Insect pathogens have adopted an array of mechanisms to subvert the immune pathways of their respective hosts. Suppression may occur directly at the level of host–pathogen interactions, for instance phagocytic capacity or phenoloxidase activation, or at the upstream signaling pathways that regulate these immune effectors. Insect pathogens of the family Baculoviridae, for example, are known to produce a UDP-glycosyltransferase (UGT) that negatively regulates ecdysone signaling. Normally, ecdysone positively regulates both molting and antimicrobial peptide production, so the inactivation of ecdysone by glycosylation results in a failure of host larvae to molt, and probably a reduced antimicrobial response. Here, we examine a putative ecdysteroid glycosyltransferase, Hba_07292 (Hb-ugt-1), which was previously identified in the hemolymph-activated transcriptome of the entomopathogenic nematode Heterorhabditis bacteriophora. Injection of recombinant Hb-ugt-1 (rHb-ugt-1) into Drosophila melanogaster flies resulted in diminished upregulation of antimicrobial peptides associated with both the Toll and Immune deficiency pathways. Ecdysone was implicated in this suppression by a reduction in Broad Complex expression and reduced pupation rates in r Hb-ugt-1-injected larvae. In addition to the finding that H. bacteriophora excreted-secreted products contain glycosyltransferase activity, these results demonstrate that Hb-ugt-1 is an immunosuppressive factor and that its activity likely involves the inactivation of ecdysone.
Similar content being viewed by others
Baculoviruses that infect insects, such as Autographa californica, have been shown to express a UDP-glycosyltransferase (UGT) that acts as a virulence factor through interference with ecdysone signaling1. The baculovirus UGT is believed to have been acquired from Lepidopteran spp. through horizontal gene transfer2, which implies that it should be ideally suited for activity in an insect host. During an infection, the A. californica UGT conjugates UDP-glucose present in the host with the ecdysteroid hormone 20-hydroxyecdysone (20E), effectively eliminating the molecule’s ability to activate its receptor3. Because 20E is the primary molting signal in insects, infected larvae will not molt, but instead continue to feed and accumulate mass that will be converted to higher viral output4,5,6. The set of effects controlled by 20E is broader than molting alone, however. Ecdysone has also been shown to regulate the immune response in Drosophila melanogaster7. In S2 cell cultures, 20E is known to enable Imd signaling and sensitize cells to the Toll pathway ligand Spätzle, in both cases promoting antimicrobial peptide (AMP) expression8,9.
Here, we examined a putative ecdysteroid glycosyltransferase Hba_07292 (Hb-ugt-1), from Heterorhabditis bacteriophora, that shares amino acid sequence similarity to baculovirus and insect UGTs. Further, we hypothesize that this glycosyltransferase functions in a similar manner to viral UGT virulence factors. A nematode UGT is unlikely to have been acquired by horizontal gene transfer, but nematodes do share an ecdysozoan lineage with insects, and therefore likely regulate molting in a similar way10. Interestingly, evidence exists that 20E might even be the signaling molecule responsible for regulating molting in a number of helminths. In the filarial parasite Brugia malayi, 20E elicits a transcriptional response featuring genes related to embryogenesis, and a homolog of the insect ecdysone receptor has been identified in its genome11. The genome of the nematode Pristionchus pacificus also contains a putative ecdysone receptor homolog, and another has been postulated in H. bacteriophora, though this has yet to be confirmed12. If this is indeed the case, H. bacteriophora would require a negative regulatory element that is active against 20E to moderate its own development. Alternatively, this molecule could function as a virulence factor similar to that of baculoviruses if it were secreted and diminished host 20E signaling. It is currently not possible to indicate whether Hb-ugt-1 participates in development, but the known relationships between nematodes and 20E does allow for a plausible explanation of why the H. bacteriophora genome would contain a UGT active against 20E.
To further examine whether Hb-ugt-1 functions as a virulence factor, we performed a number of assays related to its expression and in vivo activity in the model host D. melanogaster. In agreement with the RNA sequencing assay that initially identified this glycosyltransferase13, Hb-ugt-1 was upregulated by H. bacteriophora infective juveniles (IJs) in response to hemolymph from multiple insect species, including D. melanogaster, though interestingly not in response to the nematode’s symbiotic bacterium Photorhabdus luminescens. A recombinant version of Hb-ugt-1 (rHb-ugt-1) suppressed the upregulation of AMP gene expression following microinjection, as do total excreted-secreted (ES) products of H. bacteriophora14. ES products also contained in vitro glycosyltransferase activity. Expression of the transcription factor Broad Complex and molting ability were subsequently characterized following rHb-ugt-1 injection to determine whether ecdysone signaling might play a role in the observed AMP suppression. Both were significantly diminished, suggesting a decreased amount of active insect 20E after exposure to rHb-ugt-1. Generally, this collection of evidence is consistent with a role for Hb-ugt-1 as an ecdysone-inactivating virulence factor.
Results
Hb-ugt-1 is upregulated in response to host factors
Originally, Hb-ugt-1 was identified as part of the transcriptomic response to Manduca sexta hemolymph following a 9-h exposure15. To further explore the transcriptional response and examine expression in response to a variety of insect factors or cues, we measured Hb-ugt-1 transcript levels in response to 25% hemolymph plasma from D. melanogaster and M. sexta, as well as an in vivo exposure to Galleria mellonella through injection of infective juveniles (IJs) directly into the hemocoel. Each of these treatments resulted in the upregulation of Hb-ugt-1 between 1 and 9 h (Fig. 1A–C), indicating that the transcript is broadly upregulated in response to insect factors. Additionally, to distinguish between a role for the glycosyltransferase in infection rather than development, IJs were exposed to a lawn of P. luminescens. This treatment sustains the growth and development of H. bacteriophora but should not induce the expression of virulence factors if these are responsive strictly to insect host signals. Interestingly, P. luminescens induced a downregulation of Hb-ugt-1 through 24 h, after which expression returned to a level comparable to 0-h expression (Fig. 1D). It should be noted as well that while expression of Hb-ugt-1 is significantly higher at 48 h as compared to 24 h, the 48 h expression level is not statistically discernable from 0-h expression. Together, these results indicate that Hb-ugt-1 is upregulated upon exposure to an insect host, but is unlikely to play a role in recovery of the nematode IJ from its diapause state.
