Abstract
Fusarium head blight (FHB) caused by Fusarium graminearum (Fg) is a devastating disease of crops, especially wheat and barley, resulting in significant yield loss and reduced grain quality. Fg infection leads to the production of mycotoxins, whose consumption is toxic to humans and livestock. The Arabidopsis DMR6 gene encodes a putative 2-oxoglutarate Fe(II)-dependent oxygenase (2OGO) and has been identified as a susceptibility factor to downy mildew. We generated site-specific mutations in Arabidopsis At2OGO by CRISPR/Cas9 gene editing. The resulting At2OGO knock-out (KO) mutants display enhanced resistance to Fg in a detached inflorescence infection assay. Expression profiling of defense genes revealed that impairment of At2OGO function resulted in the upregulation of defense genes that are regulated by salicylic acid (SA), jasmonic acid (JA) and ethylene (ET) pathways. Complementation of the At2OGO-KO lines with a barley (cv. Conlon) orthologue, Hv2OGO, restored susceptibility to Fg. This result indicates that the Hv2OGO gene is functionally equivalent to its Arabidopsis counterpart and, hence, may have a similar role in conditioning susceptibility to FHB in barley. These results provide a molecular basis for proposing 2OGO as a plant immunity suppressor in Arabidopsis and potentially in barley plants and establish a rationale and strategy for enhancing FHB resistance in barley.
Similar content being viewed by others
Introduction
Fusarium head blight (FHB) or scab is primarily caused by Fusarium graminearum (Fg) (teleomorph Gibberella zeae) and is a devastating disease of cereal crops, particularly wheat, barley and maize. FHB disease has been identified as a major limiting factor in cereal crop production throughout the world1. Fg is an important hemibiotrophic fungal pathogen whose growth is favored by warm conditions and high humidity and whose dispersal is aided by frequent rainfall during flowering season2.The most noticeable disease symptom of FHB is bleaching of floral spikelets, which often leads to cell death or the production of non-viable seeds3. Since the early 1990s, it has been estimated that FHB infection in grain crops exerts an annual loss of US$3 billion4,5. In addition to yield reduction, FHB also lowers grain quality by producing mycotoxins, including deoxynivalenol (DON), which accumulate in infected grains6,7. DON-contaminated grains represent a serious health concern for both humans and livestock. Deployment of FHB-resistant wheat and barley cultivars provides the most direct way to address this problem as it simultaneously reduces yield loss and grain contamination.
Genetic engineering offers a promising approach for manipulating plant disease resistance and susceptibility. A number of different transgenic approaches have been explored in order to generate plants that display enhanced FHB resistance or tolerance. These include the overexpression of pathogenesis-related (PR) or defense response genes8,9,10, overexpression of antifungal11 or antimicrobial peptides12 introduced from other species, the inhibition of DON synthesis13,14,15, modifying the host cellular target of DON16 and detoxifying DON to less toxic compounds17,18. These transgenic plants are considered to be genetically modified organisms (GMO), which may confer an undue regulatory burden on producers and processors as well as creating a barrier to consumer acceptance. The emergence of CRISPR technology allows precise and efficient genome editing by introducing mutations at specific target sites19. CRISPR has been used intensively in mammalian cells and many whole organisms20,21,22,23,24 including both model and crop plants25,26,27,28,29,30,31,32. The potential for generating transgene-free and gene-edited disease resistant plants by CRISPR gene editing prompted us to use CRISPR technology to generate plants with enhanced FHB resistance33,34,35.
