascorbic acids, flavonoids, terpenoids and proteins) play an essential role in the mechanism of metal ions uptake, reduction of precursor salt as well as capping agents and some of them with inherent antimicrobial properties.
L. acapulcensis is an endemic perennial tree of the south of Mexico, is abundant and has been empirically used in traditional medicine because of its properties, curative for respiratory, gastrointestinal, urinary and skin infections24. Few reports describe the chemical composition of L. acapulcensis extracts25. The extract has abundant tannin compounds which grant access to antimicrobial properties26,27. AgNPs are one of the most promising antimicrobial materials applied in nanomedicine. AgNPs can interact with the microorganism cell wall, generating reactive oxygen species that ultimately leads to cell death. Therefore, we can hypothesize that the use of L. acapulcensis extract can form AgNPs with improved antimicrobial activity. The combination of AgNPs with L. acapulcensis can then make a promising alternative against infectious diseases with reduced cytotoxicity.
In this work, we report the in vitro antimicrobial activity of AgNPs synthesized by a green method using a L. acapulcensis extract. The AgNPs were tested against three bacterial strains (Escherichia coli, Pseudomonas aeruginosa and Staphylococcus aureus) and one yeast (Candida albicans) of clinical interest. Morphology, size distribution, elemental analysis and electron diffraction pattern of nanoparticles were characterized. Minimum inhibitory concentration (MIC) and Minimum Biocidal Concentration (MBC) for the biogenic AgNPs were established and compared with those obtained with AgNPs synthesized by a chemical method. Less cytotoxic effects were found for biogenic AgNPs.
Results
Biophysical characterization
The FTIR spectra of the L. acapulcensis aqueous extract was recorded to identify functional groups. Their phytochemical profile revealed the presence of alkyl halides, proteins, phenolic and aromatic compounds with transmission peaks at 592, 1631, 2340 and 1620 cm−1, respectively (Fig. 1). The FTIR band around 3422 cm−1 is assigned to intramolecular H bonds, most probably from water molecules (Fig. 1). Moreover, LC–MS suggests the presence of different molecular components (Figure S1 & Table S1–S2) with either antimicrobial, surfactant and reducing character. Other molecular compounds were also identified to be part of the extract (Figure S1 & Table S1–S2).
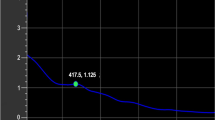
AgNPs were synthesized after the identification of the molecular structure and functional groups present in the L. acapulcensis extract. The AgNPs were first identified by a visual color change in the reaction mixture containing AgNO3 and L. acapulcensis extract over different incubation times (15, 30 and 60 min). Figure 2 shows the UV–Vis spectra (wavelengths ranging: 300–600 nm) recorded from each synthesized AgNPs. The color intensity increased with the duration of incubation time, turned from yellow to dark brown (Fig. 2). Biogenic AgNPs showed the maximum absorbance at 400 nm for solutions incubated for 15, 30 and 60 min, and the absorbance units increased in intensity (0.892, 1.125 and 1.488 a.u.) with time. No wavelength shift of the 400 nm peak was observed for the different reaction times. A color change of the suspension helped to determine AgNPs presence. Therefore, we use 15 min synthesis time for the experiments presented next.
X-ray diffraction (XRD) analysis
The crystalline nature of biogenic nanoparticles was confirmed by X-ray crystallography, recorded on a Panalitycal X’pert Pro MRD X-ray diffraction instrument, with Cu Kα radiation (λ = 0.15418 nm) over the scanning range 2θ = 30°–80°, with a step of 0.02 degree. The XRD pattern of the synthesized AgNPs (Fig. 3), shows several peaks, where the four main peaks located at 38.10°, 44.20°, 64.41° and 77.39°, corresponding to the (111), (200), (220) and (311) planes, respectively, to the facets of face-centered cubic (fcc) crystal structure of silver (JCPDS, No. 04-0783). An intense peak, located at 32.15° could be indexed to a cubic structure of AgK3 (PDF 50-1435). The presence of K was further confirmed during XPS and EDS experiments.
TEM characterization of biogenic AgNPs
The morphology and size of biogenic AgNPs were analyzed by transmission electron microscopy (TEM). Figure 4a shows the presence of spherical and quasi-spherical nanoparticles. Their particle size distribution was obtained from a histogram, considering 200 particles, measured using JMicroVision version 1.2.7 (Fig. 4b). Biogenic nanoparticles shows a size range of 1.2–62 nm with an average size of 5 nm. High-resolution TEM (HRTEM) analysis was used to determine the structure of biogenic Ag nanoparticles (Fig. 4c). HRTEM shows the crystalline structure of single biogenic nanoparticle, with visible lattice fringes. A lattice spacing of 0.238 nm was calculated, corresponding to the plane family (111) of fcc silver. Additionally, Fig. 4d shows selected area electron diffraction (SAED) pattern, which indicates their polycrystalline nature and each of the diffraction rings has been indexed to (111), (200), (220) and (311), corresponding fcc crystal structure of metallic silver (JCPDS, No. 04-0783) and a diffraction ring corresponding to (200) cubic structure of AgK3 (PDF 50-1435). The results are consistent with the XRD diffractogram obtained in Fig. 3.