Hb-ugt-1 is upregulated in response to host-associated factors, but not development-inducing symbiotic bacteria. The expression of Hb-ugt-1 in response to assorted host factors and Photorhabdus luminescens bacteria was assessed by qPCR at the indicated time points. Approximately 1,000 axenic infective juveniles (IJs) were exposed to 25% Drosophila melanogaster line Oregon-R (A) or Manduca sexta (C) hemolymph plasma while 5,000 nematodes were injected directly into the hemocoel of live Galleria mellonella larvae (B). One thousand IJs were plated on a lawn of P. luminescens as a virulence-neutral control (D). Average fold change drawn from three independent trials and calculated according to the 2−ΔΔCT method is shown, where error bars represent standard error (*p < 0.05, **p < 0.01, ***p < 0.001). Graphs were generated using GraphPad Prism version 5.03 (www.graphpad.com).
Hb-ugt-1 contains a conserved ecdysteroid glycosyltransferase domain
Using BlastP search, we identified Hb-ugt-1 as a putative glycosyltransferase containing an ecdysteroid glycosyltransferase domain (Interpro domain: IPR016224). An alignment of Hb-ugt-1 with several related glycosyltransferases indicates a substantial degree of similarity between the proteins, especially with reference to the signature sequence associated with the binding of the nucleotide sugar (Fig. 2A). The maximum likelihood phylogenetic tree (Fig. 2B) for these sequences produces an intuitive clustering of insects, nematodes, and viruses. This is likely due to stretches of low-similarity in the full-length alignment (Supplementary Fig. S1) that are nonetheless more similar for species within the same phylum. Because of the marked conservation across relevant groups of organisms and the known function of the included viral UGTs, this was considered a sufficient justification for investigating whether Hb-ugt-1 might function similarly.
Hb-ugt-1 is a putative UDP glycosyltransferase (UGT). (A) The amino acid sequence for Hb-ugt-1 displays sequence similarity to ecdysone glycosyltransferases. A region containing the domain associated with sugar binding is shown, where color intensity represents percent identity. The full alignment was produced with sequences from three insects; Drosophila melanogaster (NP_001246082.2), Bombyx mori (NP_001243972.1), and Manduca sexta (XP_030029659.1), three nematodes; Ancylostoma ceylanicum (EYC08379.1), Brugia malayi (XP_001894161.1), and Caenorhabditis elegans (NP_500913.1), and three insect-infective viruses; Spodoptera frugiperda multiple nucleopolyhedrovirus (AAP79109.1), Agrotis ipsiolon nucleopolyhedrovirus (YP_002268062.1), and Autographa californica nucelopolyhedrovirus (NP_054044.1). (B) A maximum-likelihood phylogenetic tree was generated for the aligned sequences with MEGA-X software. The consensus tree shown was developed from 200 bootstrap replicates and numbers next to branches indicate the percentage of replicates in which the associated taxa clustered together. The alignment image was produced with Jalview version 2.11.1.0 (www.jalview.org) and the image for the phylogenetic tree was exported from MEGA-X Version 10.1.5 (megasoftware.net).
Activated H. bacteriophora excreted-secreted (ES) products facilitate glycosylation in the presence of 20-hydroxyecdysone and a UDP-glucose donor molecule
The presence of a secretion signal at the N-terminus of Hb-ugt-1, as determined by SignalIP-5.0, suggests that the molecule may be secreted. Therefore, ES products of hemolymph-activated IJs were assessed for glycosyltransferase activity using a commercially available assay that detects phosphate liberated from the UDP donor molecule (UDP-glucose) following transfer of the UPD sugar to an acceptor molecule, which was 20E in this case. Glycosyltransferase activity was significantly higher (p < 0.001) in activated products as compared to both non-activated products and a Ringer’s buffer control (Fig. 3A). As a negative control, no signal was detected from activated products in the absence of 20E substrate. To further confirm the presence of a glycosyltransferase, the activated products, which contain a variety of proteins14, were assayed by western blot with a polyclonal antibody that recognizes human GTDC1 (Fig. 3B). A single band of approximately 60–70-kDa was detected in these activated products, consistent with the approximately 60-kDa molecular weight calculated from the Hb-ugt-1 predicted protein sequence (Fig. 3B). Additionally, the GTDC1 antibody was assessed for its ability to bind rHb-ugt-1, and this western blot was found to be positive (Supplementary Fig. S2). These results indicate that H. bacteriophora secretes a glycosyltransferase when exposed to an insect host, and that the activity is likely the result of a single protein, but more work is required to confirm that this protein is Hb-ugt-1.
Heterorhabditis bacteriophora activated ES products glycosylate 20-hydroxyecdysone and contain a GTDC1 domain-containing protein. (A) Activated ES products (Act) were assayed for glycosyltransferase activity against Ringer’s Buffer (R) and non-activated ES product (NA) controls using a colorimetric assay. UDP-glucose and 20-hydroxyecdysone were used as the donor sugar and sugar acceptor, respectively. All values were normalized to a negative control containing only substrate and the manufacturer-provided reaction buffer. The experiment was performed in triplicate and results are presented as phosphate input liberated from the UDP molecule (***p < 0.001). (B) ES products were separated by SDS-PAGE and labeled with anti-GTDC1 antibody to identify glycosyltransferases secreted by the nematode. The size in kilodaltons (kDa) of ladder markers is indicated on the left side of the image. The phosphate output graph was produced with GraphPad Prism version 5.03 (www.graphpad.com).
