Abstract
Monilinia blight disease caused by Monilinia vaccinii-corymbosi (Reade) Honey (M.vc) causes severe damage and economic losses in wild blueberry growing regions. Molecular mechanisms regulating defence responses of wild blueberry phenotypes towards this causal fungus are not yet fully known. A reliable quantification of gene expression using quantitative real time PCR (qPCR) is fundamental for measuring changes in target gene expression. A crucial aspect of accurate normalisation is the choice of appropriate reference genes. This study evaluated the expression stability of seven candidate reference genes (GAPDH, UBC9, UBC28, TIP41, CaCSa, PPR and RH8) in floral tissues of diploid and tetraploid wild blueberry phenotypes challenged with M.vc. The expression stability was calculated using five algorithms: geNorm, NormFinder, BestKeeper, deltaCt and RefFinder. The results indicated that UBC9 and GAPDH were the most stable reference genes, while RH8 and PPR were the least stable ones. To further validate the suitability of the analyzed reference genes, the expression level of a pathogenesis related protein gene (i.e., PR3) was analysed for both phenotypes at four time points of infection. Our results may be beneficial for future studies involving the quantification of relative gene expression levels in wild blueberry species.
Similar content being viewed by others
Introduction
Monilinia blight caused by Monilinia vaccinii-corymbosi (Reade) Honey, is a destructive disease in commercial wild blueberry fields in the Atlantic Provinces of Canada and Maine, US. Under favourable weather conditions, infections may cause considerable losses in yield and post-harvest quality of berries1,2. The pathogen infects almost all blueberry species, such as V. angustifolium (sweet lowbush blueberry), V. myrtilloides (sour top lowbush blueberry), and V. corymbosum L. (northern highbush blueberry). The wild blueberry fields comprise tetraploid, V. angustifolium, V. a. f. nigrum, and diploid, V. myrtilloides phenotypes. Commercial wild blueberry fields mostly consist of Vaccinium angustifolium (tetraploid) (~ 70–80% on a surface area basis), V. myrtilloides (diploid) (~ 10–20%) and a few hybrids3.
Monilinia damage is often variable in commercial fields due to varying levels of genotypic resistance, differences in morphological features, and bud growth and development among phenotypes. Over the years, minimal damage from monilinia blight has been reported in V. myrtilloides and has been identified as the potential source of Monilinia blight resistance4,5. In addition to genetic and morphological variability, sufficient variations in yield parameters (berry size, number of berries per cluster, stem density) have been reported between the two phenotypes6. Although the Monilinia–Vaccinium pathosystem is an important phenomenon, a detailed molecular understanding of gene expression profile during the infection process is lacking within and between the phenotypes in wild blueberries.
Analyzing stress responsive genes from diverse biological samples under experimental conditions provides insights into the underlying signalling and regulatory mechanisms7. Quantitative real time PCR (qRT-PCR) has become an effective approach to examine and validate the changes in gene expression owing to its accuracy, specificity and sensitivity8,9,10,11. However, the reliability of the technique is largely influenced on the normalization strategy such as the usage of one or more stable reference genes. Ideally, the reference genes used for normalisation should have a uniform expression regardless of the experimental conditions11,12. Hence, the selection and validation of stable reference genes for each experimental condition is a prerequisite for performing qRT-PCR studies13,14,15.
In the present study, seven reference genes (GAPDH, CaCSa, TIP41, UBC28, PPR, UBC9 and RH8) were selected as candidate reference genes based on previous reports in Vaccinium spp16,17. Their expression stabilities in floral tissues of diploid and tetraploid wild blueberry phenotypes challenged with Monilinia vaccinii- corymbosi was evaluated at four time-points after infection. Five different statistical software programs (geNorm8, NormFinder18, BestKeeper19, delta Ct20 and RefFinder21) were used to analyse the stability of the candidate reference genes and to select the most appropriate ones. This study will lay a foundation for future gene expression research in wild blueberry.