Chemical characterization by XPS and EDS
The general XPS survey showed the presence of C, O, N, Ag and K with traces of chlorine. A high-resolution spectrum was collected for the Ag 3d in Fig. 5. Our Gaussian–Lorentzian curves showed two chemical states of Ag. The curve assigned to Ag0 NPs resulted in 368.5 eV for the electrons arising from the Ag 3d5/2 level. AΔ = 6.0 eV was estimated between the Ag 3d5/2 and Ag 3d3/2. Then, the second chemical state is assigned to Ag–O (366.3 eV)28, with almost 2.2 eV, shifted to the lower binding energy when compared to Ag0. The presence of Ag–O can be related to Ag+ species, which might suggest the presence of Ag+ in suspension from unreacted Ag or AgNP leaching. In this region, we also observed the presence of the K 2s emission. The SEM–EDS chemical bulk quantification for the dry sample of NPs displayed the peaks related to the Ag, O, C and K clearly and some traces of P see supplementary information (Figure S2). Cl was not quantified by SEM–EDS analysis.
Antimicrobial susceptibility testing
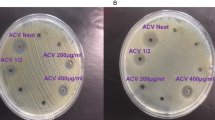
The antimicrobial effect of biogenic AgNPs was investigated on four clinical pathogenic organisms (E. coli, P. aeruginosa, S. aureus and C. albicans) using the agar well diffusion method and by determining the MIC and MBC, the results were compared to those obtained by AgNPs synthesized by the chemical method. The disk diffusion method shows the magnitude of the susceptibility of the pathogenic microorganisms (Fig. 6). The aqueous extract produced a diffuse ring for all microorganisms. Biogenic AgNPs possessed higher antimicrobial activity than chemical AgNPs. The mean of three replicates of the diameter of inhibition zones (in millimeters) containing AgNPs suspension is presented in Table 1. Biogenic AgNPs showed a higher inhibitory effect against C. albicans. For this type of AgNPs, the inhibition zone reached 18.0 ± 1.3 mm for E. coli, 16.0 ± 1.0 mm for S. aureus and 15.0 ± 0.5 mm for P. aeruginosa. Lower inhibition zones were found for the chemical prepared AgNPs. The MIC and MBC were found between 2.5 µg/mL to 5.0 µg/mL for chemical NPs and between 0.06 and 0.25 µg/mL for biogenic NPs. These results confirm the high antimicrobial potency of biogenic NPs compared to chemical NPs (Table 1). The lowest MIC of biogenic NPs at 0.06 µg/mL was obtained against E. coli, P. aeruginosa and S. aureus. The growth of C. albicans inhibited at 0.13 µg/mL. MBC of chemical AgNPs were not recorded for all pathogenic microorganisms in the tested concentration range (≥5 µg/mL). Biogenic AgNPs inhibited at 0.25 µg/mL for C. albicans and 0.13 µg/mL for E. coli, S. aureus and P. aeruginosa.
Cytotoxicity assay of biogenic AgNPs
Lymphocytes are an important part of the immune system and they have already been used to evaluate NPs cytotoxicity29,30,31. In this work, to determine the cytotoxic effect of biogenic AgNPs, human peripheral blood lymphocytes (HPBL) were treated with 1.3 µg/mL for 24 h. Viability was assayed by trypan blue exclusion test (Figure S3), no decrease in the viability of HPBL was observed. However, to determine if the biogenic AgNPs can lead to an earlier apoptosis death pathway, apoptosis/necrosis test was done using the Alexa fluor 488 cell death evaluation kit (Invitrogen, USA). Micrographs in Figure S3 show that exist more cells AV+ comparing to a negative control (untreated cells). This marker interacts with the phosphatidylserine in the plasma membrane, indicating an apoptotic cell death pathway. Moreover, no PI+ lymphocytes were found in the cultures treated (Fig. 7) that suggests these AgNPs are in an early stage of apoptosis that could be resolved by the cell, so these AgNPs are not harmful to the first line of immune defense of the organism.
Micrographs of lymphocytes culture evaluated. Images show 40x bright-field of control (a), and apoptotic (b) and necrotic (c) lymphocytes exposed to 1.3 µg/mL AgNPs. (d–f) show the corresponding image on fluorescent microscopy. Black, green and red circles show AV-/PI-, AV+/PI-stained lymphocytes and AV-/PI+ micronucleus.