Recombinant Hb-ugt-1 suppresses the upregulation of antimicrobial peptide (AMP) genes
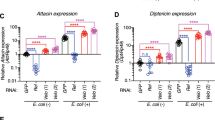
Recombinant Hb-ugt-1 produced via an Sf9 expression system was used to examine immune-related effects in D. melanogaster. Adult flies injected with approximately 7 ng of rHb-ugt-1 or BSA in PBS were collected at 6-h post-injection to determine the expression of AMP genes representing both the Toll and Imd pathways by qPCR analysis. The H. bacteriophora putative i-type lysozyme rHba_19909, which was expressed and purified according to the same protocol as Hb-ugt-1, and BSA were injected separately as controls. The upregulation of Diptericin, Attacin, and Metchnikowin were all significantly lower following an injection of rHb-ugt-1 as compared to BSA, whereas injection of rHba_19909 had no effect (Fig. 4A,C,E). Although Hba_19909 does appear to have a mild inhibitory effect on the upregulation of Diptericin as compared to BSA, this difference was not statistically significant (p = 0.08).
Recombinant Hb-ugt-1 suppresses a subset of antimicrobial peptide genes in Drosophila melanogaster. (A–E) Adult flies of the Oregon-R line of D. melanogaster were injected with approximately 7 ng of recombinant protein or BSA prior to homogenization and RNA extraction at 6-h post injection. Expression for the indicated antimicrobial peptide genes was normalized to rp49 and fold change was calculated relative to the 0-h time point. Average expression for three trials is shown where each trial consisted of two replicates with five flies each, three males and two females. Error bars indicate standard error and significance was assessed using a one-way ANOVA (**p < 0.01). (F) Alternatively, survival for injected RelE20 (Rel) flies was assessed every 12 h and compared to that of the wild type (wt) w1118 line that serves as their genetic background. Curves consist of average values for three trials at two replicates of 10 flies each per trial. Bars represent standard error and significance was assessed with a Mantel–Cox test (***p < 0.001). All graphs were generated with GraphPad Prism version 5.03 (www.graphpad.com).
To further examine the physiological significance of the rHb-ugt-1-reduced AMP gene expression, rHb-ugt-1 was injected into D. melanogaster Relish mutants, which are incapable of mounting an Imd-based response due to a lack of the terminal transcription factor in the Imd pathway15. While these mutants were capable of surviving a BSA injection at a rate comparable to wild type, an injection of rHb-ugt-1 resulted in significantly lower survival over a six-day period (Fig. 4F). Conversely, wild type flies injected with rHb-ugt-1 survived at a similar rate as buffer-injected controls. The suppressed AMP upregulation demonstrates that rHb-ugt-1 has immunosuppressive effects, and the mortality seen in the recombinant-injected mutants indicates that the degree of this immunosuppression is consequential to the survival of the insect.
Hb-ugt-1 suppresses the expression of broad-complex, an ecdysone-responsive transcription factor
The binding of 20E to its receptor is known to induce the expression of several transcription factors, including Broad-Complex (Br-C), which subsequently upregulates components of the immune response, including the Peptidoglycan Recognition Protein LC (PGRP-LC) and some AMPs, even in the absence of Imd pathway activity8. To assess whether ecdysone signaling might be impaired in rHb-ugt-1-injected D. melanogaster, larvae and adult flies were injected with 5 ng and 7 ng of rHb-ugt-1, respectively. They were then processed for Br-C expression measurements via qPCR (Fig. 5). In both cases, Br-C expression was significantly reduced, at a 6-h time point in adult flies (Fig. 5A) and at a 30-min time point in larvae (Fig. 5B). Because of the closely associated transcriptional response of Br-C to 20E signaling, this suggests that 20E signaling is diminished in insects injected with the recombinant protein. This reduction may also be responsible for the decreased upregulation of AMPs seen in response to injection of rHb-ugt-1, as disruption of Br-C alone has been found to be sufficient for a significant reduction in upregulation of AMPs, including Diptericin8.
Hb-ugt-1 recombinant protein suppresses Broad-Complex expression in Drosophila melanogaster adults and larvae. (A) Adult D. melanogaster Oregon-R were injected with approximately 7 ng of recombinant Hb-ugt-1 or BSA and assessed for Broad-Complex expression normalized to rp49 at a 6-h post injection. (B) Third instar larvae of the same line were co-injected with approximately 5 ng of recombinant protein and 1.5 ng of 20-hydroxyecdysone prior to collection at 0.5-h for RNA extraction and qPCR. Fold change relative to the 0-h time point is shown for three independent trials consisting of 10 larvae total across two replicates. Error bars indicate standard error and significance was assessed using a student’s t-test (**p < 0.01). Graphs were exported from GraphPad Prism version 5.03 (www.graphpad.com).
Pupation is delayed in larvae injected with rHb-ugt-1
To support the observed effect on Br-C expression, we injected D. melanogaster third-instar larvae (Fig. 6C) with 5 ng of rHb-ugt-1 or BSA and measured the timing of their commitment to pupation, as this process is regulated positively by ecdysone signaling. Commitment to pupation, based on immobility and spiracle inversion (Fig. 6B), was found to be approximately 50% lower following injection of rHb-ugt-1 as compared to BSA (Fig. 6A) at 8-h post injection. The total rate of pupation at a 24-h time point and the rate of eclosion were unaffected (Supplementary Fig. S3). This effect on pupation further implicates ecdysone signaling as a target of Hb-ugt-1. The fact that pupation is delayed, but not disabled also agrees with the prediction that a transient injection of rHb-ugt-1 will temporarily decrease the concentration of active 20E signaling hormone.
Hb-ugt-1 reduces the pupation rate of third instar Drosophila melanogaster larvae. (A) Third instar larvae of Oregon-R D. melanogaster injected with 5 ng of recombinant Hb-ugt-1 or BSA were surveyed at 8-h post-injection for immobility and spiracle inversion, which were used to indicate a commitment to pupation. Significantly fewer larvae begin pupation when injected with rHb-ugt-1 as compared to BSA. (B and C) Representative images are shown for a pupated larva featuring the characteristic spiracle inversion and a larva that has not begun pupation, respectively. Melanization is present at the wounding site for both larvae. Bars indicate the average rate collected from five independent trials and a total of 73 and 77 larvae, respectively, for BSA and rHb-ugt-1 injections. Error bars represent standard error and significance was assigned by Chi-Square analysis (*p < 0.05). The graph was generated using GraphPad Prism version 5.03 (www.graphpad.com).