Results
Selection of reference genes, amplification efficiency and specificity
A total of seven candidate reference genes were selected to identify suitable RGs for gene expression studies using qPCR in wild blueberry. Additionally, PR3 gene was used to validate the accuracy of identified RGs in wild blueberry phenotypes infected with M. vaccinii-corymbosi. The specificity of the analysed primer pairs was confirmed via detecting single fragment of the expected size on 2% agarose gel electrophoresis and a single peak with no signals on the negative controls in the melt curve analysis (Table 1, Supplementary Fig S1 & S2). All the tested RGs had efficiency (E %) values ranged from 95 to 105%, with regression coefficient (R2) varying from 0.994 to 0.999 (Table 1, Supplementary Fig S3). The results showed that all the primer pairs were suitable for RT-qPCR analysis.
Expression profiling of candidate reference genes
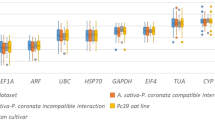
The raw quantification cycle (Cq) values were used to quantify the expression levels of candidate reference genes where lower Cq values mean higher expression levels. Cq values for each of the seven candidate reference genes in V. myrtilloides and V. a. f. nigrum are listed in Supplementary Table S1 (there were no Cq values in the negative controls), and a box and whiskers plot were used to describe the raw Cq value distribution (Fig. 1). In V. myrtilloides, the Cq values varied from 19.03 to 26.65, while it was 18.81 to 29.18 in V. a. f. nigrum. GAPDH was the most expressed gene in both phenotypes with a mean of 19.94 and 20.32 respectively. The stability was analysed in comparison with the time-course of disease infection.
Stability ranking of the candidate reference genes
Expression stabilities of the seven candidate reference genes were determined using geNorm, NormFinder, ΔCq, and BestKeeper and their overall stabilities were ranked by RefFinder across all the timepoints and phenotypes.
geNorm analysis
The expression stability rankings based on the M-values for the seven candidate reference genes on wild blueberry phenotypes is displayed (Table 2). The gene with the lowest M-value (cut-off 1.5) was the most stable reference gene in terms of gene expression and vice versa. The M-values for the tested genes in all samples and groups were lower than the default limit of 1.5. Genorm analysis for V. myrtilloides illustrated that CaCSa (0.385) and GAPDH (0.386) were the more stable genes and PPR (0.888) exhibited the least stability. On contrary, UBC9 (0.239) and GAPDH (0.239) exhibited high expression stability in V. a. f. nigrum and RH8 (0.432) were the least stable one (Table 2). Among the total samples, GAPDH and UBC9 were the most stable genes with M values of 0.487 and 0.512, whereas RH8 exhibited least stability (0.701). Finally, the pairwise variation (Vn/Vn+1) for both the phenotypes and entire group resulted in V2/3 < 0.15 (Fig. 2) which indicated that two reference genes were sufficient for accurate normalisation of RT-qPCR data.
NormFinder analysis
The expression stability of the seven candidate genes was analyzed using NormFinder, which is an excel based mathematical tool that measures gene expression stability by comparing the variation within and between user-defined sample groups. NormFinder ranks the control genes on the basis of their stability value (SV), lower value indicates higher gene expression stability and vice versa. The NormFinder algorithm results agreed with GeNorm analysis (Table 2). NormFinder selected CaCSa (SV = 0.043) and UBC9 (SV = 0.047) as the most stable genes for V. myrtilloides, whereas, GAPDH (0.014) and PPR (0.047) for V. a. f. nigrum. For overall analysis, UBC9 (0.080) and CaCSa (0.104) exhibited the most stable genes and RH8 the least stable one (0.195).
BestKeeper analysis
BestKeeper ranks the stabilities of the candidate reference genes based on their standard deviation (SD) and the coefficient of variation (CV). Genes with SD > 1 were considered unacceptable reference genes. Based on the results from the BestKeeper analysis for V. myrtilloides, all genes except RH8 were calculated to have an SD value lower than 1 (Table 2). The rankings by BestKeeper analysis for V. myrtilloides showed that the most stable reference genes were CaCSa (0.62) followed by UBC28 (0.75) and UBC9 (0.75). For V. a. f. nigrum UBC28 (0.42) and GAPDH (0.43) were observed as the most stable ones (Table 2). In the total dataset, UBC28 and CaCSa were the most stable genes, with SD values of 0.59 and 0.62 respectively (p < 0.001). For all the three groups RH8 exhibited the least stability.