Discussion
This work shows the synthesis of biogenic AgNPs using an L. acapulcensis aqueous extract as a reducing agent for the first time. The L. acapulcensis extract has important biological components like proteins and ethylene groups detected by FTIR and could act as a capping/stabilization agents32,33,also, alkyl halides act as reducing agents. LC–MS reveals the presence of antimicrobial agents (18α-Glycyrrhetinic acid and ruspolinone) along with the presence of antioxidants (e.g., members of the azoles group) and alkaloids act as reducing agent (ruspolinone). These molecules and others can end-capping the AgNPs, enhancing AgNP antimicrobial activity greatly. Visual observation and UV–Vis spectroscopy easily followed the formation of biogenic AgNPs by reducing Ag+ ions into Ag0 ions. The surface quantification showed that Ag0 NPs resulted in 68% with 32% of Ag–O species. The Ag–O correspond with the remaining cations of the Ag precursor possibly from the unreacted Ag precursor during synthesis. It is important to note that leaching of Ag might play an important role, but the cytotoxicity assay was rather low for the green synthesized NPs. It was observed that upon the addition of the L. acapulcensis aqueous extract into the volumetric flask containing the AgNO3 solution, the color of the aqueous extract changed from yellow to dark brown within 15 min of incubation, this showed the formation of AgNPs. Its formation was also confirmed by the absorption peak at 400 nm in the UV–Vis spectra for all different incubation times. The intensity of brown color and absorbance units increases with the increase of the incubation period. Therefore, 15 min was used for the synthesis of biogenic AgNPs, providing an easy way and rapid protocol, which was an important advantage of biological methods compared to other methods using different primary biological material because they are normally synthesized within 30 min or more21,22,34,35,36,37. TEM showed a widespread distribution of biogenic AgNPs with spherical and quasi-spherical shapes. Some of them were elongated due to the aggregation of two or more nanoparticles. Their sizes were in the range 1.2–62 nm, with an average of 5 nm and 50% of the particles were in the 1 to 4 nm range. Similar results have been reported for AgNPs with sizes from 2 to 75 nm using plant extracts as reducing agents34,35,38,39,40,41.
There are studies and reports that plant extract NPs have strong antimicrobial activity. These AgNPs were effective in killing a range of bacterial pathogens involved in different infectious diseases, e.g., S. aureus, P. aeruginosa, E. coli and C. albicans are related to most common hospital-acquired infections36,42. Our biogenic AgNPs possessed higher antimicrobial activity than other biogenic AgNPs40,41. Reports on the mechanism of antimicrobial AgNPs action implies the dimorphic transition, DNA loses its ability to replicate and membrane disruption inhibiting bacterial growth43.
Navarro and collaborators reported the antimicrobial evaluation L. acapulcensis extracts with dichloromethane (D), hexane (H), water (W) and methanol (M). They used higher amounts of biomass in order to evidence the MIC values (S. aureus: 4.0 mg/mL (H), 1.0 mg/mL (D) and 1.0 mg/mL (M); E. coli: >8.0 mg/mL (H), 4.0 mg/mL (D) and 4.0 mg/mL (M); C. albicans: >8.0 mg/mL (H), 4.0 mg/mL (D) and 2.0 mg/mL (M)). Regarding the results of water extracts of L. acapulcensis, the authors did not mention why they did not present the MIC results27. In our case, the aqueous extract in the disk diffusion studies showed a diffuse ring, which is evidence of low antimicrobial activity. This low antimicrobial activity could be due to a lower amount of biomass (~3-fold lower than the biomass required by Navarro et al. 26 and Navarro et al. 27 used to obtain an aqueous extract of L. acapulcensis). The antimicrobial activity was enhanced using AgNPs synthesized from an aqueous extract of L. acapulcensis due to the combination with their antimicrobial substances present in the extract. Our MIC results were lower for S. aureus, E. coli and C. albicans than those obtained by Navarro and collaborators (S. aureus: 0.06 µg/mL vs 4.0 (H), 1.0 (D) and 1.0 mg/mL (M); E. coli: 0.06 µg/mL vs >8.0 (H), 4.0 (D) and 4.0 mg/mL (M) and C. albicans: 0.13 µg/mL vs >8.0 (H), 4.0 (D) and 2.0 mg/mL (M)) 27. For P. aeruginosa, a large zone of inhibition was compared to the one reported by Gopinath et al.44. Results obtained in the MIC and evidence of pronounced antibacterial activity of our biogenic AgNPs on P. aeruginosa can be compared to those obtained by other researches using different aqueous extract mediated AgNPs of different carnivorous plant tissue (Drosera binata, Drosera indica, Drosera spatulata and Dionaea muscipula)45. All the bacteria were eliminated with 0.13 µg/mL for E. coli, S. aureus and P. aeruginosa and 0.25 µg/mL for C. albicans. Several reports showed that lower MIC values had stronger antibacterial potencies, but their stability and biocompatibility are related to small sizes between 10 and 15 nm46,47,48. Additionally, the MIC and MBC values of biogenic AgNPs confirmed their greater antimicrobial potency than chemical AgNPs.
In short, we showed that it is possible to obtain biogenic AgNPs using an aqueous extract of L. acapulcensis, which provides a quick, efficient and simple technique for the green synthesis of nanomaterials. The presence of alkyl halides and other reducing agents in the extract of L. acapulcensis allows the reduction of Ag+ ions into AgNPs. The biosynthesized AgNPs showed a significant antimicrobial effect against C. albicans, E. coli, S. aureus and P. aeruginosa. Nevertheless, the microbicidal activity of biogenic AgNPs remains at lower concentrations than the chemically synthesized AgNPs. Our AgNPs did not induce a decrease in cell viability in human peripheral blood lymphocytes at the concentration and time evaluated. Prospective studies are needed to demonstrate the long-term efficacy and the potential beneficial impact on infectious diseases.