Discussion
Based on its protein sequence, the candidate virulence factor gene Hb-ugt-1 was predicted to possess a conserved ecdysteroid glycosyltransferase domain. This by itself does not necessarily indicate that this glycosyltransferase functions as a virulence factor, as glycosyltransferases are involved in a wide variety of processes16. Still, targeting ecdysone signaling with a glycosyltransferase is apparently such an effective strategy for undermining a host that most baculoviruses maintain an ecdysteroid UGT in their genome17. With this in mind, the possibility of a similar role for Hb-ugt-1 warrants further investigation.
Typically, pathogens upregulate the expression of their virulence factors in response to host entry or physiological cues. Notably, we found that Hb-ugt-1 gene expression is rapidly increased and sustained in response to insect hemolymph, both in vitro and in vivo. The magnitude of this increase varied between treatments, but this is to be expected given that the degree of IJ activation in other entomopathogenic nematodes depends on a collection of factors, including host species18,19. Each treatment showed significant upregulation by one hour, which by itself indicates that Hb-ugt-1 could be involved in either virulence or the growth and development of the nematode, although the latter is made less likely by the observed downregulation following exposure to P. luminescens bacteria. Interestingly, the expression level of Hb-ugt-1 on P. luminescens returns to a level comparable to the 0-h time point at 48 h. This time point is developmentally relevant in that this is approximately when H. bacteriophora nematodes will transition from recovering juveniles to fourth stage pre-hermaphrodites20 and at which point they will cease development without their symbiotic bacteria21. This developmental stage is also the point at which ecdysteroids reach their highest concentration in Haemonchus contortus22. An important avenue of future research would be to examine a potential role for UGTs in H. bacteriophora development and uncover the modes of regulation that make this possible.
Another important characteristic of a virulence factor is that it is secreted to the exterior of the nematode so that it can interact with the host environment, as documented in other species23. To this end, we tested H. bacteriophora ES products for glycosyltransferase activity using UDP-glucose as a sugar donor and 20E as the acceptor molecule, as these are the prevalent substrates for glycosyltransferases of invertebrates and plants24,25. Glycosyltransferase activity was observed only from the ES products of hemolymph-activated IJs, indicating that H. bacteriophora does produce and externally secrete a glycosyltransferase in response to host exposure. It should be noted, however, that while this assay does demonstrate glycosyltransferase activity, additional work is required to confirm 20E as the substrate due to the fact that glycosyltransferases can act on other proteins and lipids present in the ES products26. The presence of glycosyltransferase activity was further supported by the identification of a GTDC1-labeled protein in the activated ES products of H. bacteriophora. While this protein is larger than the predicted size, it is within Hb-ugt-1 a range that could be attributed to post-translational modification. Other helminths also produce multiple isoforms of UDP-glycosyltransferases27, which may be the case with H. bacteriophora and could alternatively explain the size discrepancy.
To test the immunosuppressive capacity of the recombinant protein, we examined the induction of AMP encoding genes following injection of rHb-ugt-1, where the tissue damage caused by injection alone is known to activate both the humoral and cellular immune responses28,29. The AMP genes Diptericin, Attacin, and Metchnikowin were upregulated to a significantly lower degree following rHb-ugt-1 injection as compared to BSA injections. Drosomycin and Defensin (Fig. 4B and D) were induced at comparable levels for each treatment, but this may be at least partially due to a lower baseline responsiveness to injection injury. In the case of Defensin, this AMP would also not be expected to have an impact on the course of an infection as neither Steinernema nor Heterorhabditis nematodes induce Defensin expression in D. melanogaster larvae30,31. With regard to toxicity, rHb-ugt-1 was not found to induce mortality in wild type flies, but the lower survival rate in immunocompromised flies injected with recombinant protein does indicate that the immune or otherwise physiological effects are relevant to the survival of the fly. An exact cause of mortality in these flies unfortunately cannot be determined from these data alone, but this question represents an important point to resolve in future work.
Two separate ecdysone-responsive factors were tested to determine if the immunosuppressive effect of rHb-ugt-1 was related to ecdysone signaling. Expression of the transcription factor Br-C was selected due to its documented role as a link between ecdysone signaling and innate immunity32,33,34, and concordantly its expression was reduced in both D. melanogaster adults and larvae following injection of rHb-ugt-1. Furthermore, a delay in the onset of pupation was observed in third-instar larvae injected with the recombinant, similarly indicating diminished ecdysone signaling, as 20E is the primary signal for this process35. This indicates that the H. bacteriophora glycosyltransferase may be acting in a manner similar to the baculovirus UGTs, namely that its observed effects appear to stem from a capacity to deactivate the 20E signaling hormone.
Additional work must be done to fully elucidate the role of Hb-ugt-1 as a virulence factor with activity against 20E, but the findings presented here offer strong foundational support for this notion. Future work could expand on the information presented here by further clarifying the role of Hb-ugt-1 as it pertains to development within the nematode as well as virulence, and also the collection of immune impacts Hb-ugt-1 might have, given that ecdysone is involved in a variety of immune mechanisms, including the nodulation response36,37. The in vitro reaction properties of the recombinant enzyme may also be examined to investigate how this protein participates in the dynamics of a natural infection. These questions are of great interest for the description of host-parasite interactions, but this knowledge could have broader impacts as well. Information about individual virulence factors can be used to develop stronger parasites for the biocontrol of insect pests, which is a widespread application of Heterorhabditis species38, and to combat nematode virulence strategies, as steroid hormones likewise play a role in vertebrate immunity39, and may be targeted by glycosyltransferases of vertebrate-infective nematode parasites.