Identification of host susceptibility genes provides a foundation for developing disease resistant plants through genome editing. A susceptibility gene called DMR6 (downy mildew resistance 6) was discovered and characterized while screening Arabidopsis ethyl methane sulfonate and T-DNA insertion mutants for the loss-of-susceptibility to downy mildew disease caused by the Hyaloperonospora arabidopsidis36,37. Map-based gene cloning revealed that DMR6 encodes a putative 2-oxoglutarate Fe(II)-dependent oxygenase (2OGO) that belongs to the 2-oxoglutarate-dependent dioxygenase superfamily. The dmr6-1 null and dmr6-2 T-DNA mutations result in incorrect intron splicing and the resulting inability to produce a functional 2OGO protein. Such mutants display a decreased susceptibility to downy mildew and this is thought to result from the observation that 2OGO is a negative regulator of defense-associated genes. It was later shown that DMR6 and its paralog DLO1 (DMR6-like oxygenase 1) were co-expressed during pathogen infection and redundantly suppressed plant immunity through a mechanism of salicylic acid (SA) homeostasis36. Hence, mutations in DMR6 and DLO1 resulted in an increase of SA levels and the upregulation of pathogenesis related (PR)-1, PR-2 and PR-5 genes. Interestingly, the dlo1 mutant showed a lower level of resistance to H. arabidopsidis compared to the dmr6 mutant, whereas, the dmr6dlo1 double mutant showed complete resistance to downy mildew, albeit with an accompanying dwarf phenotype. It was concluded that dmr6 mutant alone provides broad spectrum disease resistance to H. parasitica, Pseudomonas syringae and Phytophthora capsici38. It was also shown the dmr6 mutant was resistant to Fusarium culmorum39. The biochemical mechanism of DMR6 was further elucidated by a recent report40 showing that DMR6 encodes a salicylic acid 5-hydroxylase (S5H) which catalyzes the conversion of SA to 2,5-dihydroxybenzoate (2,5-DHBA) by hydroxylating SA at the C5 position of its phenyl ring. It was shown that the DMR6/S5H expression is induced by SA and that conversion of SA to 2,5-DHBA provides a feedback mechanism which maintains SA homeostasis in Arabidopsis cells. Considering the broad-spectrum resistance to different pathogens induced by DMR6/S5H/2OGO mutation, generating CRISPR-edited dmr6 mutants represents a promising solution to restrict diseases in crop plants.
Arabidopsis has been shown to be infected by F. graminearum and used to study the interaction between the host plant and the pathogen41,42. In this study, we generated 2OGO-knock out (KO) mutant Arabidopsis plants using the CRISPR/Cas9 system. The CRISPR-edited Arabidopsis 2OGO-KO mutants carry frameshift mutations that are predicted to produce non-functional 2OGO enzymes. These edited plants, termed At2OGO-KO, show enhanced resistance to Fg in inflorescence tissues while remaining normal with respect to plant growth and development. We also identified a 2OGO orthologue of barley (cv. Conlon) and introduced the corresponding Hv2OGO cDNA into an Arabidopsis 2OGO-KO mutant line. This complementation experiment fully restored susceptibility of Arabidopsis to Fg, indicating that Hv2OGO is able to function as a suppressor of FHB resistance in Arabidopsis, most likely through conserved biochemical and molecular mechanisms. This result suggests that Hv2OGO may play a similar role in conditioning FHB susceptibility and other infectious diseases in barley. These studies provide a foundation for the creation of CRISPR/Cas9-edited barley plants that are similarly and significantly enhanced for resistance to FHB.
Results
Generating 2OGO-KO (dmr6) mutants in Arabidopsis using CRISPR/Cas9 system
2OGO genomic (At5g24530) and mRNA (NM_122361.4) sequences were obtained from the TAIR and NCBI databases respectively. The active site of 2OGO (oxoglutarate/iron-dependent dioxygenase domain), which resides within the C-terminal portion of its protein sequence (from a.a.188 to a.a.288) corresponds to exon 3 of the genomic sequence and is responsible for the 2OGO-dependent susceptibility phenotype in A. thaliana. We designed a guide RNA (gRNA) that specifically targets exon 3 (target region: 5′- GGTCTCCAGATCTTGATCGA-3′) and that also flanks a BglII restriction site (Fig. 2, underlined) which could be used for mutant screening. The gRNA target was ligated into the psgR-At vector (kindly provided by Dr. J. Zhu28) which contains an Arabidopsis U6 promoter to express the gRNA and a Cas9 gene whose expression is controlled by the 2 × 35 S promoter and the nopaline synthase (NOS) terminator. For Agrobacterium tumefaciens transformation, the gRNA/Cas9 cassettes were subcloned into EcoRI- and HindIII-digested pCAMBIA1300, resulting in the plant expression vector pRD207. Flowering Arabidopsis Col-0 plants were transformed via Agrobacterium-mediated transformation using Agrobacterium strain EHA105 and the floral-dip Arabidopsis transformation method. The collected T0 seeds were screened for germination on 50 μg/L hygromycin. A total of 14 putative T1 mutant plants were recovered from antibiotic selection, all of which were phenotypically similar to the WT plant and produced viable seeds.
