Abstract
Yersinia enterocolitica is an enteric bacterium which can cause severe gastroenteritis. Beta-lactams are the most widely used antibiotics against Y. enterocolitica. Y. enterocolitica produces two chromosomal β-lactamases, BlaA and BlaB. BlaB is an Ambler Class C inducible broad spectrum cephlaosporinase which showed differential enzyme activity in different biotypes of Y. enterocolitica. The expression of blaB is mainly regulated by ampR- the transcriptional regulator and, ampD - which helps in peptidoglycan recycling. The aim of this study was to identify and characterize genetic determinants underlying differential enzyme activity of BlaB in Y. enterocolitica biotypes 1 A, IB, 2 and 4. Thus, ampR, blaB and ampD were PCR-amplified and modeled in silico. The intercistronic region containing promoters of ampR and blaB was also investigated. Our results indicated that blaB was more inducible in biotypes 2 and 4, than in biotypes 1 A and 1B. Superimposition of in silico modeled proteins suggested that variations in amino acid sequences of AmpR, BlaB and AmpD were not responsible for hyper-production of BlaB in biotypes 2 and 4. Analysis of promoter regions of ampR and blaB revealed variations at −30, −37 and −58 positions from blaB transcription start site. Studies on relative expression levels of blaB in different biotypes by qRT-PCR indicated that nucleotide variations at these positions might contribute to a higher enzyme activity of BlaB in biotypes 2 and 4. However, this is a preliminary study and further studies including more strains of each biotype are required to strengthen our findings. Nevertheless, to the best of our knowledge, this is the first study which has investigated the genetic determinants underlying differential inducible production of BlaB in different biotypes of Y. enterocolitica.
Similar content being viewed by others
Introduction
Infections due to Yersinia enterocolitica have been reported from almost all the countries around the world. In developed countries, Y. enterocolitica ranks third among the etiological agents of bacterial gastroenteritis (after Campylobacter and Salmonella)1. This species comprise a heterogeneous population of bacteria divided into more than sixty serotypes and six biotypes, which show varied ecological niches, pathogenic properties and geographical distribution2. Strains of Y. enterocolitica can be classified as controversial pathogenic (biotype 1 A), highly pathogenic (biotype 1B) and less pathogenic (biotypes 2–5)2.
Beta-lactams are among the most widely used classes of antibiotics which are effective against several bacteria, including Y. enterocolitica. Bacterial resistance against this class of antibiotics has evolved primarily due to elaboration of β-lactamases, the β-lactam hydrolyzing enzymes. More than 1300 unique β-lactamases have been reported in clinical isolates3. Most strains of Y. enterocolitica produce two chromosomal β-lactamases, BlaA and BlaB4. BlaA is a broad-spectrum constitutively expressed Ambler class A penicillinase and BlaB is an Ambler Class C “AmpC-type” broad spectrum cephlaosporinase5. Chromosomal AmpC β-lactamases are usually inducible, while, except for DHA enzymes, plasmid-mediated AmpC enzymes are not6,7.
As reported for the AmpC enzymes present in other members of the family Enterobacteriaceae expression of “AmpC-type” enzymes in Y. enterocolitica is mainly regulated by ampR which is a transcriptional regulator of ampC and, ampD which participates in recycling of peptidoglycan8. The ampR located at the 5′side of ampC encodes a transcriptional regulator of LysR family. It is transcribed in an opposite orientation and is necessary for induction of ampC expression9. The ampD encodes a cytoplasmic N-acetyl-anhydromuarmoyl-L-alanine amidase that participates in recycling of peptidoglycan. AmpC β-lactamases are normally produced in bacteria at low levels (repressed state). But, during the course of medical therapy, exposure of bacteria to antibiotics like imipenem and cefoxitin results in generation and accumulation of large quantities of cell wall degradation products (muropeptides) which cannot be recycled by AmpD. The un-recycled muropeptides bind to ampR and inhibits its normal activity. This results in ampC de-repression i.e activation of ampC expression10,11. Thus, bacterial strains which were initially susceptible to the third generation cephalosporins, during the course of treatment develop resistance for them12. In Y. enterocolitica the ampR-ampC system was studied in a strain IP97, serotype O: 5b and was reportedly similar to the ampR-ampC system of Citrobacter freundii8.