ΔCq analysis
The ranking order of the seven candidate genes evaluated using ΔCq method were listed in Table 2. For V. myrtilloides, UBC28 and CaCSa ranked as the most two stable RGs, whereas, for V. a. f. nigrum UBC9 and GAPDH ranked as the top two. For the entire dataset, the results were similar to geNorm and NormFinder, with UBC9 as the consistently stable reference gene, only differing in the ranking order. RH8 ranked as the least stable gene as demonstrated by other algorithms.
RefFinder analysis
A comprehensive ranking was performed to confirm the stability ranking of the seven candidate reference genes. In the comparative analysis of reference genes from V. myrtilloides and V. a. f. nigrum, UBC9 and UBC28 were ranked as the most stable RGs (Fig. 3). However, due to the differences in ploidy as well as defense response level, a comprehensive ranking order of reference genes was generated for each phenotype. The expression of CaCSa, UBC9 and UBC28 were found to be the most stable for gene expression normalization in V. myrtilloides. While, GAPDH, UBC9 and PPR ranked as the best for V. a. f. nigrum. By contrast, RH8 and TIP41 ranked as the least stable genes for both the phenotypes.
Validation of the best and least ranked reference genes
To validate the effectiveness of the selected reference genes, the expression pattern of PR3 (MK292725) was analyzed in wild blueberry-Monilinia pathosystem. The relative transcript abundances of PR3 gene was normalized to two most stable genes (UBC9 and GAPDH) for V. a. f. nigrum and (UBC9 and UBC28) for V. myrtilloides as resulted from the comprehensive analysis.
In V. myrtilloides, the tolerant phenotype, data normalizations using the two most stable reference genes (UBC9 and UBC28) resulted in consistent PR3 expression pattern with gradual increase in expression over time points (Fig. 4A). However, in V. a. f. nigrum, the susceptible phenotype, the expression was high at day 0, then reduced the expression and not significantly elicited compared to control.
Relative expression of PR3 gene at four time points (0, 3, 6 and 10 days) after inoculation using M. vaccinii-corymbosi. (A) V. myrtilloides normalized using the two most stable reference gene (UBC9 and UBC28) and (B) V. a. f. nigrum normalized using the stable reference genes (UBC9 and GAPDH). Values represent the means ± standard errors, where n = 3 biological replicates (with each replicate comprising tissue pooled from 15 stems).
The expression data was also normalized using the least stable gene (RH8) reported for the two phenotypes (Fig. 4B). For both V. a. f. nigrum and V. myrtilloides no remarkable expression observed for all the analysed timepoints. These results indicated that the least stable gene, RH8 failed to standardize the expression data effectively. Our results confirm that using different reference genes for normalisation causes great differences among the expression patterns.
Discussion
RT-qPCR has emerged as a powerful tool to study transcript abundance of a specific gene in distinct biological samples owing to its precision, accuracy and sensitivity11. However, accurate normalization of gene expression remains a major criterion during qPCR analysis, as various steps during qPCR analysis can introduce variations arising from RNA extractions, cDNA synthesis, PCR procedure and sample loading13. Using a stably expressed reference gene is a prerequisite to obtain accurate interpretation of the transcript abundance results. Ideally, a reference gene should have constant expression in samples irrespective of the experimental conditions, developmental stages or species22,23. Several studies have reported the observation of variable expression of traditional reference genes in different plant species, as they have differential expression under different experimental conditions24,25,26. For example, Czechowski et al.27 reported that Arabidopsis have demonstrated variations in reference gene stability under different experimental conditions. Thus, the most appropriate reference genes should be properly evaluated and confirmed in all biological samples.
Many reliable reference genes have been determined in Vaccinium spp16,17,28 however, this study is the first to assess reference genes in wild blueberry-monilinia pathosystem. Also, the uniqueness of this study is that it was performed under field conditions. Field conditions differ from controlled environmental conditions, as plants are exposed to adverse environmental factors and multiple stresses. As stated by Samarth and Jameson29, the selection of appropriate reference genes from field conditions is more complex than controlled environmental studies. Tashiro et al.30 also pointed out the necessity of reference gene validation in studies involving non-model plant species from heterologous plant population. In this study, we assessed seven RGs for their use as internal controls in gene expression studies of the wild blueberry phenotypes upon monilinia blight infection under field conditions.