Screening of At2OGO mutants created by pRD207 CRISPR gene editing vector. (A) RFLP assay by BglII on WT and T1 plants. (B) T7E1 assay on WT and T1 plants. (C) RFLP assay by BglII to screen for homozygous T2 mutants. The 420 bp band indicates mutation of the target site while the 144 bp and 276 bp bands are consistent with products expected from the WT locus. Due to the sample size, the images were grouped from different gels, but all samples in the same image were electrophoresed at the same voltage for the same length of time. The 1Kb+ molecular weight marker and the WT sample were from the same gel.
Molecular characterization of At2OGO-KO (dmr6) mutants and identification of homozygous progeny
The presence of the Cas9 gene in putative At2OGO (pRD207) CRISPR-edited transformants was confirmed by PCR using Cas9 specific primers and genomic DNA extracted from each T1 line as the template. Putative CRISPR-edited lines were further characterized by restriction fragment length polymorphism (RFLP) for loss of the BglII site that resides within the edited target (Figs. 1, 2). Sanger sequencing of PCR products amplified from around the edited region was used to characterize the nature of mutations present in the T1 progeny. A 420 base pair (bp) genomic DNA (gDNA) region, flanking the target region, was PCR-amplified and analyzed for the presence or absence of the BglII restriction digestion (Fig. 1A) and by T7E1 heteroduplex hybridization assay (Fig. 1B). Figure 1A shows that BglII digestion of PCR products amplified from all of the putative mutant lines gave rise to three DNA fragments. In contrast, BglII digestion of amplicons from wild type (WT) plants gave rise to only two DNA fragments. The lower two bands (276 bp and 144 bp) represent the digested PCR fragments generated by BglII digest of unedited alleles while the upper band (420 bp) represents the undigested PCR fragments amplified from edited alleles. The presence of both BglII-digested and BglII-undigested products in the lines analyzed indicates that all the RD207 mutants recovered are monoallelic, each containing one edited allele and one WT allele. The complete digestion of the WT amplified PCR product indicates that BglII was able to completely digest these templates and that the presence of three bands in the mutant lines is not the result of incomplete DNA digestion. That the RD207 plants carry monoallelic mutations was further supported by the results of the T7E1 assay, which revealed three DNA fragments in all mutants, characteristic of both digested and undigested assay products. To confirm the presence and nature of CRISPR/Cas9-edited mutations in the RD207 plants, the undigested PCR amplified fragments straddling the target mutation site were purified and their DNA sequences were determined by Sanger sequencing. T1 mutants were selfed and the resulting T2 generation was analyzed by RFLP assay to screen for homozygous, biallelic mutants. As shown in Fig. 1C, 13 homozygous T2 plants were identified. The presence of a single BglII-undigested PCR fragment in these T2 plants indicate that they all contain biallelic mutations at the At2OGO target site.
Figure 2 shows the DNA sequences flanking the gene editing target site for RD207 (At2OGO-KO) mutants in both the T1 and T2 generations. Nine different mutation patterns were observed in the T1 mutants. Thirteen out of fourteen T1 mutants had nucleotide deletions around the target site while one mutant had an insertion within the target site. 57% (8 out of 14) of the T1 mutants were confirmed to have frameshift (f.s.) mutations that would disrupt the active site of At2OGO. Most of the mutation patterns from T1 generation were retained in the T2 homozygous mutants. 31% (4 out of 13) of the identified homozygous T2 mutants carried stable frameshifting deletions at the target site. PCR analysis with Cas9-specific primers showed that the 13 frameshifted T2 homozygous mutants had lost the gRNA and the Cas9 cassettes through meiotic segregation, indicating that these plants were edited at the target site but otherwise transgene-free.