Methods
Infective juvenile activation for Hb-ugt-1 expression analysis
For in vitro activation experiments in insect hemolymph, 20 third-instar D. melanogaster larvae were collected, rinsed with water, and pinched with forceps near the mouth hooks to create a small incision. Before releasing the forceps, larvae were submerged in a 5 μl aliquot of a 2.5 μg/ml solution of phenylthiourea (PTU) in PBS, contained in a 0.5 ml centrifuge tube nested in a 1.5 ml centrifuge tube. Critically, the 0.5 ml tube featured an incision at the bottom of the tube allowing for passage of fluid, but not larvae from the 0.5 ml tube to the 1.5 ml tube. Hemolymph was collected from the larva by centrifugation at 4 °C for 30 s at 17,900 × g. The resulting approximately 10 μl of extracted hemolymph was diluted two-fold with PTU-PBS, filtered with a 10 μm polyethylene filter, and kept on ice to prevent melanization. This process was repeated three times to produce a sufficient volume of hemolymph for each activation trial. To collect Manduca sexta hemolymph, the posterior horn of a fifth instar larvae was surface sterilized with 70% ethanol and cut to release the hemolymph. The extracted hemolymph was diluted 1:4 in ice-cold Ringer’s buffer (100 mM NaCl, 1.8 mM KCl, 2 mM CaCl2, 1 mM MgCl2, and 5 mM HEPES, pH 6.9) supplemented with PTU to a final concentration of 0.33 mM. Before use, M. sexta hemolymph was centrifuged for 5 min at 4,000 × g and filtered with a 0.45 μm syringe filter. Approximately 1,000 H. bacteriophora IJs were transferred to a 1.5 ml centrifuge tube and washed twice with water by centrifugation for 30 s. After the second wash, the supernatant was discarded and a 40 μl aliquot of the extracted hemolymph solution was pipetted into the pellet of IJs and mixed before incubating at 28 °C for the specified time points. Axenic IJs for these experiments were generated as previously described14, where IJs were propagated in fifth or sixth instar G. mellonella larvae that had been infected with the RET16 derivative of P. temperata NC1.
In vivo activation of IJs was carried out by the injection of IJs directly into G. mellonella larvae. The injected aliquot of IJs was prepared by pelleting 5,000 nematodes as in the in vitro experiments, though with the addition of a 5-min axenization step using a 3% hypochlorite solution. These surface-sterilized IJs were washed three times with PBS and concentrated into a 100 μl volume of PBS. Nematodes were injected into Galleria larvae using an 18.5G needle and 1 ml syringe after the larvae had been surface-sterilized with 70% ethanol and anesthetized on ice for 15 min. The injected larvae were returned to the ice for a period of five minutes before being transferred onto slightly moist filter paper in a petri dish. After the allotted incubation time, the injected Galleria were dissected in PBS and an average of approximately 2,000 IJs were collected from the solution by pipette.
Gene expression changes in response to P. luminescens exposure were measured by directly exposing H. bacteriophora IJs to a bacterial lawn. Photorhabdus luminescens subspecies laumondii, strain TT01, was grown overnight in LB at 28 °C with shaking at 220 rpm. A 50 μl aliquot of overnight culture was spread in the center of LB agar plates, and the plates were incubated for 48 h at 28 °C. Approximately 2,000 washed H. bacteriophora IJs in a 100 μl aliquot of water were applied to the P. luminescens lawn and allowed to incubate at room temperature in the dark on a moist paper towel until the desired time point. In order to recover nematodes from the plate, 5 ml of M9 buffer were pipetted directly onto the plate and swirled gently before being collected and dispensed into a 15 ml Falcon tube. This process was performed three times in total for each plate to ensure optimal recovery.
Sequence alignment and phylogenetics
The sequence alignment was produced with BioEdit software (version 7.2.5) following the ClustalW algorithm and organized graphically with JalView software (version 2.11.0). The gap opening and extension penalties for pairwise alignments were set to 1 and 0.1, respectively, while the corresponding penalties for the multiple alignment were 3 and 0.2. Minor manual adjustment was permitted where the alignment could be improved unambiguously. The phylogenetic tree was constructed using MEGA software (version 10.1.5) according to the Maximum Likelihood method with 200 bootstrap replicates.
Glycosyltransferase activity in H. bacteriophora secreted products
Excretory-secretory products were collected from axenic H. bacteriophora IJs as described previously14. Briefly, 200,000 IJs were soaked in 25% M. sexta hemolymph for a period of 20 h, washed in Ringer’s buffer, and placed in fresh Ringer’s buffer for a 5-h ES product collection period. The resulting supernatants were filtered and concentrated such that supernatants from three preparations of 200,000 IJs each were pooled and concentrated to a final volume of 100 μl. This allowed standardization of ES products to equal numbers of IJs so that despite activated and non-activated products containing different concentrations of protein, they both contain 2,000 IJ equivalents/μl. Glycosyltransferase activity was assayed with a colorimetric assay specific for phosphate-coupled glycosyltransferase reactions (R&D Systems Glycosyltransferase Activity Kit EA001) according to the manufacturer’s instructions. The glycosyltransferase activity of 10,000 IJ equivalents (5 μl) was measured using 5 mM UDP-glucose (Abcam) and 5 mM 20-hydroxyecdysone (Sigma) in DMSO as the sugar donor and acceptor for the reaction, respectively. Reactions were incubated at 37 °C for 3 h, at which point the samples were diluted with 100 μl of deionized water in a 96-well flat bottom plate (Greiner) and 30 μl each of the provided Malachite Green reagents were added sequentially. Each sample was incubated for an additional 20 min at room temperature before absorbance was measured at 620 nm. Absorbance measurements collected from three independent experiments were analyzed for statistical significance by one-way ANOVA.