Earlier studies have shown that Y. enterocolitica might produce two β-lactamases (BlaA and BlaB), only one or none of them13,14,15. Also, β-lactamases were reportedly differentially inducible in different biotypes of Y. enterocolitica13,14,16. Thus, the aim of the present study was to identify and characterize genetic determinants underlying differential inducible expression of blaB in Y. enterocolitica strains of biotypes 1 A, IB, 2 and 4. Besides modifications/mutations in the β-lactamase genes, mutations in ampR and ampD might also result in variable production of chromosomal β-lactamases17,18,19. Hence, gene sequences of ampR, blaB and ampD were investigated and, their 3D structures were in silico modeled to understand their role in differential inducible expression of blaB in Y. enterocolitica biotypes 1 A, IB, 2 and 4. Since, mutations in the promoters can also play an important role in the expression levels of β-lactamases; hence, the intercistronic region containing promoters of ampR and blaB were also investigated. To the best of our knowledge, this is the first study which has investigated the genetic determinants underlying differential inducible expression of blaB in Y. enterocolitica strains of different biotypes.
Results
Specific activity of BlaB before and after induction with imipenem and phenotypic detection of AmpC production
All the strains showed an increase in the β-lactamase specific activity after induction with imipenem. Strains of biotypes 1 A and 1B showed a ca. 2 fold increase, while of biotypes 2 and 4 showed ca. 5 fold increase in BlaB production after induction (Table 1). All the strains tested negative for AmpC production using AmpC E-test strips.
PCR amplification, sequencing and multiple sequence alignment (MSA) of the inter cistronic region of ampR and blaB
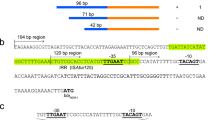
The primer pair B11f and B12r resulted in the desired amplicon of 1076 bp in strains of all the biotypes. BLAST analysis of the sequenced PCR amplicons confirmed that these encoded the intercistronic region of ampR and blaB, along with partial gene sequences of ampR and blaB. As expected, blaB and its regulator ampR were linked in opposite orientations. MSA of the intercistronic region containing the promoter sequences of blaB and ampR of strains of different biotypes revealed that the −10 and −35 regions of ampR and blaB promoters were similar in all biotypes. However, variations viz. G → A at −30, A → G at −37 and C → T at −58 positions from the ampC transcription start site were observed in biotypes 2 and 4 (Fig. 1).
Multiple sequence alignment of the intercistronic region of ampR and blaB of Y. enterocolitica biotypes 1 A, 1B, 2 and 4. The −10 and −35 regions of ampR and blaB are enclosed in boxes. CAT and ATG (shown in bold faces) denote the transcription start site of ampR and blaB, respectively. Nucleotide variations observed at −30, −37 and −58 positions from blaB transcription start site are shown in red colour and in bold faces. Arrows indicate the orientation of transcription of ampR and blaB.
PCR amplification, sequencing and multiple sequence alignment (MSA) of complete coding sequence (CCDS) of ampR, blaB and ampD
Primer pairs RF and RB resulted in the desired amplicons of 730 bp, B15 and B16 in 1002 bp, DF and DR in 521 bp in strains of all the biotypes. Sequencing of the PCR amplicons followed by BLAST analysis confirmed that these encoded for AmpR, BlaB and AmpD, respectively.
Analysis of MSA of amino acid sequences of AmpR revealed that AmpR sequences were similar in all the biotypes, except for a few variations. The three critical amino acid positions of AmpR, viz. G-102, D-135, and Y-264, were conserved in all the strains. In Y. enterocolitica biotype 1B, D was replaced by H at amino acid position 82, T by A at position 103 and S by P at position 207. In Y. enterocolitica biotype 1 A, D was replaced by N at amino acid position 178 and, R was replaced by K at amino acid position 185. In both Y. enterocoallitica strains of biotypes 1 A and 1B, I was replaced by M at amino acid position 92. Amino acid sequences of AmpR of strains of biotypes 2 and 4 were identical (Table 2, Supplementary Figure 1).