In the present study, four algorithms (geNorm8, NormFinder18, BestKeeper19 and delta CT20) was used to evaluate the best suited reference gene in wild blueberry phenotypes, V. myrtilloides and V. a. f. nigrum, for studying gene expression pattern during Monilinia blight infection. Several studies indicated the use of multiple statistical algorithms which not only minimize the errors associated with reference gene selection but ensures a more reliable evaluation31,32,33,34. This is probably due to the differences in algorithm programs exhibited by each method23,33. Moreover, variability in the stability ranking of reference genes based on algorithms has been reported in several studies35,36. Interestingly, the ranking order generated by using four programs roughly the same for both the phenotypes, with the first three most stable genes differing only on the ranking order. In addition, the pairwise variation determined using geNorm could be used as an indicator for estimating the optimal number of reference genes for normalization. In this study, the pairwise variation values for both V. myrtilloides and V. a. f. nigrum was V2/3 < 0.15, indicates that two reference genes were sufficient for gene expression normalization.
Even though the ranking order of the candidate reference genes may have differed, all the statistical programs showed UBC9 as the most stable reference gene in the analysed phenotypes having different ploidy level. In this study, GAPDH and UBC9 were observed as most stable reference genes for V. angustifolium. f. nigrum and UBC9, UBC28 and CaCSa for V. myrtilloides. GAPDH was used for normalization in studies with V. myrtillus- Botrytis pathosystem37. Vashisth et al.16 and Die and Rowland17 ranked reference genes on vegetative and reproductive organs of rabbiteye and southern highbush blueberry. Vashisth et al.16 reported CaCSa, RH8, and UBC28 as the most stably expressed gene in southern highbush blueberry across multiple organs analysed. Die and Rowland17 also reported RH8, CaCSa, PPR, GAPDH and UBC9 (M = 0.483 and CV = 0.210) as the most stably expressed reference genes for floral bud tissues evaluated in highbush blueberry. However, in our study, RH8 and TIP41 were found to be the least stable reference gene for all the assessed phenotypes. Several studies have reported variable expression levels in reference genes among closely related species38. Moreover, several studies demonstrated variations in expression profiles of reference genes in different pathosystems33,39.
PR3 was used as a target gene to validate the credibility of the selected reference genes. PR3 which include chitinases of classes Ia, Ib, II, IV, VI, and VII are important weaponry of plants against pathogens40. According to Thomma et al.41, PR3 which belong to the pathogenesis-related (PR) protein family play an important role in plant defense response to necrotrophic pathogens. Several studies reported the up-regulation of PR genes against many phytopathogenic fungi42,43,44. When comparing the most stable and least stable reference genes, the expression of target gene was consistent and upregulated in V. myrtilloides (UBC9 and UBC28) and V. a. f. nigrum (UBC9 and GAPDH) even though differences in expression observed between the phenotypes. However, no response observed in both phenotypes when analysed using the least stable gene (RH8). Our results agree with the findings of Cardot et al.45, where elicitation of chitinase genes observed in tolerant than in susceptible varieties.
In conclusion, this is the first study in which a set of candidate reference genes was analysed in terms of their expression stability in wild blueberry phenotypes infected with Monilinia vaccinii-corymbosi. Five different statistical algorithms showed slight differences in the final ranking of reference gene, however by combining and analysing the data together, we demonstrated that UBC9 is the most stably expressed transcript in wild blueberry phenotypes regardless of ploidy level.