Protein electrophoresis and western blotting
As described previously14, the concentration of protein in activated ES products was determined using a Pierce BCA Protein Assay Kit (Thermo Scientific). As the activated products were the only preparation to produce a readable signal from the BCA assay, a 10 μl aliquot corresponding to 150 ng of activated products and an equivalent 10 μl volume of non-activated products were reduced in 50 mM DTT and loaded into a Novex WedgeWell 4–20% Tris–Glycine Gel (Invitrogen). The composition of each well was adjusted to 26 μl of sample and water, 4 μl of DTT, and 10 μl of Laemmli buffer. Additionally, 5 μl of PageRuler Plus Prestained Protein Ladder (Thermo Scientific) were loaded separately for size estimation. To label glycosyltransferases in the ES products, the gel was transferred to nitrocellulose with a Trans-Blot Turbo Transfer System (Bio Rad) and blocked overnight at 4 °C with 5% powdered milk in Tris-NaCl-Tween (TNT, 25 mM Tris pH 7.4, 0.5 M NaCl, 0.1% Tween 20)40. The membrane was then incubated with a 1:2,000 dilution of GTDC1 rabbit polyclonal antibody (Proteintech) in 5% powdered milk for 2 h at room temperature. Secondary antibody labeling was achieved with a 1-h incubation with a 1:2,000 dilution of anti-rabbit IgG, HRP-linked antibody (Cell Signaling Technology) in 5% dry milk at room temperature. Signal for the HRP substrate was generated by exposure to Supersignal West Femto Maximum Sensitivity Substrate (Thermo Fisher) for 5 min.
Recombinant expression of H. bacteriophora candidate genes
Two candidate virulence factor genes were expressed in Sf9 cells using a pMIB/V5-His A expression vector. Initial amplicons were generated using primers specific for Hb-ugt-1 (F: 5′ CTTGGTACCTAAAATCCTAGTCTTTAGCC 3′; R: 5′ AGACTCGAGTTCGGATTTCATTTTTTTCTCCG 3′) and Hba_19909 (F: 5′ CTTGGTACCCAATTGCCTTCATTGTATTTG 3′; R: 5′ AGACTCGAGCGAACAGCCACAGCACTTTTTGA 3′), each modified to contain restriction sites for KpnI and XhoI at the 5′ and 3′ ends, respectively, of the amplicons. Following cloning of the amplicons, One Shot TOP10 chemically competent E. coli were transformed with these constructs and selected on LB plates containing 100 μg/mL ampicillin. The same primers were used to confirm successful transformations via colony-PCR prior to growth in liquid culture, plasmid isolation, and sequencing. Plasmids featuring correct sequences were transfected into Sf9 insect cells using Cellfectin II reagent (Invitrogen) and selectively allowed to colonize wells in a 6-well plate by propagating in Sf-900 II SFM (Thermo Fisher) containing 10 μg/ml blasticidin and 15 μg/ml gentamycin. Upon sufficient growth and differentiation from untransfected cells, cultures were suspended in 300 ml of liquid medium and allowed to grow at 27 °C with shaking at 130 rpm until they reached a concentration of 6 × 106 cells/ml. Culture supernatants were queried for the presence of recombinant protein by western blot, targeting the V5 epitope (Anti-V5-HRP antibody, Invitrogen), and if positive, the total culture supernatants were prepared for purification, first through concentration driven by 8 kDa polyethylene glycol flakes, and then dialysis in 150 mM sodium phosphate at 4 °C for approximately 16 to 20 h. The resulting solution was centrifuged to remove debris, diluted 1:3 in low-stringency wash buffer (50 mM NaH2PO4, 500 mM NaCl, 40 mM Imidazole, 0.5% Tween 20), and co-incubated with 1 ml Ni-NTA agarose (Qiagen) for 1 h rotating at 90 rpm at room temperature. The recombinant-bound matrices were loaded onto an affinity purification column and washed with 10 equivalents of low-stringency wash buffer. Following this washing step, elution proceeded with two individual washes of high-stringency buffer (50 mM NaH2PO4, 500 mM NaCl, 100 mM imidazole, 0.5% Tween 20) followed by 6 individual washes with elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole). Each aliquot was assessed again by western blot and those fractions containing recombinant protein were further concentrated against polyethylene glycol and dialyzed overnight in PBS. Before storage at − 20 °C, protein concentration was determined with a Pierce BCA Protein Assay Kit (Thermo Fisher) and the solution containing the purified recombinant protein was supplemented with protease inhibitor cocktail (Sigma).
Injection of flies and larvae for the assessment of gene expression, survival, and pupation rate
All injections were performed with a Drummond Nanoject III Programable Nanoliter Injector, where treatments were transmitted through an oil-filled pulled glass capillary that had been opened with forceps to a degree that would allow delivery of the treatments while causing minimal damage to the organism. Adult flies were injected between 7 and 10 days after eclosion, whereas larvae were selected once they had reached the wandering third instar phase. In both cases, D. melanogaster stocks were maintained on a yeast—supplemented cornmeal-soy-based diet (Meidi Laboratories) at 25 °C on a 12-h day-night cycle. Oregon-R flies were used for AMP and Br-C expression studies while survival rates were collected in reference to the RelE20 line and its associated background, w1118. All injections were preceded by a period of anesthetization with carbon dioxide, though generally the technique was adapted to optimize the recovery of each life stage. Adults were injected intramesothoracially with an approximately 69 nl aliquot containing 7 ng of recombinant protein or BSA in PBS-protease inhibitor solution before placement in vials containing instant Drosophila medium (Carolina Biological) incubated under the same conditions. Survival was assessed once every 12 h, or flies were collected at the 6-h time point for gene expression analysis. Alternatively, Oregon-R larvae were anesthetized with carbon dioxide for 2 to 3 min before being placed on slightly moist filter paper and injected with 5 ng of each treatment. Injections were delivered at a shallow angle between segments in the dorsal side of the abdomen, such that the probability of damaging the imaginal discs or organs would be low. Injected larvae were placed on fresh filter paper moistened with Ringer’s buffer and incubated at 25 °C for a period of 8 h, at which point larvae were observed for characteristics indicative of pupation.