Analysis of MSA of amino acid sequences of BlaB revealed that the sequences were similar, except for a few variations. Analysis of MSA of amino acid sequences of BlaB revealed that the sequences were similar, except for a few variations. In Y. enterocolitica 1B, S was replaced by A at amino acid position 14, T by S at position 22, S by at T at position 26, N by K at position 39, V by I at position 57, A by T at position 75 and G by A at position 271. In Y. enterocolitica biotypes 1 A and 1B, S was replaced by T at amino acid position 25, S by N at 301, R by G at 308. In biotype 1 A, A was replaced S at amino acid position 13, T by S at position 21, M by I at position 199 and E by A at position 277. In biotype 4 an aminoacid was missing at amino acid position 166 and T was replaced by P at position 251(Table 2, Supplementary Figure 2).
Analysis of MSA of amino acid sequences of AmpD revealed that amino acid sequences were similar in all the biotypes, except for a few variations. In Y. enterocolitica biotype 1B, Q was replaced by R at amino acid position 55, A by G at position 72 and T by A at position 106. In biotype 1 A, V was replaced by A at amino acid position 140. In both biotypes 1 A and 1B, T was replaced by A and E by G at amino acid positions 34 and 73, respectively. In Y. enterocolitica strains of biotypes 1 A and 2, S was replaced by N at amino acid position 145 (Table 2, Supplementary Figure 3).
modeling of AmpR, AmpC and AmpD, evaluation and superimposition of the protein models
The top three templates (PDB ID: 5mmh_A, 5y2y, 5z50_A) which exhibited a sequence identity of more than 90% with AmpR of Y. enterocolitica were used as templates for modeling AmpR with I-TASSER. The C-scores of the final selected models for AmpR of Y. enterocolitica strains 1 A, 1B and 2/4 were, −0.78, −0.76 and −0.90, respectively. The selected models were further validated for accuracy of the prediction. The PROCHECK results indicated that more than 75% of the residues of the modeled AmpR proteins were in the allowed regions of the Ramachandran map. The average ERRAT scores and Verify-3D scores further confirmed that the predicted 3-D models were reliable and within the acceptable range. The results of the AmpR model validation are presented in Supplementary Table 1. The superimposed structure representing 3D model of the AmpR present in Y. enterocolitica biotypes 1 A, 1B, 2/4 is shown in Supplementary Figure 4 and the root mean square deviation (RMSD) values in Supplementary Table 2.
The top five templates (PDB ID: 2zc7_A, 1ga0_A, 5ggw_B) which exhibited a sequence identity of more than 90% with BlaB of Y. enterocolitica were used as templates for modeling BlaB with I-TASSER. The C-scores of the final selected models for BlaB of Y. enterocolitica biotypes 1 A, 1B, 2 and 4 were −0.73,−1.06, −0.16 and −1.86, respectively. The results of the BlaB model validation are presented in Supplementary Table 3. The superimposed structure representing 3D model of the BlaB present in Y. enterocolitica strains of biotypes 1 A, 1B, 2 and 4 is shown in Supplementary Figure 5 and the RMSD values in Supplementary Table 4.
The top three templates (PDB ID: 6fhg_A, 1j3g_A, 6j3w_A) which exhibited a sequence identity of more than 90% with AmpD of Y. enterocolitica were used as templates for modeling AmpD. The C scores of the final selected models for AmpD of Y. enterocolitica biotypes 1 A, 1B, 2 and 4 were −1.13, −1.59, −1.39 and −1.162, respectively. The results of the AmpD model validation are presented in Supplementary Table 5. The superimposed structure representing the 3D model of AmpD in Y. enterocolitica biotypes 1 A, 1B, 2 and 4 is shown in Supplementary Figure 6 and the RMSD values in Supplementary Table 6.
Relative expression of ampR and blaB as determined by qRT–PCR
The relative change in expression levels of mRNA of ampR after induction was observed to be non significant in all the strains. However, all the strains showed an increase in the expression levels of mRNA of blaB after induction. The fold change in relative expression of blaB was more in strains of biotypes 2 and 4 (~3–4 times) than in biotypes 1 A and 1B (~1.3 times) (Fig. 2).