Methods
Plant material, M.vc inoculations and experimental design
The wild blueberry diploid (V. myrtilloides) and tetraploid (V. angustifolium f. nigrum) phenotypes were selected from a commercial wild blueberry field, NS, Canada. Three biological replicates were selected for each phenotype and each replicate was separated into two, 0.5 × 1 m sample areas. One day before inoculation, one sample area within each replicate was sprayed with the fungicide Proline (a.i. prothioconazole) at a rate of 315 ml product·ha-1 using a CO2 powered, Bell spray Inc. hand-held research sprayer with 2 m boom with 4 Tee Jet Visiflow 8003VS nozzles at a pressure of 220 kPa to serve as control plots. Monilinia vaccinii-corymbosi inoculum was prepared from four-week old M.vc cultures isolated from monilinia blighted shoots and mummy berries. Floral buds at F3 stage (floral bud scale separation and appearance of new growth) was tagged for each phenotype and inoculum (2 × 105 ascospores·mL−1) was sprayed at all angles until runoff. Each phenotype sample area was immediately covered with 2 mm plastic film and row cover to provide incubating conditions (100% RH), which created conditions required for Monilinia infection46. After 72 h, the plastic film and row cover were removed and floral bud tissue from 15 random stems in each plot (control and inoculated) was harvested for RNA extraction and immediately flash frozen in liquid and stored at − 80 °C until further use. Floral bud tissues were collected as day 0 (before inoculation), 3, 6 and 10 days after inoculation.
Total RNA isolation and cDNA synthesis
Total RNA was isolated from V. angustifolium f. nigrum and V. myrtilloides floral buds inoculated with M.vc as well as from control buds using RNeasy plant mini kit (Qiagen, US) following manual instructions. Residual genomic DNA was digested by RNase-free DNase (Qiagen, US) according to the manufacturer’s instructions. The concentration and purity of RNA samples was assessed using Nanodrop ND 1,000 spectrophotometer. RNA samples with an OD260/280 value (1.9–2.1) and OD260/230 (≥ 2.0) was used to determine the quality and purity of the extracted RNA34. RNA integrity was assessed by using 1.2% (w/v) agarose gel electrophoresis. cDNA was synthesized from 1 µg of total RNA in a final reaction volume of 20 µl using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manual instructions and stored at − 20 °C until use.
Candidate reference genes: selection, primer design and amplification efficiency
Seven candidate genes were selected based on previous studies on V. corymbosum (highbush blueberry)16 and V. myrtillus (European blueberry)37 to identify the most suitable reference genes for gene expression analysis in wild blueberry. V. corymbosum ESTs (https://www.vaccinium.org) were mined to design primers for CaCSa, UBC9, TIP41 and PPR using Primer Premier 5.0 (Premier Biosoft International, Palo Alto, California, USA) (Table 1). The primer sequences were blasted on NCBI database (https://blast.ncbi.nlm.nih.gov) to determine their homology with respective genes. A cDNA pool representing all the samples per phenotype was used to determine the amplification efficiency of each target/reference gene31. A tenfold cDNA dilution series (10, 102, 103, 104, and 105) was used to generate a standard curve for estimation of amplification efficiency (E = (10[−1/slope] − 1) × 100%) and correlation coefficient (R2)31.
Quantitative real-time PCR (qPCR)
qPCR assay was performed using a CFX Connect Real-time Detection System (Biorad, US) to analyze the specific expression of reference/target gene. Each PCR reaction mixture (10 µl) contained 2 µl of diluted cDNA (20-fold dilution), 5 µl SsoAdvanced SYBR Green Supermix (Biorad), and 1 µl (10 nM) of each forward and reverse primer. The amplification conditions were as follows: an initial denaturation at 95 °C for 180 s, followed by 40 cycles at 95 °C for 10 s, 60 °C for 20 s. Each run was completed with a melting curve analysis (65–95 °C with at increments of 0.5 °C) to verify the specificity of the amplification. The cycling conditions were based on the method described by Petriccione et al.33. A no-template control (NTC) was included for each gene assay to confirm the absence of non-specific products47.
Determination of reference gene expression stability
The Cq value (quantification value) of each reference gene under the four different time points for both V. myrtilloides and V.a.f nigrum was recorded using the qPCR system. Four widely used software: geNorm8, NormFinder18, BestKeeper19, and ΔCq20 method was used to rank the expression stability of the reference genes. Finally, we used RefFinder21, a web-based user-friendly comprehensive tool, which integrates all four algorithms providing an overall ranking of the used genes.