Conclusions
The present work showed that it is possible to obtain biogenic AgNPs using an aqueous extract of L. acapulcensis. The biosynthesized AgNPs shows a significant antimicrobial effect against C. albicans, E. coli, S. aureus and P. aeruginosa at lower concentrations than chemically synthesized AgNPs. In terms of toxicity, biogenic AgNPs did not induce a decrease in cell viability in human peripheral blood lymphocytes.
Methods
Preparation of L. acapulcensis aqueous extract
The stem and roots of L. acapulcensis were dried in a laboratory oven at 60 °C for 2 h until constant weight was reached. Afterward, 2 g of biological material were immersed in distilled water (100 mL) and were kept in a heated plate and allowed to boil for 15 min. The extract was filtered and kept refrigerated at 4 °C.
HPLC-ESI-QTOF-MS analysis of L. acapulcensis extract
The plant extract was filtered through 0.2 µm PVDF filters (Sterivex, Millipore, Bedford, MA, USA). Determination of phytochemical composition was performed on a UHPLC (model 1260) coupled to a 6530 model Accurate-Mass QTOF LC/MS; Agilent Technologies (Palo Alto, CA, USA) equipped with an ESI interface operating in positive ion mode and an Agilent XDB-C8 2.7 μm 3 × 50 mm, 2.7 column. The mobile phase was 0.2% formic acid in water as eluent A and 0.1% formic acid in acetonitrile as eluent B, with the following set of operation parameters: Capillary voltage, 3500 V; nebulizer pressure, 35 psi; dry gas flow, 8l/min; dry gas temperature, 300 °C ; LC–MS mass spectra were recorded across the range mass 100–1,700 m/z; fragmentor 135 V; column temperature 40 oC; solvent gradient conditions: 0 min, 0% B; 5 min, 10% B; 10 min, 80% B; 12 min, 100% B and then 15 min, 0%B. Compound identification was performed through MassHunter Workstation software using libraries G3874-60007 Massahunter METLIN PCDL B.08.00.
Synthesis of biogenic nanoparticles
AgNPs were synthesised by reducing a silver nitrate solution at 0.001 M in the presence of the L. acapulcensis extract. A volume of 2.5 mL of AgNO3 was added to 2.5 mL of L. acapulcensis aqueous extract (ratio 1:1) and incubated at ambient conditions for 2 min. Then the reaction solution was prepared to a final volume of 10 mL with distilled water and the solutions were exposed under white light during 15, 30 and 60 min. The synthesis progress was monitored using UV–Vis spectroscopy (JENWAY, model 6505, UK) with a wavelength range from 300 to 600 with a resolution of 1 nm. The aqueous extract was used as a blank. Experiments were carried out in triplicate. FTIR spectroscopy analysis was carried out to reveal the functional group of biomolecules present in the L. acapulcensis aqueous extract using the instrument BRUKER Tensor 27 at room temperature with a range of resolution of 400–4000 cm−1.
Synthesis of AgNPs by chemical method
For the synthesis of AgNPs by a chemical method, silver nitrate solution (0.001 M) and sodium borohydride (0.1 M) were used as a metal salt precursor and a reducing agent, respectively. Polyethylene glycol (PEG) 80 at 50 mM was used as a stabilizing agent.
Characterization of AgNPs
The strutural characterization of biogenic AgNPs was analized by X-ray difraction, recorded on a Panalitycal X’pert Pro MRD X-ray diffraction instrument, with Cu Kα radiation (λ = 0.15418 nm) over the scanning range 2θ = 30–80°, with a step of 0.02 degree. The synthesized biogenic AgNPs were examined on a Hitachi H-7500, JEOL 2010 and a HRTEM, JEM-2200FS, JEOL, transmission electron microscopes. Samples were prepared by placing a 10 µL aliquot of biogenic AgNPs on carbon-coated copper grids (300 mesh, Ted Pella Inc.). At least 200 particles were measured using the software JMicroVision version 1.2.7 (www.jmicrovision.com) to characterize the size distribution of AgNPs. The SEM–EDS chemical bulk analysis was carried in a High-resolution SEM microscope from JEOL model JSM-S300 equipped with an energy dispersive X-ray system at 35 kV for 100 s. The sample was placed in a carbon doble-face tape. Surface analysis of the the AgNPs material was carried out by XPS using a commercial instrument SPECS spectrometer equipped with a PHOIBOS® 150 WAL hemispherical electron analyzer and an AlKα X-ray source. The sample was placed in a molybdenum holder with a micro indium foil to avoid adventitious carbon. Gaussian–Lorentzian curve fitting was done using the CasaXPS software Ver. 2.3.23 in the high-resolution emission-line regions. Semi-quantitative analysis of the chemical states was calculated for each contribution based on its peak area percentages.