Gene expression analysis
As previously described14, RNA was extracted from five flies or larvae, or the entirety of the activated nematodes. Isolation was achieved via homogenization in TRIzol reagent (Ambion, Life Technologies), where IJs were freeze-thawed at − 80 °C for four repetitions prior to pestle homogenization to enhance yield. Reverse transcription was performed on 1 μg of total RNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative PCR (qPCR) reactions were conducted and monitored with a CFX96 Real-Time System, C1000 Thermal Cycler (Bio-Rad) with the following cycling conditions: 95 °C for 2 min, 40 repetitions of 95 °C for 15 s followed by 61 °C for 30 s, and then one round of 95 °C for 15 s, 65 °C for 5 s, and finally 95 °C for 5 s. Each reaction well contained 10 μl GreenLink No-ROX qPCR Mix (BioLink), 40 ng of cDNA template, forward and reverse primers at a final concentration of 200 nM and ultrapure water to 20 μl total. Primers were as follows: Diptericin (F: 5′ GCTGCGCAATCGCTTCTACT 3′; R: 5′ TGGTGGAGTTGGGCTTCATG 3′), Defensin (F: 5′ CGCATAGAAGCGAGCCACATG 3′; R: 5′ GCAGTAGCCGCCTTTGAACC 3′), Drosomycin (F: 5′ GACTTGTTCGCCCTCTTCG 3′; R: 5′ CTTGCACACACGACGACAG 3′), Metchnikowin (F: 5′ TCTTGGAGCGATTTTTCTGG 3′; R: 5′ AATAAATTGGACCCGGTCTTG 3′), Attacin (F: 5′ CAATGGCAGACACAATCTGG 3′; R: 5′ ATTCCTGGGAAGTTGCTGTG 3′) Hb-ugt-1 (F: 5′ TTCTTAACGACACGCGACTG 3′; R: 5′ CTCGTGCTGCAGATTCTTGA 3′), Broad-Complex (F: 5′ GAGCACACCCTGCAAACAC 3′; R: 5′ GCTGCGTGAGTCCAGAGAC 3′), rp49 (F: 5′ GATGACCATCCGCCCAGCA 3′; R: 5′ CGGACCGACAGCTGCTTGGC 3′), and rpl32 (F: 5′ ATCGGATAGATACCACCGCC 3′; R: 5′ TTGTGGGCATAGCACG 3′).
Statistical analysis for gene expression was performed on values derived from qPCR measurements processed according to the 2−ΔΔCT method41,42 with all values being normalized to rp49 in the case of flies and rpl32 for H. bacteriophora. The expression of Hb-ugt-1 in H. bacteriophora was assessed using a one-way ANOVA comparing dCt values gathered from three independent experiments. A Bonferroni multiple comparisons test was used to assign significance to comparisons between specific treatments. For all other gene expression assays, ddCt values were collected from three independent trials at two replicates per trial. Antimicrobial peptide gene expression was assessed with a one-way ANOVA as above while Broad-Complex expression was analyzed with a student’s t-test.
References
O’Reilly, D. R. & Miller, L. K. A baculovirus blocks insect molting by producing ecdysteroid UDP-glucosyl transferase. Science 245, 1110–1112 (1989).
Hughes, A. L. Origin of ecdysosteroid UDP-glycosyltransferases of baculoviruses through horizontal gene transfer from lepidoptera. Coevolution 1, 1–7 (2013).
Evans, O. P. & O’reilly, D. R. Purification and kinetic analysis of a baculovirus ecdysteroid UDP-glucosyltransferase. Biochem. J. 330, 1265–1270 (1998).
Kaplanis, J. N., Thompson, M. J., Dutky, S. R. & Robbins, W. E. The ecdysteroids from the tobacco hornworm during pupal-adult development five days after peak titer of molting hormone activity. Steroids 34, 333–345 (1979).
Yamanaka, N., Rewitz, K. F. & O’Connor, M. B. Ecdysone control of developmental transitions: lessons from Drosophila research. Annu. Rev. Entomol. 58, 497–516 (2013).
O’Reilly, D. R. Baculovirus-encoded ecdysteroid UDP-glucosyltransferases. Insect Biochem. Mol. Biol. 25, 541–550 (1995).
Verma, P. & Tapadia, M. G. Early gene Broad complex plays a key role in regulating the immune response triggered by ecdysone in the Malpighian tubules of Drosophila melanogaster. Mol. Immunol. 66, 325–339 (2015).
Rus, F. et al. Ecdysone triggered PGRP-LC expression controls Drosophila innate immunity. EMBO J. 32, 1626–1638 (2013).
Tanji, T., Hu, X., Weber, A. N. R. & Ip, Y. T. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol. Cell Biol. 27, 4578–4588 (2007).
Aguinaldo, A. M. A. et al. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature 387, 489–493 (1997).
Liu, C., Enright, T., Tzertzinis, G. & Unnasch, T. R. Identification of genes containing ecdysone response elements in the genome of Brugia malayi. Mol. Biochem. Parasitol. 186, 38–43 (2012).
Parihar, M. et al. The genome of the nematode Pristionchus pacificus encodes putative homologs of RXR/Usp and EcR. Gen. Comp. Endocrinol. 167, 11–17 (2010).
Vadnal, J. et al. Identification of candidate infection genes from the model entomopathogenic nematode Heterorhabditis bacteriophora. BMC Genom. 18, 8 (2017).
Kenney, E., Hawdon, J. M., O’Halloran, D. & Eleftherianos, I. Heterorhabditis bacteriophora excreted-secreted products enable infection by Photorhabdus luminescens through suppression of the Imd pathway. Front. Immunol. 10, 2372 (2019).
Hedengren, M. et al. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol. Cell. 4, 827–837 (1999).
Huang, F.-F. et al. The UDP-glucosyltransferase multigene family in Bombyx mori. BMC Genomics 9, 563 (2008).
Ahn, S.-J., Vogel, H. & Heckel, D. G. Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect Biochem. Mol. Biol. 42, 133–147 (2012).
Lu, D., Sepulveda, C. & Dillman, A. R. Infective juveniles of the entomopathogenic nematode steinernema scapterisci are preferentially activated by cricket tissue. PLoS ONE 12, e0169410 (2017).