Discussion
The aim of the present study was to understand reasons underlying differential inducible production of BlaB, an “AmpC-type” β-lactamase in Y. enterocolitica strains of biotypes 1 A, 1B, 2 and 4. Our results indicated that BlaB was inducible in strains of biotypes 1 A, 1B, 2 and 4. Interestingly, the level of production of BlaB after induction was more in biotypes 2 and 4 (~5 times) than in biotypes 1 A and 1B (~2 times). Several studies have reported that the level of production of BlaB after induction varied among different biotypes of Y. enterocolitica13,14. Another interesting observation was that the AmpC E-test strips failed to detect BlaB production, while the spectrophotometric enzyme assays and PCR-amplification confirmed that “AmpC-type” inducible cephalosporinases were present in Y. enterocolitica biotypes 1 A, 1B, 2 and 4. Thus, our results suggest that E-test strips of cefotetan/cefotetan+cloxacillin should not be used for phenotypic detection of AmpC production in Y. enterocolitica.
It was reported that increase in the level of β-lactamase activity in Y. enterocolitica when cultivated in the growth medium containing imipenem indicated that production of these enzymes might be subject to regulatory control20. There can be several reasons underlying variations in the expression/production of chromosomal β-lactamase like, mutations in the gene and/or promoter regions, modifications in the regulatory regions19 etc. Mutations in ampR - the transcriptional regulator of ampC are less frequent, but might also result in hyperinducibility or constitutive hyperproduction of AmpC17. In clinical isolates, mutations in ampD were frequently associated with hyperinducibility or hyperproduction of AmpC18. Thus, genes encoding AmpR, BlaB and AmpD including the intercistronic region of ampR and blaB were PCR-amplified, compared and analyzed in Y. enterocolitica strains of biotypes 1 A, 1B, 2 and 4.
Analysis of AmpR sequences of biotypes 1 A, 1B, 2 and 4 revealed that two domains were present - a substrate binding domain of LysR-type transcriptional regulators (LTTRs) of PBP2_LTTR_substrate super family (accession- cl25412) and, a domain of bacterial regulatory helix-turn-helix protein, LysR family - HTH_1 (pfam00126). Also, the three amino acid positions viz. G-102, D-135, and Y-264 that are important for the biological activity of AmpR were found to be conserved in AmpR of strains of all biotypes8. MSA analysis revealed that the AmpR sequence of biotypes 2 and 4 were identical, while a few variations were present in AmpR sequences of other biotypes. Hence, AmpR of different biotypes were in silico modeled and superimposed. The 3D protein models of AmpR variants showed a strong structural alignment and significantly lower RMSD values. This indicated that variations in AmpR sequences might not be responsible for differential inducible production of BlaB in different biotypes.
Analysis of amino acid sequences of BlaB revealed that the two significant motifs conserved in the Ambler class C β-lactamases - 151SXXK154 and 342KTG344, (X can be any amino acid) were conserved in BlaB of all the biotypes. Chen et al.21 reported that the key catalytic residues of the AmpC enzymes are: S-64, K-67, Y-150, N-152, K-315 and A-318 and, substitutions at these sites decreased the enzymatic activity of AmpC. Though, all the key catalytic residues were found to be present in BlaB of biotypes 1 A, 1B, 2 and 4 but their respective positions were S-89, K-92, Y-175, N-177, K-342 and A-345. The 24 amino acids long signal peptide at the N-terminal was excluded from comparative analysis. Though, no variations were observed in the catalytic residues, a few variations were observed at some other sites. The results of in silico protein modeling and superimposition of 3D models of BlaB variants revealed a strong structural alignment and significantly lower RMSD values. This indicated that variations in amino acid sequences of BlaB might not be responsible for differential inducible production BlaB in different biotypes.
The critical amino acid residue positions in Y. enterocolitica AmpD are A-43,H-123H (the amidase catalytic sites), H-123, D-133 (Zn binding sites), and K-131, D-133, A-43, V-57, W-64 (substrate binding sites)22. Though, no variations were observed at these sites, a few variations were observed at some other positions. The results of the superimposition of the 3D protein models of AmpD variants indicated that variations in AmpD sequences might not be responsible for differential inducibility of blaB in different biotypes.