Antimicrobial sensitivity testing
The microorganisms used for antimicrobial susceptibility evaluation were: E. coli (ATCC 25922), P. aeruginosa (ATCC 27853), S. aureus (ATCC 49476) and C. Albicans (ATCC 49476). The disk diffusion method was used to determine a zone of inhibition. Aliquots of 0.1 mL of each test organism were spread on LB agar for bacteria and PDA agar for C. albicans. Each Petri plates were dried. Paper discs loaded with 10 µL of biogenic AgNPs, chemical AgNPs and L. acapulcensis aqueous extract were placed on the surface of agar plates and incubated at 37 °C during 24 h after which diameters of inhibition zones were measured. All the tests were run in triplicate and the average result was taken.
Determination of minimum inhibitory concentration and minimum biocidal concentration
The Minimal Inhibitory Concentration (MIC) and Minimum Biocidal Concentration (MBC) were determined using the agar dilution method. Inoculums of bacteria (1–1.5 × 108 CFU/ml) and C. albicans (1–5 × 106 CFU/ml) were prepared according to the 0.5 McFarland standard. Stock solutions of AgNPs were two-fold concentrated for bacteria and ten-fold concentrated for yeast in concentrations in the range of 0.1–5 µg/mL for chemical nanoparticles and 0.02–1 µg/mL for biogenic nanoparticles. Then 0.5 mL of treatment (bacteria or yeast) and 0.5 mL of AgNPs were mixed and incubated at 37 °C during 24 h at 250 rpm (Orbit Environ Shaker). After incubation, 10 μL of each tube were transferred to LB agar for bacteria and YPD agar for C. albicans and incubated at 37 °C for 24 h. The MIC was the lowest concentration of an antimicrobial that inhibits the visible growth and the MBC was the lowest concentration that killed ≥ 99.9% of cells. The MIC and MBC were determined in triplicate and were carried out on at least three different days. Positive control was used for each microorganism (without treatment).
Isolation of human peripheral blood lymphocytes
Human Peripheral Blood Lymphocytes (HPBL) were isolated from a blood sample obtained by venipuncture of a healthy donor using heparinized tubes. Blood samples were collected from healthy donors with prior informed consent. Protocols to collect and disposed blood samples were according to the mexican normativity (NOM-253-SSA1-2012, NOM-003-SSA1-1993) and approved by the Ethical Committee of the Health Sciences School from Autonomous University of Baja California, Mexico with file number 003/2019. Blood was first diluted 1:1 (v/v) in physiological saline solution and subsequently separated by ficoll density gradient centrifugation49, consistent with the providers protocol (Ficoll® Paque Plus GE Healthcare, USA). Briefly, 12 mL of diluted blood was carefully placed over 2 mL of ficoll, then centrifuged at 400G during 30 min. The layer of mononuclear cells was transferred to a centrifuge tube under sterile conditions. Cells were washed with physiological saline solution and seeded on a petri dish with RPMI 1640 media supplemented with 10% of autologous serum and incubated 2 h at 37 °C with 70% of relative humidity and 5% CO2. After the incubation time, lymphocytes (non-adherent cells) were taken from the supernatant and monocytes (adherent cells) were discarded.
Cell viability assay
Lymphocytes isolated from human peripheral blood were used for cytotoxicity determination. 5 × 105 lymphocytes per well were seeded in 96 well tissue culture plate and subsequently treated with AgNPs at a final concentration of 1.3 µg/mL. Cells were incubated for 24 h at 37 °C and 5% CO2. Viability was determined by the Trypan blue exclusion test using a Neubauer’s chamber under optical microscope. Untreated cells were used as negative control and 3 experiments by triplicate were done.
Apoptosis and necrosis determination
Cells were obtained and treated as for the cell viability assay. In this case, after incubation, cells were stained using the Alexa Fluor™ 488 kit (Invitrogen, USA) according to the providers protocol. Briefly, cells treated and controls placed in a 96 well plate were added with 20 µL of binding buffer 1x (BD Pharmingen™, USA) and 1 µL Anexin V, incubated 15 min and stained whit 1 µL of Propidium lodide followed by 15 more min of incubation under a 5% CO2 atmosphere at 37 °C to identify apoptotic and necrotic cells. Samples were then observed through a 40x objective using AxioCam ICc5 on 0.63 × C-Mount 60 N-C interface camera using a fluorescence microscope (Axio Lab A1, Carl ZEISS) for live cellular imaging. Excitation wavelengths of 455 nm were used to excite AV and PI.
Data analysis
All the measurements were replicated three times for each assay and the results are presented as mean ± SD. Data were analyzed using Minitab version 18.0 and the descriptive statistic was used.
References
Li, Y. et al. Nanocrystalline silver particles: synthesis, agglomeration and sputtering induced by electron beam. J. Colloid Interface Sci. 209, 347–349. https://doi.org/10.1006/jcis.1998.5879 (1999).
Singh, J. et al. ‘Green’ synthesis of metals and their oxide nanoparticles: applications for environmental remediation. J. Nanobiotechnol. 16, 1–24. https://doi.org/10.1186/s12951-018-0408-4 (2018).