We chose the homozygous At2OGO-KO mutant line RD207-15-5 for subsequent disease assessment and barley gene complementation experiments. The RD207-15-5 mutant carries an 8 bp frameshifting deletion in exon 3 of the DMR6 gene resulting in the premature termination of the gene product. It is expected that this truncated gene product would lack an intact oxoglutarate/iron-dependent dioxygenase domain at the C-terminal, resulting in the loss of enzymatic activity, congruent with the similar disruption of function in the original dmr6 point mutant. The reduction of Hyaloperonospora parasitica sporangiophores in the dmr6 mutant37 suggested that the RD207-15-5 mutant might also display reduced susceptibility to Fg infection.
Identification of DMR6 (2OGO) orthologue gene from barley cultivar Conlon for complementation test
We searched the PGSB barley genome database for DMR6 orthologues in barley (Hordeum vulgare, Hv). Currently, high quality reference genome assembly and automated gene annotation have been carried out only on barley cv. Morex43. One copy of the DMR6 orthologue, Hv2OGO gene was identified in cv. Morex. The mRNA and gDNA sequences were annotated as MLOC_59596.1 and HORVU4Hr1G084810.2, respectively, in the database. We focused on DMR6 orthologues from Conlon because this cultivar is more susceptible to Fg than is Morex, and provides a better baseline for determining the effects on susceptibility of inactivating this gene in barley, which is our ultimate goal. In order to characterize the Hv2OGO gene sequence in cv. Conlon, as compared to cv. Morex, total RNA from cv. Conlon was extracted and sequenced. The RNAseq data were assembled and aligned with the cv. Morex Hv2OGO gene sequence to identify the corresponding Conlon 2OGO gene sequence. The Conlon Hv2OGO nucleotide sequence was shown to be 99.9% identical to the Morex Hv2OGO nucleotide sequence except for a single substitution at position 396 of the cDNA sequence, changing from cytosine (C) to thymine (T) base. This nucleotide change changes the corresponding amino acid from phenylalanine to leucine, which is a conservative change and maintains side-chain hydrophobicity. The Conlon Hv2OGO gene was cloned into the binary vector pEL103 (kindly provided by Dr. E. Lam44,), resulting in pRD331, which was then transformed into EHA105 strain of Agrobacterium tumefaciens. The homozygous At2OGO-KO (RD207-15-5) plants were transformed with pRD331 using the Agrobacterium-mediated floral dip method. Collected T0 seeds were screened for germination on 70 μg/L kanamycin-containing medium. A total of 7 putative T1 Hv2OGO-complement-transformants were recovered from antibiotic selection and integration of the Hv2OGO transgene confirmed by PCR amplification of gDNA using Hv2OGO gene-specific primers (data not shown). Homozygous At2OGO-KO/Hv2OGO plants were obtained in the resulting T2 generation.