Mutations in the promoter sequences and/or insertions in the promoter regions have been reportedly associated with hyper production of β-lactamases in the family Enterobacteriaceae23. Hence, the intercistronic region between the start codons of ampR and blaB were investigated for their role in differential regulation of expression of blaB. The intercistronic region between ampR and blaB start codons, known as the control region is 135 bp long and, contains promoters for both and, the AmpR binding site8. A previous study reported that the −35 and −10 regions of the promoters of ampR and ampC of Y. enterocolitica were similar to the ampR and ampC promoters of Citrobacter freundii8,24. Our results indicated that the sequence of the ampR promoters including the −35 and −10 regions were identical in all biotypes. Hence, these might not be responsible for higher inducible expression of blaB in biotypes 2 and 4. Results of the MSA of blaB promoters revealed that the −35 and −10 regions of the blaB promoters were identical in all biotypes. However, variations viz. G → A at −30, A → G at −37, C → T at −58 positions from the ampC transcription start site were observed in biotypes 2 and 4. Several researchers have reported that in Enterobacteriaceae, mutations and insertions at sites other than the −35 (Pribnow box) and −10 region (TATA box) of the ampC promoters resulted in a hyper expression of ampC in clinical isolates23,25,26,27,28.
To validate the role of these variations, if any, in differential inducible expression of blaB in strains of biotypes 2 and 4, expression levels of ampR and blaB were measured before and after induction using qRT-PCR. The relative expression levels of mRNA of ampR after induction were found to be non-significant in all biotypes. This might be attributed to the fact that the ampR promoter regions were conserved in all biotypes. However, the difference in the relative expression levels of mRNA of blaB after induction was significant. The fold change in relative expression of blaB in strains of biotypes 2 and 4 was ca. 3–4 times, while it was ca. 1.3 times in biotypes 1 A and 1B, which broadly reiterated the results obtained by measurement of β-lactamase specific activity. This suggested that variations in the promoter regions might be responsible for higher inducible production of BlaB which was observed in strains of biotypes 2 and 4. It is pertinent to mention here that though, the fold changes (2–∆∆CT) in mRNA levels of blaB were similar, these were not identical with the fold changes observed in the β-lactamase specific activity after induction. Such small variations in the protein production and mRNA expression studies might be attributed to the differences in the post-translational modifications and post-transcriptional processing of proteins and mRNAs, respectively. Also, the differences in the degradation rates of proteins and mRNA during bacterial growth might also contribute to similar, but non-identical levels of bacterial mRNA and proteins.
To summarize, our results indicated that blaB was more inducible in biotypes 2 and 4, than in biotypes 1 A and 1B. Though, a few variations were present in amino acid sequences of AmpR, BlaB and AmpD, superimposition of the 3D protein models of AmpR, BlaB and AmpD suggested that these variations were not responsible for hyper production of BlaB in biotypes 2 and 4. Analysis of the promoter regions of ampR and blaB revealed variations at −30, −37 and −58 regions of the blaB promoter. Studies on relative expression levels of blaB in different biotypes by qRT-PCR suggested that nucleotide variations at these positions might be important for higher levels of transcription and, consequently a higher enzyme activity in biotypes 2 and 4, after induction. The results of this study are expected to help in devising novel intervention strategies against yersiniosis. However, this is a preliminary study and further experiments on promoter strength incorporating more strains of each biotype are required to strengthen these findings.
Materials and Methods
Bacterial strains
In the present study, four clinical strains of Y. enterocolitica representing biotypes 1 A, 1B, 2 and 4 were used. Y. enterocolitica strain representing biotype 1 A was isolated from India, while strains of biotypes 1B, 2 and 4 were isolated from different parts of the world and were received as kind gifts from foreign laboratories. The details of these strains viz. serotypes, laboratory and reference laboratory accession numbers and country of origin are given in Table 1. All the strains were maintained on tryptone soy agar at 4 °C.
Induction of blaB expression, preparation of cell lysates and spectrophotometeric assay of β -lactamases
Y. enterocolitica strains were induced for production of BlaB by cultivating bacteria in tryptic soy broth (TSB) containing imipenem (concentration − 0.5 mg/l). The methods for induction and preparation of cell lysates have been described earlier15. The enzyme activity of BlaB was assessed spectrophotometrically by hydrolysis of nitrocefin. The contents of the assay mixture and the methods have been described earlier15,22. The enzyme specific activity was expressed as µmol of nitrocefin hydrolyzed/min/mg of protein. The experiments were repeated for each strain in triplicates and the average results were reported ± standard error mean (SEM).
Phenotypic detection of AmpC production using E-test strips
Phenotypic detection of AmpC production after induction with imipenem was done using E-test strips of cefotetan/cefotetan+cloxacillin (bioMerieux Inc., MO, USA) following the methods described earlier29. Following AmpC E-test, if cefotetan/cefotetan + cloxacillin (CN/CNI) ration was ≥8 a strain was considered as AmpC producer.