Yilmaz, A. & Yilmaz, M. Bimetallic core-shell nanoparticles of gold and silver via bioinspired polydopamine layer as surface-enhanced Raman spectroscopy (SERS) platform. Nanomaterials 10, 688. https://doi.org/10.3390/nano10040688 (2020).
Scholes, G. D. Controlling the optical properties of inorganic nanoparticles. Adv. Funct. Mater. 18, 1157–1172. https://doi.org/10.1002/adfm.200800151 (2008).
Susarrey-Arce, A. et al. Inception and trapping of ZnO nanoparticles within desilicated mordenite and ZSM-5 zeolites. Part. Part. Syst. Char. 27, 100–111. https://doi.org/10.1002/ppsc.201000027 (2011).
Chen, H., Roco, M. C., Li, X. & Lin, Y. Trends in nanotechnology patents. Nat. Nanotechnol. 3, 123–125. https://doi.org/10.1038/nnano.2008.51 (2008).
Smetana, A. B., Klabunde, K. J. & Sorensen, C. M. Synthesis of spherical silver nanoparticles by digestive ripening, stabilization with various agents and their 3-D and 2-D superlattice formation. J. Colloid Interface Sci. 284, 521–526. https://doi.org/10.1016/j.jcis.2004.10.038 (2005).
Lee, H., Chou, K. S. & Huang, K. C. Inkjet printing of nanosized silver colloids. Nanotechnology 16, 2436–2441 (2005).
Wakuda, D., Kim, K. S. & Suganuma, K. Room temperature sintering of Ag nanoparticles by drying solvent. Scripta Mater. 59, 649–652. https://doi.org/10.1016/j.scriptamat.2008.05.028 (2008).
Anna, Z. et al. Preparation of silver nanoparticles with controlled particle size. Procedia Chem. 1, 1560–1566. https://doi.org/10.1016/j.proche.2009.11.004 (2009).
Kholoud, M. M. et al. Synthesis and applications of silver nanoparticles. Arab. J. Chem. 3, 135–140. https://doi.org/10.1016/j.arabjc.2010.04.008 (2010).
Toisawa, K., Hayashi, Y. & Takizawa, H. Synthesis of highly concentrated Ag nanoparticles in a heterogeneous solid-liquid system under ultrasonic irradiation. Mater. Trans. 51, 1764–1768. https://doi.org/10.2320/matertrans.MJ201005 (2010).
Iravani, S., Korbekandi, H., Mirmohammadi, S. V. & Zolfaghari, B. Synthesis of silver nanoparticles:chemical, physical and biological methods. Res. Pharm. Sci. 9, 385–406. https://doi.org/10.1155/2013/639725 (2014).
Pallavicini, P. et al. Fast dissolution of silver nanoparticles at physiological pH. J. Colloid Interface Sci. 563, 117–188. https://doi.org/10.1016/j.jcis.2019.12.081 (2020).
Gade, A. K. et al. Exploitation of Aspergillus niger for synthesis of silver nanoparticles. J. Biobased Mater. Bioenergy 2, 243–247. https://doi.org/10.1166/jbmb.2008.401 (2008).
Khadri, H. et al. Green synthesis of silver nanoparticles with high fungicidal activity from olive seed extract. Adv. Nanopart. 2, 241–246. https://doi.org/10.4236/anp.2013.23034 (2013).
Prabu, H. J. & Johnson, I. Plant-mediated biosynthesis and characterization of silver nanoparticles by leaf extracts of Tragia involucrata, Cymbopogon citronella, Solanum verbascifolium and Tylophora ovata. Karbala Int. J. Mod. Sci. 1, 237–246. https://doi.org/10.1016/j.kijoms.2015.12.003 (2015).
Shakeel, A. H. et al. Green synthesis of silver nanoparticles using Azadirachta indicaaqueous leaf extract. J. Radiat. Res. Appl. Sci. 9, 1–7. https://doi.org/10.1016/j.jrras.2015.06.006 (2016).
Khatoon, A. H. et al. Silver nanoparticles from leaf extract of Mentha piperita: eco-friendly synthesis and effect on acetylcholinesterase activity. Life Sci. 15, 430–434. https://doi.org/10.1016/j.lfs.2018.08.046 (2018).
Annu, et al. Fruit waste (peel) as bio-reductant to synthesize silver nanoparticles with antimicrobial, antioxidant and cytotoxic activities. J. Appl. Biomed. 16, 221–231. https://doi.org/10.1016/j.jab.2018.02.002 (2018).
Padalia, H., Moteriya, P. & Chanda, S. Green synthesis of silver nanoparticles from marigold flower and its synergistic antimicrobial potential. Arab. J Chem. 8, 732–741. https://doi.org/10.1016/j.arabjc.2014.11.015 (2015).
Lakshmanan, G., Sathiyaseelan, A., Kalaichelvan, P. T. & Murugesan, K. Plant-mediated synthesis of silver nanoparticles using fruit extract of Cleome viscosa L.: assessment of their antibacterial and anticancer activity. Karbala Int. J. Mod. Sci. 4, 61–68. https://doi.org/10.1016/j.kijoms.2017.10.007 (2018).