The GeNorm algorithm, which is a module of qbase + software package (Biogazelle), was used to evaluate the candidate reference genes based on their expression stability values (M-values) and pairwise variations (Vn/Vn + 1)48. The default set value was 1.5; gene with the lowest M-value was the most stably expressed one. The computed pairwise variation (Vn/n + 1), was used to determine the optimal number of reference genes required for normalisation of the data. A (Vn/n + 1) value < 0.15 indicated the appropriate number of reference genes required for analysis8.
For Normfinder, the raw Cq values were converted into relative quantities (RQ) using the formula RQ = 2(Cq min − Cq sample), where Cq min is the lowest Cq value across the sample pool. Normfinder evaluated the expression stability of candidate reference genes at inter-group and intra-group levels. Ideally, the two genes with the lowest stability values were the most appropriate genes to be used for normalisation18.
BestKeeper was performed using the original Microsoft Excel-based formulas19. It calculates the standard deviation of the Cq value between the whole data set, and the gene with the lowest standard deviation (SD) is proposed as most suitable. The comparative ΔCq method manually compares relative expression of pairs of genes within each sample.
Validation of reference genes
To confirm the reliability of selected reference genes, the relative expression profiles of PR3 gene was determined and normalized with the two most stable and two least stable genes. The relative expression levels were calculated by 2−△△Ct method49. For each qPCR experiment, three technical replicates were performed for each biological replicate. A one-way analysis of variance (ANOVA) was performed using the PROC GLIMMIX procedures of SAS (version 9.3, SAS institute, Inc., Cary, NC) for each time-point. Fisher’s LSD was used for multiple means comparison at the level of α = 0.05.
Data availability
The data that support the findings of this study are available in the article and Supplementary Files.
References
Hildebrand, P. D. & Braun, P. G. Factors affecting infection of lowbush blueberry by ascospores of Monilinia vaccinii-corymbosi. Can. J. Plant Pathol. 13, 232–240 (1991).
Percival, D., Jose, S., Guo, L., Schilder, A. & Olson, R. A. Monilinia vaccinii-corymbosi sensitivity to demethylation inhibitor fungicides and its effect on Monilinia blight control in wild blueberry fields. In North American Blueberry Research and Extension Workers Conference. Vol. 18 (2018). https://digitalcommons.library.umaine.edu/nabrew2018/proceedingpapers/proceedingpapers/18.
Janes, D. E. & Percival, D. C. Trends in lowbush blueberry cultivar development. J. Am. Pomol. Soc. 57(2), 63–69 (2003).
Galletta, G. J. Blueberries and cranberries. In Advances in Fruit Breeding (eds. J. Janick, J. N. Moore) 154–196 (Purdue University Press, West Lafayette, 1975).
Ehlenfeldt, M. & Stretch, A. Resistance to blighting by Monilinia vaccinii-corymbosi in diploid and polyploid vaccinium species. HortScience 36. https://doi.org/10.21273/HORTSCI.36.5.955 (2001).
Morrison, S., Smagula, J. M. & Litten, W. Morphology, growth, and rhizome development of Vaccinium angustifolium Ait. seedlings, rooted softwood cuttings, and micropropagated plantlets. HortScience 35, 738–741 (2000).
Wise, R. P., Moscou, M. J., Bogdanove, A. J. & Whitham, S. A. Transcript profiling in host–pathogen interactions. Annu. Rev. Phytopathol. 45, 329–369 (2007).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, 34–41 (2002).
Gachon, C., Mingam, A. & Charrier, B. Real-time PCR: What relevance to plant studies?. J. Exp. Bot. 55, 1445–1454. https://doi.org/10.1093/jxb/erh181 (2004).
Kozera, B. & Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 54, 391. https://doi.org/10.1007/s13353-013-0173-x (2013).
Bustin, S. A. Quantification of mRNA using real-time reverse transcription PCR RT-PCR: Trends and problems. J. Mol. Endocrinol. 29, 23–29 (2002).
Radonić, A. et al. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 313, 856–862 (2004).
Huggett, J., Dheda, K., Bustin, S. & Zumla, A. Real-time RT-PCR normalisation. Strategies and considerations. Genes Immun. 6, 279–284 (2005).