Isolation of genomic DNA
The genomic DNA was isolated from overnight grown bacterial culture in TSB at 28 °C. The total genomic DNA was isolated using DNeasy Tissue kit (Qiagen, Hilden, Germany), eluted in sterile water and quantitated spectrophotometrically at 260 nm.
PCR amplification of the intercistronic region containing promoters of blaB and ampR and, complete coding sequences (CCDS) of ampR, blaB and ampD
The intercistronic region containing promoters of ampR and blaB along with the partial gene regions of ampR and blaB was amplified using primers B11f and B12r. Primer pairs RF and RB were used for amplification of CCDS of ampR, B15 and B16 for amplification of CCDS of blaB, and DF and DR for amplification of CCDS of ampD. The components of the PCR reaction mixture and PCR conditions, except the annealing temperatures have been described earlier30. The details of the primer sequences and the annealing temperatures are presented in Table 3. The PCR amplicons were electrophoresed and visualized under UV transilluminator.
Sequencing of the intercistronic regions of ampR and blaB and, CCDS of ampR, blaB and ampD
PCR amplicons containing intercistronic regions of ampR and blaB along with their promoter regions and, CCDS of ampR, blaB and ampD were purified and sequenced at a commercial facility (Invitrogen BioServices India Pvt. Ltd., Bangalore, India). The sequences were identified by homology search using NCBI-BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The GenBank accession number of ampR of Y. enterocolitica strains 20, 8081, IP134, W22703 were MK511112, MN242777- MN242779 respectively, of blaB of Y. enterocolitica strains 20, 8081, IP134, W22703 were MN172161- MN172164 respectively and, of ampD of strains 20, 8081, IP134, W22703 were MK511120, MN172158-MN172160 respectively. The CCDS of ampR, blaB and ampD were translated using ExPASy (https://www.expasy.org/) and aligned by Clustal Omega (http://www.ebi.uc.ak/clustal).
3-D structure predictions of AmpR, BlaB and AmpD: modeling and validation
Since the protein structures of AmpR, BlaB and AmpD of Y. enterocoloitica are not known; the 3D structures of AmpR, BlaB and AmpD of strains of different biotypes were predicted using the web interface iterative threading assembly refinement (I-TASSER) (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). Since the amino acid sequences of AmpR of strains of biotype 2 and 4 were identical, hence only one sequence representing AmpR of both the biotypes were modeled. Five models were predicted by I-TASSER for AmpR, BlaB and AmpD each, of which the best model was selected on the basis of the confidence score (C-score). The selected models were further validated for accuracy using the programs PROCHECK31 ERRAT32 and Verify 3D33. The protein models of AmpR, BlaB and AmpD of each biotype were superimposed and visualized using PyMol (https://pymol.org/2/).
Determination of expression levels of ampR and blaB by real time PCR (qRT-PCR)
The effect of promoter variations on relative expression levels of ampR and blaB in Y. enterocolitica strains before and after induction with imipenem was studied by qRT-PCR. Total RNA was extracted from the bacterial cultures before and after induction with imipenem using SV Total RNA isolation system (Promega, Madison, WI, USA). The concentration of RNA was quantified spectrophotometrically. The cDNA was prepared from each sample (template −1 μg RNA) using a commercial kit (cDNA synthesis kit, TaKaRa, Shiga, Japan). Primers were designed for amplification of ampR and blaB using the software Primer3 (http://simgene.com/Primer3). One of the housekeeping genes, gapA was included as the reference gene34. The details of the primers are given in the Supplementary Table 7. The qRT-PCR was performed at a commercial facility (Genotypic Technology Pvt. Ltd. Bengaluru, India) using SYBR Green chemistry (Brilliant II SYBR Green qPCR master mix, Agilent Technologies, USA) in Stratagene mx3005P instrument (Agilent Technologies, USA). The cycling conditions for amplification were as follows: initial denaturation for 95 °C for 10 min followed by 40 cycles at 95 °C for 30 sec, 58 °C for 30 sec. The mean Ct value of technical replicates was used to calculate the relative expression level of genes. The experiments were performed in triplicate and the average results were reported ± SD. The relative quantification of genes was performed using the standard 2−ΔΔCt method, also known as the delta-delta CT method, as described by Pfaffl35,36.