Benakashani, F., Allafchian, A. & Jalali, S. A. H. Green synthesis, characterization and antibacterial activity of silver nanoparticles from root extract of Lepidium draba weed. Green Chem. Lett. Rev. 10, 324–330. https://doi.org/10.1080/17518253.2017.1363297 (2017).
Argueta, A., Cano, L. & Rodarte, M. Atlas de las Plantas de la Medicina Tradicional Mexicana, Tomo 1–3. Instituto Nacional Indigenista, México D.F., 1994; pp. 1786.
Gonzáles-Cortazar, M. et al. Lysiloma acapulcensis leaves contain anthelmintic metabolites that reduce the gastrointestinal nematode egg population in sheep faeces. Comp. Clin. Pathol. 27, 189–197. https://doi.org/10.1007/s00580-017-2577-1 (2018).
Garcı́a, V.M.N., , et al. Antifungal activities of nine traditional Mexican medicinal plants. J. Ethnopharmacol. 87, 85–88. https://doi.org/10.1016/S0378-8741(03)00114-4 (2003).
Navarro García, V. M. et al. Antifungal and antibacterial activity of four selected mexican medicinal plants. Pharm. Biol. 44, 297–300. https://doi.org/10.1080/13880200600715837 (2006).
Lopez-Salido, I., Chan-Lim, D. & Dok-Kim, Y. Ag nanoparticles on highly ordered pyrolytic graphite (HOPG) surfaces studied using STM and XPS. Surf. Sci. 588, 6–18. https://doi.org/10.1016/j.susc.2005.05.021 (2005).
Orta-García, S. T. et al. Analysis of cytotoxic effects of silver nanoclusters on human peripheral blood mononuclear cells “in vitro”. J. Appl. Toxicol. 35, 1189–1199. https://doi.org/10.1002/jat.3190 (2015).
Vergallo, C. et al. Cytotoxicity of β-D-glucose coated silver nanoparticles on human lymphocytes. AIP Conf. Proc. 1603, 78–85. https://doi.org/10.1063/1.4883045 (2014).
Ruiz-Ruiz, B. et al. Cytokinesis-block micronucleus assay using human lymphocytes as a sensitive tool for cytotoxicity/genotoxicity evaluation of AgNPs. ACS Omega 5, 12005–12015. https://doi.org/10.1021/acsomega.0c00149 (2020).
Li, J., Zhu, J. & Liu, X. Ultrafine silver nanoparticles obtained from ethylene glycol at room temperature: catalyzed by tungstate ions. Dalton Trans. 43, 132–137. https://doi.org/10.1039/c3dt52242c (2014).
Iravani, S., Korbekandi, H., Mirmohammadi, S. V. & Zolfaghari, B. Synthesis of silver nanoparticles: chemical, physical and biological methods. Res. Pharm. Sci. 9, 385–406 (2014).
Chandran, S. P. et al. Synthesis of gold nanotriangles and silver nanoparticles using aloevera plant extract. Biotechnol. Prog. 22, 577–583. https://doi.org/10.1021/bp0501423H (2006).
Tripathy, A. et al. (2009) Process variables in biomimetic synthesis of silver nanoparticles by aqueous extract of Azadirachta indica (Neem) leaves. J. Nanoparticle Res. 12, 237–246. https://doi.org/10.1007/s11051-009-9602-5 (2009).
Anbazhagan, S. H. et al. Synthesis, characterization and biological applications of mycosynthesized silver nanoparticles. 3 Biotech 5, 333. https://doi.org/10.1007/s13205-017-0961-9 (2007).
Różalska, B. et al. Biogenic nanosilver synthesized in Metarhizium robertsii waste mycelium extract: as a modulator of Candida albicans morphogenesis, membrane lipidome and biofilm. PLoS ONE 13, e0194254. https://doi.org/10.1371/journal.pone.0194254 (2018).
Huang, J. et al. Biosynthesis of silver and gold nanoparticles by novel sundried Cinnamomum camphora leaf. Nanotechnology 18, 105104. https://doi.org/10.1088/0957-4484/18/10/105104/pdf (2007).
Hernández-Morales, L. et al. Study of the green synthesis of silver nanoparticles using a natural extract of dark or white Salvia hispanica L. seeds and their antibacterial application. Appl. Surf. Sci. 489, 952–961. https://doi.org/10.1016/j.apsusc.2019.06.031 (2019).
Jahan, I., Erci, F. & Isildak, I. Microwave-assisted green synthesis of non-cytotoxic silver nanoparticles using the aqueous extract of Rosa santana (rose) petals and their antimicrobial activity. Anal. Lett. 52, 1–14. https://doi.org/10.1080/00032719.2019.1572179 (2019).
Salari, S., Bahabadi, S. E., Samzadeh-Kermani, A. & Yosefzaei, F. In-vitro evaluation of antioxidant and antibacterial potential of green synthesized silver nanoparticles using Prosopis farcta fruit extract. Iran. J. Pharm. Res. 18, 430–445 (2019).