Alonso, V., Nasrolahi, S. & Dillman, A. R. Host-specific activation of entomopathogenic nematode infective juveniles. Insects 9, 59 (2018).
Johnigk, S.-A. & Ehlers, R.-U. Juvenile development and life cycle of Heterorhabditis bacteriophora and H. indica (Nematoda: Heterorhabditidae). Nematology 1, 251–260 (1999).
Han, R. & Ehlers, R.-U. Pathogenicity, development, and reproduction of Heterorhabditis bacteriophora and Steinernema carpocapsae under axenic in vivo conditions. J. Invertebr. Pathol. 75, 55–58 (2000).
Fleming, M. W. Ecdysteroids during development in the ovine parasitic nematode, Haemonchus contortus. Comp. Biochem. Physiol. Part B Comp. Biochem. 104, 653–655 (1993).
Chang, D. Z., Serra, L., Lu, D., Mortazavi, A. & Dillman, A. R. A core set of venom proteins is released by entomopathogenic nematodes in the genus Steinernema. PLoS Pathog. 15, e1007626 (2019).
Caradoc-Davies, K. M., Graves, S., O’Reilly, D. R., Evans, O. P. & Ward, V. K. Identification and in vivo characterization of the Epiphyas postvittana nucleopolyhedrovirus ecdysteroid UDP-glucosyltransferase. Virus Genes 22, 255–264 (2001).
Bock, K. W. The UDP-glycosyltransferase (UGT) superfamily expressed in humans, insects and plants: animal plant arms-race and co-evolution. Biochem. Pharmacol. 99, 11–17 (2016).
Breton, C., Fournel-Gigleux, S. & Palcic, M. M. Recent structures, evolution and mechanisms of glycosyltransferases. Curr. Opin. Struct. Biol. 22, 540–549 (2012).
Matoušková, P. et al. UDP-glycosyltransferase family in Haemonchus contortus: phylogenetic analysis, constitutive expression, sex-differences and resistance-related differences. Int. J. Parasitol. Drugs Drug Resist. 8, 420–429 (2018).
Kenmoku, H., Hori, A., Kuraishi, T. & Kurata, S. A novel mode of induction of the humoral innate immune response in Drosophila larvae. Dis. Model Mech. 10, 271–281 (2017).
Márkus, R., Kurucz, É, Rus, F. & Andó, I. Sterile wounding is a minimal and sufficient trigger for a cellular immune response in Drosophila melanogaster. Immunol. Lett. 101, 108–111 (2005).
Hallem, E. A., Rengarajan, M., Ciche, T. A. & Sternberg, P. W. Nematodes, bacteria, and flies: a tripartite model for nematode parasitism. Curr. Biol. 17, 898–904 (2007).
Peña, J. M., Carrillo, M. A. & Hallem, E. A. Variation in the susceptibility of Drosophila to different entomopathogenic nematodes. Infect. Immun. 83, 1130–1138 (2015).
Xiong, X.-P. et al. miR-34 modulates innate immunity and ecdysone signaling in Drosophila. PLoS Pathog. 12, e1006034 (2016).
Zheng, W. et al. Dehydration triggers ecdysone-mediated recognition-protein priming and elevated anti-bacterial immune responses in Drosophila Malpighian tubule renal cells. BMC Biol. 16, 60 (2018).
Ma, H. et al. 20-Hydroxyecdysone regulates the transcription of the lysozyme via broad-complex Z2 gene in silkworm, Bombyx mori. Dev. Comp. Immunol. 94, 66–72 (2019).
Kaieda, Y. et al. Glue protein production can be triggered by steroid hormone signaling independent of the developmental program in Drosophila melanogaster. Dev. Biol. 430, 166–176 (2017).
Sorrentino, R. P., Carton, Y. & Govind, S. Cellular immune response to parasite infection in the Drosophila lymph gland is developmentally regulated. Dev. Biol. 243, 65–80 (2002).
Franssens, V. et al. 20-Hydroxyecdysone and juvenile hormone regulate the laminarin-induced nodulation reaction in larvae of the flesh fly, Neobellieria bullata. Dev. Comp. Immunol. 30, 735–740 (2006).
Labaude, S. & Griffin, C. T. Transmission success of entomopathogenic nematodes used in pest control. Insects 9, 72 (2018).
Benagiano, M., Bianchi, P., D’Elios, M. M., Brosens, I. & Benagiano, G. Autoimmune diseases: role of steroid hormones. Best Pract. Res. Clin. Obstet. Gynaecol. 60, 24–34 (2019).
Sherman, L. S., Schrankel, C. S., Brown, K. J. & Smith, L. C. Extraordinary diversity of immune response proteins among sea urchins: nickel-isolated Sp185/333 proteins show broad variations in size and charge. PLoS ONE 10, e0138892 (2015).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 (2008).
Acknowledgements
We thank members of the Department of Biological Sciences at The George Washington University (GWU) for critical reading of the manuscript, and Kyle Devine for rearing the flies. This work was supported by a George Washington University facilitating fund awarded to D.O'H., J.H., and I.E., and a Harlan summer fellowship to E.K.
Author information
Authors and Affiliations
Contributions
E.K. conceived and designed the experiments, performed the experiments, analyzed the data, wrote the paper, prepared the figures, and reviewed drafts of the paper. A.Y. provided guidance on recombinant expression and reviewed drafts of the paper. J.H., D.O'H., L.G. and I.E. conceived and designed the experiments, supervised the project, reviewed drafts of the paper, and prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kenney, E., Yaparla, A., Hawdon, J.M. et al. A putative UDP-glycosyltransferase from Heterorhabditis bacteriophora suppresses antimicrobial peptide gene expression and factors related to ecdysone signaling. Sci Rep 10, 12312 (2020). https://doi.org/10.1038/s41598-020-69306-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-69306-2
This article is cited by
-
Dauer juvenile recovery transcriptome of two contrasting EMS mutants of the entomopathogenic nematode Heterorhabditis bacteriophora
World Journal of Microbiology and Biotechnology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.