References
European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union one health 2018 zoonoses report. EFSA J. 17, 5926 https://doi.org/10.2903/j.efsa.2019.5926. (2019).
Bottone, E. J. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10, 257–276 (1997).
Bush, K. Proliferation and significance of clinically relevant β-lactamases. Ann. N. Y. Acad. Sci. 1277, 84–90 (2013).
Cornelis, G. In Antibiotic resistance in Y. enterocolitica (ed. Bottone, E. J.) 55-71, (CRC Press 1981), Boca Raton, Fla.
Pham, J. N., Bell, S. M., Martin, L. & Carniel, E. The β-lactamases and β-lactam antibiotic susceptibility of Yersinia enterocolitica. J. Antimicrob. Chemother. 46, 951–957 (2000).
Coudron, P. E., Moland, E. S. & Thomson, K. S. Occurrence and detection of AmpC beta-lactamases among Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates at a veterans medical center. J. Clin. Microbiol. 38, 1791–1796 (2000).
Fortineau, N., Poirel, L. & Nordmann, P. Plasmid-mediated and inducible cephalosporinase DHA-2 from Klebsiella pneumonia. J. Antimicrob. Chemother. 47, 207–101 (2001).
Seoane, A., Francia, M. V. & Garcia Lobo, J. M. Nucleotide sequence of the ampC-ampR region from the chromosome of Yersinia enterocolitica. Antimicrob. Agents Chemother. 36, 1049–1052 (1992).
Lindquist, S., Lindberg, F. & Normark, S. Binding of the Citrobacter freundii AmpR regulator to a single DNA site provides both autoregulation and activation of the inducible ampC β-lactamase gene. J. Bacteriol. 171, 3746–3753 (1989).
Normark, S. β-Lactamase induction in Gram-negative bacteria is intimately linked to peptidoglycan recycling. Microb. Drug Resist. 1, 111–114 (1995).
Dietz, H., Pfeifle, D. & Wiedemann, B. The signal molecule for β-lactamase induction in Enterobacter cloacae is the anhydromuramyl-pentapeptide. Antimicrob. Agents Chemother. 41, 2113–2120 (1997).
Livermore, D. M. & Brown, D. F. Detection of beta-lactamase-mediated resistance. J. Antimicrob. Chemother. 48, 59–64 (2001).
Stock, I. & Wiedemann, B. J. An in-vitro study of the antimicrobial susceptibilities of Yersinia enterocolitica and the definition of a database. J. Antimicrob. Chemother. 43, 37–45 (1999).
Stock, I., Heisig, P. & Wiedemann, B. Beta-lactamase expression in Yersinia enterocolitica biovars 1A, 1B, and 3. J. Med. Microbiol. 49, 403–408 (2000).
Sharma, S., Ramnani, P. & Virdi, J. S. Detection and assay of beta-lactamases in clinical and non-clinical strains of Yersinia enterocolitica biovar 1A. J. Antimicrob. Chemother. 54, 401–405 (2004).
Bonke, R. et al. Antimicrobial susceptibility and distribution of β-lactamase A (blaA) and β-lactamase B (blaB) genes in enteropathogenic Yersinia species. Microb. Drug Res. 17, 575–581 (2011).
Hanson, N. D. & Sanders, C. C. Regulation of inducible AmpC β -lactamase expression among Enterobacteriaceae. Curr. Pharm. Des. 5, 881–94 (1999).
Schmidtke, A. J. & Hanson, N. D. Model system to evaluate the effect of ampD mutations on AmpC-mediated β-lactam resistance. Antimicrob. Agents Chemother. 50, 2030–37 (2006).
Pfeifer, Y., Cullik, A. & Witte, W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int. J. Med. Microbiol. 300, 371–379 (2010).
Bent, Z. W. & Young, G. M. Contribution of BlaA and BlaB beta-lactamases to antibiotic susceptibility of Yersinia enterocolitica biovar 1B. Antimicrob. Agents Chemother. 54, 4000–2 (2010).
Chen, Y., Minasov, G., Roth, T. A., Prati, F. & Shoichet, B. K. The deacylation mechanism of AmpC-lactamase at ultrahigh resolution. J. Am. Chem. Soc. 128, 2970–2976 (2006).