Guerra, J. D. et al. Selective antifungal activity of silver nanoparticles: a comparative study between Candida tropicalis and Saccharomyces boulardii. Colloid Interface Sci. 37, 100280. https://doi.org/10.1016/j.colcom.2020.100280 (2020).
Dakal, T. C., Kumar, A., Majumdar, R. S. & Yadav, V. Mechanistic basis of antimicrobial actions of silver nanoparticles. Front. Microbiol. 7, 1831. https://doi.org/10.3389/fmicb.2016.01831 (2016).
Gopinath, V. et al. Biogenic synthesis, characterization of antibacterial silver nanoparticles and its cell cytotoxicity. Arab. J. Chem. 10, 1107–1117. https://doi.org/10.1016/j.arabjc.2015.11.011 (2017).
Banasiuk, R. et al. Carnivorous plants used for green synthesis of silver nanoparticles with broad-spectrum antimicrobial activity. Bioproc. Biosyst. Eng. 32, 79–84. https://doi.org/10.1007/s00449-008-0224-6 (2009).
Yacaman, M. J., Ascencio, J. A., Liu, H. B. & Gardea-Torresdey, J. Structure shape and stability of nanometric sized particles. J. Vac. Sci. Technol. B Microelectron. Nanometer. Struct. Process. Meas. Phenom. 19, 1091–1103. https://doi.org/10.1116/1.1387089 (2001).
Song, J. Y. & Kim, B. S. Rapid biological synthesis of silver nanoparticles using plant leaf extracts. Bioproc. Biosyst. Eng. 32, 79–84. https://doi.org/10.1007/s00449-008-0224-6 (2009).
Kim, S. H. et al. Antibacterial activity of silver-nanoparticles against Staphylococcus aureus and Escherichia coli. Korean J. Microbiol. Biotechnol. 39, 77–85 (2011).
Boyum, A. Isolation of mononuclear cells and granulocytes from human blood (Paper IV). Scand. J. Clin. Lab. Invest. Suppl. 97, 77–89 (1968).
Acknowledgements
The authors are grateful to Arturo Flores Tavira for providing biological material and Eloisa Aparicio, Francisco Ruiz, I. Gradilla and D. Domínguez for technical support on XRD,TEM, SEM-EDS and XPS, respectively. D. Garibo acknowledges the Cátedras CONACyT program (Project Number: 111). A.S.-A. would like to thank Pioneers in Health Care Innovation Fund (PIHC)-2019. PIHC was financially supported by an unrestricted research grant from the Pioneers in Health Care Innovation Fund, established by the University of Twente, Saxion University of Applied Sciences, Medisch Spectrum Twente, ZiekenhuisGroep Twente and Deventer Hospital.
Author information
Authors and Affiliations
Contributions
Participated in research design: Diana Garibo. Conducted experiments: Diana Garibo, Iván Estrada, Hugo Tiznado, Alberto Blanco, José A. Rodriguez, Yanis Toledano-Magaña, Hugo A. Borbón-Nuñez, Jorge N. Díaz de León, Ernesto García Mendoza, Marcela Ovalle-Marroquin, Alicia G. Soto-Ramos, Oscar A. Romo and Luis A. Chávez-Almazán. Performed data analysis: Diana Garibo, Iván Estrada, Hugo Tiznado, Luis A. Chávez-Almazán, Yanis Toledano-Magaña, Hugo A. Borbón-Nuñez, Jorge N. Díaz de León, Ernesto García Mendoza, Marcela Ovalle-Marroquin, Alicia G. Soto-Ramos, Alberto Blanco, José A. Rodriguez, Oscar A. Romo and Arturo Susarrey-Arce. Wrote the manuscript: Diana Garibo, Hugo A. Borbón-Nuñez, Jorge N. Díaz de León, Ernesto García Mendoza, Yanis Toledano-Magaña, Alberto Blanco, José A. Rodriguez, Luis A. Chávez-Almazán and Arturo Susarrey-Arce.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Garibo, D., Borbón-Nuñez, H.A., de León, J.N.D. et al. Green synthesis of silver nanoparticles using Lysiloma acapulcensis exhibit high-antimicrobial activity. Sci Rep 10, 12805 (2020). https://doi.org/10.1038/s41598-020-69606-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-69606-7
This article is cited by
-
In situ and bio-green synthesis of silver nanoparticles immobilized on zeolite as a recyclable catalyst for the degradation of OPDs
Scientific Reports (2024)
-
Characterization, antibacterial, and cytotoxic activities of silver nanoparticles using the whole biofilm layer as a macromolecule in biosynthesis
Scientific Reports (2024)
-
Plackett–Burman screening of physico-chemical variables affecting Citrus peel-mediated synthesis of silver nanoparticles and their antimicrobial activity
Scientific Reports (2024)
-
Environmentally friendly synthesis of silver nanoparticles using Prickly Pear extract and their antimicrobial and antioxidant activities
Biomass Conversion and Biorefinery (2024)
-
Synthesis and characterization of bioactive silver nanoparticles: evaluation of their anticancer potential against MCF7 cell line and antibacterial activity against foodborne pathogens
Journal of Sol-Gel Science and Technology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.