Characterization of disease phenotype on At2OGO-KO and At2OGO-KO/Hv2OGO homozygous plants
The homozygous At2OGO-KO mutant (line RD207-15-5), which contains an 8 bp frameshifting deletion in exon 3 of the DMR6 gene, as well as homozygous At2OGO-KO/Hv2OGO-complemented plants (line RD207-15-5/331-1) were used in Fg infection assays together with WT Arabidopsis plants as a control. Fg infection assays were performed by point inoculation of the sGFP-tagged Fg macroconidia onto similarly staged inflorescences detached from 5-week old WT, At2OGO-KO and At2OGO-KO/Hv2OGO Arabidopsis plants followed by stereomicroscopic observation performed each day post inoculation (dpi) for 6 days (Fig. 3). At 2 dpi, fungal hyphae started to grow on the inflorescence surface of both WT and At2OGO-KO/Hv2OGO plants. Strikingly, At2OGO-KO inflorescences remained free from external hyphae growth at this stage. Hyphal growth on WT and At2OGO-KO/Hv2OGO inflorescences spread from the inoculated inflorescence to neighboring inflorescences starting from 3 to 4 dpi whereas At2OGO-KO inflorescences had minimal external hyphal growth on inflorescences at 4 dpi, even at the site of inoculation. At 5 to 6 dpi, the Fg hyphae on WT and At2OGO-KO/Hv2OGO inflorescences became more abundant and started to colonize the entire inflorescence tissue. At this stage the fungus formed a thick mycelial mat and host tissues started to show wilting symptoms. At2OGO-KO inflorescences remained in their initial infection condition at 6 dpi, with a minimal amount of external hyphal growth and no observable symptoms in host plant tissues. It is evident from these observations, that the inactivation of the At2OGO gene slowed down the infection progress of Fg, in concert with its previously established role in conditioning susceptibility to H. arabidopsidis. Our results showed that the At2OGO-KO inflorescences were more resistant to Fg infection than were WT inflorescences. Moreover, the introduction of Hv2OGO into RD207-15-5 (At2OGO-KO) plants fully restored susceptibility of the host to Fg. These results represent a functional complementation of the At2OGO-KO by Hv2OGO and strongly suggest that Hv2OGO may also function as an Fg susceptible factor in barley.
In order to assess the ability of Fg to colonize the host plant, we quantitated the amount of Fg that had penetrated into the host tissue by qPCR analysis of fungal DNA (Fig. 4). Inoculated inflorescences were collected daily and Fg hyphae and macroconidia were washed off plant tissue surfaces using three washes of sterile deionized water accompanied by mild shaking. The rinsed and pat-dried inflorescences were then processed for analysis by qPCR using primers specific for the sGFP gene present in the Fg strain45 used in these studies. WT and At2OGO-KO/Hv2OGO inflorescences showed a similar time course of infection with the amount of Fg present within inflorescence tissues increasing steadily and consistently, starting at 48 hpi and continuing to 144 hpi. In contrast, colonization of At2OGO-KO by Fg proceeded at a significantly reduced rate, equivalent to 30–60% that observed in WT or At2OGO-KO/Hv2OGO complemented plants. Indeed, Fg levels in At2OGO-KO inflorescences never reached the levels of infection observed in WT or complemented inflorescences within the time course of the experiment. Spore production on Fg inoculated inflorescences was quantitated using a hemocytometer (Fig. 5) and provided another readout of the degree of colonization of the plants. These results resembled the qPCR assay in that the spore production increased rapidly in WT and At2OGO-KO/Hv2OGO inflorescences starting from 96 hpi. Again, in contrast, spore production in At2OGO-KO inflorescences was markedly reduced, compared to WT levels, indicating that the KO-mutation of At2OGO suppressed Fg proliferation and that this deficiency could be complemented by introduction of an orthologous gene from barley. At 144 hpi, spore production in KO and complemented plants were, respectively, 41% and 25% lower than in WT plants. It is worth nothing that the ability of the barley orthologue to complement At2OGO-KO is only partial, in that WT levels of Fg DNA were not reached until late in infection, at 144 hpi, while the number of spores produced in the complemented line never fully reached those seen in WT plants. Nonetheless, the barley orthologue was clearly able to substantially restore Fg colonization of the host, compared to At2OGO-KO plants. Together, these results show that At2OGO mutants are effective in restricting Fg growth and that the barley Hv2OGO gene can function to restore susceptibility when expressed in this heterologous plant system.
qPCR quantitative analysis of Fg growth on inoculated inflorescences. Total DNA was prepared from Fg-infected, detached Arabidopsis inflorescences and the amount of Fg determined by qPCR using primers specific for the sGFP-transgene present in the Fg genome. Experiments were repeated on three independent sample batches. The percentage reduction relative to WT detached Arabidopsis inflorescences is shown within each group of samples collected at a particular time point post inoculation. Statistical significance was determined by t-test analysis followed by Holm-Sidak method, with alpha = 0.05. Each time point was analyzed individually, without assuming a consistent SD (p-value: *≤ 0.05; **≤ 0.01; ***≤ 0.001).