Singhal, N., Pandey, D., Kumar, M. & Virdi, J. S. Molecular analysis of ampR and ampD to understand variability in inducible expression of “BlaB-like” cephalosporinase. Yersinia enterocolitica biotype 1A. Gene. 1, 704:25–30, https://doi.org/10.1016/j.gene.2019.04.031 (2019).
Siu, L. K. et al. High-level expression of ampC beta-lactamase due to insertion of nucleotides between -10 and -35 promoter sequences in Escherichia coli clinical isolates: cases not responsive to extended-spectrum-cephalosporin treatment. Antimicrob. Agents Chemother. 47, 2138–44 (2003).
Pai, H. et al. Epidemiology and clinical features of bloodstream infections caused by AmpC-type-beta-lactamase-producing Klebsiella pneumonia. Antimicrob. Agents Chemother. 48, 3720–3728 (1992).
Caroff, N., Espaze, E., Gautreau, D., Richet, H. & Reynaud, A. Analysis of the effects of −42 and −32 ampC promoter mutations in clinical isolates of Escherichia coli hyperproducing ampC. J. Antimicrob. Chemother. 45, 783–788 (2000).
Tracz, D. M. et al. Canadian Nosocomial Infection Surveillance Program. ampC gene expression in promoter mutants of cefoxitin-resistant Escherichia coli clinical isolates. FEMS Microbiol. Lett. 270, 265–71 (2007).
Haldorsen, B. et al. The AmpC phenotype in Norwegian clinical isolates of Escherichia coli is associated with an acquired ISEcp1-like ampC element or hyperproduction of the endogenous AmpC. J. Antimicrob. Chemother. 62, 694–702, https://doi.org/10.1093/jac/dkn257 (2008).
Yu, W., Bing, L. & Zhenhua, L. AmpC promoter and attenuator mutations affect function of three Escherichia coli strains. Curr. Microbiol. 59, 244–247, https://doi.org/10.1007/s00284-009-9426-7 (2009).
Singhal, N., Pandey, D., Singh, S. N., Kumar, M. & Virdi, J. S. Molecular Characteristics of “BlaB-Like” Chromosomal Inducible Cephalosporinase of Yersinia enterocolitica Biotype 1A Strains. Microb. Drug Resist. 25, 824–829, https://doi.org/10.1089/mdr.2018.0282 (2019).
Singhal, N. et al. Molecular analysis of β-lactamase genes to understand their differential expression in strains of Yersinia enterocolitica biotype 1A. Sci. Rep. 4, 5270, https://doi.org/10.1038/srep05270 (2014).
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK - a program to check the stereochemical quality of protein structures. J. App. Cryst. 26, 283–291 (1993).
Colovos, C. & Yeates, T. O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 2, 1511–1519 (1993).
Lüthy, R., Bowie, J. U. & Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature. 356, 83–85 (1992).
Kanaujia, P. K., Bajaj, P., Kumar, S., Singhal, N. & Virdi, J. S. Proteomic analysis of Yersinia enterocolitica biovar 1A under iron-rich and iron-poor conditions indicate existence of efficiently regulated mechanisms of iron homeostasis. J. Proteomics. 21, 39–49, https://doi.org/10.1016/j.jprot.2015.04.015 (2015).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 1(29), e45 (2001). (9).
Singhal, N., Pandey, D., Singh, N. S., Kumar, M. & Virdi, J. S. ampD homologs in biotypes of Yersinia enterocolitica: Implications in regulation of chromosomal AmpC-type cephalosporinases. Infect. Genet. Evol. 69, 211–215, https://doi.org/10.1016/j.meegid.2019.01.033 (2019).
Author information
Authors and Affiliations
Contributions
N.S. and J.S.V. wrote the main manuscript text. M.K. and D.P. performed the bioinformatic analysis. N.S. and N.S.S. performed the experiments. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Singhal, N., Pandey, D., Singh, N.S. et al. Exploring the genetic determinants underlying the differential production of an inducible chromosomal cephalosporinase - BlaB in Yersinia enterocolitica biotypes 1A, 1B, 2 and 4. Sci Rep 10, 10167 (2020). https://doi.org/10.1038/s41598-020-67174-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-67174-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.