Dheda, K. et al. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 37, 112–119 (2004).
Yang, Q. et al. Reference gene selection for qRT-PCR in Caragana korshinskii Kom. under different stress conditions. Mol. Biol. Rep. 41, 2325–2334 (2014).
Vashisth, T., Johnson, L. & Malladi, A. An efficient RNA isolation procedure and identification of reference genes for normalization of gene expression in blueberry. Plant Cell Rep. 30, 2167–2176 (2011).
Die, J. V. & Rowland, L. J. Superior cross-species reference genes: A blueberry case study. PLoS ONE 8(9), e73354. https://doi.org/10.1371/journal.pone.0073354 (2013).
Andersen, C. L., Jensen, J. L. & Ørntoft, T. F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250 (2004).
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515 (2004).
Silver, N., Best, S., Jiang, J. & Thein, S. L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. Bmc Mol Biol 7 (2006).
Xie, F., Xiao, P., Chen, D., Xu, L. & Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 80(1), 75–84 (2012).
Wan, H. et al. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 399, 257–261. https://doi.org/10.1016/j.ab.2009.12.008 (2010).
Kumar, K., Muthamilarasan, M. & Prasad, M. Reference genes for quantitative real-time PCR analysis in the model plant foxtail millet (Setaria italica L.) subjected to abiotic stress conditions. Plant Cell Tissue Organ. Cult. 115, 13–22. https://doi.org/10.1007/s11240-013-0335-x (2013).
Gutierrez, L., Mauriat, M., Pelloux, J., Bellini, C. & Van Wuytswinkel, O. Towards a systematic validation of references in real-time RT-PCR. Plant Cell 20, 1734–1735 (2008).
Ma, S. et al. Expression stabilities of candidate reference genes for RT-qPCR under different stress conditions in soybean. PLoS ONE 8, e75271. https://doi.org/10.1371/journal.pone.0075271 (2013).
Zhu, J. et al. Reference gene selection for quantitative real-time PCR normalization in Caragana intermedia under different abiotic stress conditions. PLoS ONE 8(1), e53196 (2013).
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K. & Scheible, W. R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17 (2005).
Zifkin, M. et al. Gene expression and metabolite profiling of developing highbush blueberry fruit indicates transcriptional regulation of flavonoid metabolism and activation of abscisic acid metabolism. Plant Physiol. 158(1), 200–224. https://doi.org/10.1104/pp.111.180950 (2012).
Samarth, A. & Jameson, P. E. Selection of reference genes for flowering pathway analysis in the masting plants, Celmisia lyallii and Chionochloa pallens, under variable environmental conditions. Sci. Rep. 9, 9767. https://doi.org/10.1038/s41598-019-45780-1 (2019).
Tashiro, R. M., Philips, J. G. & Winefield, C. S. Identification of suitable grapevine reference genes for qRT-PCR derived from heterologous species. Mol. Genet. Genomics 291, 483–492 (2016).
Monteiro, F., Sebastiana, M., Pais, M. S. & Figueiredo, A. Reference gene selection and validation for the early responses to downy mildew infection in susceptible and resistant Vitis vinifera cultivars. PLoS ONE 8, e72998 (2013).
Yang, H. et al. Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L.. Gene 538, 113–122 (2014).
Petriccione, M., Mastrobuoni, F., Zampella, L. & Scortichini, M. Reference gene selection for normalization of RT-qPCR gene expression data from Actinidia deliciosa leaves infected with Pseudomonas syringae pv. actinidiae. Sci. Rep. 5, 16961 (2015).
Shivhare, R. & Lata, C. Selection of suitable reference genes for assessing gene expression in pearl millet under different abiotic stresses and their combinations. Sci. Rep. 6, 23036. https://doi.org/10.1038/srep23036 (2016).
Andrade, L. M. et al. Reference genes for normalization of qPCR assays in sugarcane plants under water deficit. Plant Methods 13, 28. https://doi.org/10.1186/s13007-017-0178-2 (2017).
Ye, J. et al. Selection of suitable reference genes for qRT-PCR normalisation under different experimental conditions in Eucommia ulmoides Oliv. Sci Rep. 8, 15043. https://doi.org/10.1038/s41598-018-33342-w (2018).
Koskimäki, J. J. et al. Flavonoid biosynthesis and degradation play a role in early defence responses of bilberry (Vaccinium myrtillus) against biotic stress. Eur. J. Plant Pathol. 125, 629–640 (2009).
Jarosova, J. & Kundu, J. Validation of reference genes as internal control for studying viral infections in cereals by quantitative real-time RT-PCR. BMC Plant Biol. 10, 146. https://doi.org/10.1186/1471-2229-10-146 (2010).
Borges, A., Tsai, S. & Caldas, D. Validation of reference genes for RT-qPCR normalization in common bean during biotic and abiotic stresses. Plant Cell Rep. 31(5), 827–838 (2012).
Rawat, S., Ali, S., Mittra, B. & Grover, A. Expression analysis of chitinase upon challenge inoculation to Alternaria wounding and defense inducers in Brassica juncea. Biotechnol. Rep. 13, 72–79. https://doi.org/10.1016/j.btre.2017.01.001 (2017).
Thomma, B. P. H. J. et al. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95, 15107–15111. https://doi.org/10.1073/pnas.95.25.15107 (1998).
Mishina, T. E. & Zeier, J. Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 50, 500–513 (2007).
Kusajima, M. et al. Suppressive effect of abscissic acid on systemic acquired resistance in tobacco plants. J. Gen. Plant Pathol. 76, 161–167 (2010).
González-Grandío, E., Poza-Carrión, C., Sorzano, C. O. & Cubas, P. BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell 25, 834–850 (2013).
Cardot, C. et al. Comparison of the molecular responses of tolerant, susceptible and highly susceptible grapevine cultivars during interaction with the pathogenic fungus Eutypa lata. Front. Plant Sci. 10, 991. https://doi.org/10.3389/fpls.2019.00991 (2019).
Delbridge, R. & Hildebrand, P. Monilinia Blight of Lowbush Blueberry. https://cdn.dal.ca/content/dam/dalhousie/images/sites/wildblueberry/pdfs/Monilinia_Blight_Lowbush_Blueberry.pdf (1997).
Li, M. Y., Song, X., Wang, F. & Xiong, A. S. Suitable reference genes for accurate gene expression analysis in parsley (Petroselinum crispum) for abiotic stresses and hormone stimuli. Front. Plant Sci. 7, 1481. https://doi.org/10.3389/fpls.2016.01481 (2016).
Hellemans, J., Mortier, G., De Paepe, A., Speleman, F. & Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8(2), R19 (2007).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408 (2001).
Acknowledgements
The authors would like to acknowledge the funding support received from the Collaborative Research and Development Program (CRDPJ 507170-16) of the Natural Sciences and Engineering Research Council of Canada; Bragg Lumber Company and the Wild Blueberry Producers Association of Nova Scotia.
Author information
Authors and Affiliations
Contributions
S.J. conceived, designed and executed the experiments and the analysis. J.A. assisted with sample infection and collection from the field. DP conceived the overall research project, provided the M.vc. cultures and is the PI for the initiative. S.J. wrote the manuscript. D.P. and L.J. supervised the study and revised the manuscript. All authors revised and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jose, S., Abbey, J., Jaakola, L. et al. Selection and validation of reliable reference genes for gene expression studies from Monilinia vaccinii-corymbosi infected wild blueberry phenotypes. Sci Rep 10, 11688 (2020). https://doi.org/10.1038/s41598-020-68597-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-68597-9
This article is cited by
-
Modulation of defense genes and phenolic compounds in wild blueberry in response to Botrytis cinerea under field conditions
BMC Plant Biology (2023)
-
Evaluation and validation of reference genes for RT-qPCR gene expression in Naegleria gruberi
Scientific Reports (2023)
-
Evaluation of reference genes for qRT-PCR studies in the colchicine producing Gloriosa superba L.
Plant Biotechnology Reports (2023)
-
Elucidation of the molecular responses during the primary infection of wild blueberry phenotypes with Monilinia vaccinii-corymbosi under field conditions
BMC Plant Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.