Quantitative measurement of spore production on Fg inoculated inflorescences. WT, At2OGO-KO and At2OGO-KO/Hv2OGO lines were inoculated with Fg at 0 h and spores collected and determined at the indicated times. The percentage reduction of spore production (compared to WT) is indicated for each of the 72 h, 96 h, 120 h and 144 h time points. The experiment was repeated with three independent batches of samples. Statistical significance was determined by t-test analysis followed by Holm-Sidak method, with alpha = 0.05. Each time point was analyzed individually, without assuming a consistent SD (p-value: *≤ 0.05; **≤ 0.01; ***≤ 0.001).
Gene expression profiling on plant defense signaling pathway-related genes
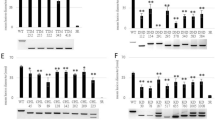
The involvement of salicylic acid (SA), jasmonic acid (JA) and ethylene (ET) in the response of WT, At2OGO-KO and At2OGO-KO/Hv2OGO plants to Fg was explored by analyzing changes in the expression of genes specific to these signaling pathways. Levels of gene expression were determined by RT-qPCR. At2OGO T-DNA-KO plants have been shown to accumulate SA, due to the inability of these mutants to metabolize SA to 2,5-DHBA36. Higher levels of SA can induce defense-related genes and this may lead to the heightened resistance observed in our studies and in previous studies of dmr6 plants inoculated with Fg. Previous studies showed that the expression of PR genes, such as PR1, in dmr6 plants was constitutively elevated ~10-fold in healthy, uninfected leaf tissues38. Consequently, we analyzed uninoculated floral and leaf samples from the At2OGO mutant plants generated here by gene editing. Figure 6A shows that in this study, basal levels of gene expression of PR1, PR2 and PR5 in At2OGO-KO leaf tissues were markedly elevated compared to WT plants. Indeed, we observed an even stronger induction for the PR2 gene, which was induced ~20-fold over WT levels, compared to the ~10-fold induction observed for this gene in dmr6 leaf tissues38. Surprisingly, uninoculated floral samples of At2OGO-KO did not show a similar elevation of basal expression of genes encoding PR-proteins, compared to WT levels (Fig. 6B).
Relative expression of plant defense-related genes by RT-qPCR on non-inoculated leaf (A) and floral (B) samples of WT, At2OGO-KO and At2OGO-KO/Hv2OGO plants. The experiment was repeated three times with samples from three individual plants. The relative expression values were averaged and standard errors calculated.
Instead, we observed a marked difference in the levels of these genes in floral tissues only when plants were inoculated with Fg. For these studies, inflorescences were point-inoculated with sGFP-tagged Fg and tissues collected at 3, 6, 12, 24, 48 hpi and processed for RT-qPCR analysis. In contrast to the elevated levels of defense-related genes observed in leaf tissues of At2OGO-KO plants, constitutive levels in floral tissue remained near to those seen for WT and complemented plants (Figs. 6 and 7 at 0 h). Surprisingly, Fg did not induce defense genes in the inoculated inflorescences from either WT or complemented plants.
Relative expression of plant defense-related genes by RT-qPCR on Fg-inoculated floral tissues of WT, At2OGO-KO and At2OGO-KO/Hv2OGO plants. Relative expression profile includes genes involved in SA signaling (A–D), JA/ET signaling (E–I) and ROS activation (J). RT-qPCR assay was repeated three times with different batch of samples. The relative gene expression was calculated using the 2−ΔΔCt formula.
In contrast, these genes were massively induced in Fg-inoculated At2OGO-KO floral tissues. All defense-related genes analyzed were significantly induced by 6 hpi in At2OGO-KO inflorescences, compared to either WT or to At2OGO-KO/Hv2OGO inflorescences (Fig. 7). Figure 7A–D shows relative expression of SA-related genes (NPR1) and pathogenesis-related genes (PR1, PR2 and PR5) which are key components of SA signaling in plant defense responses to attempted infection. The SA-related gene expression started to increase at 3 hpi and peaked at 12 hpi. PR3 and PDF1.2 (Fig. 7E,F), JA and ET responsive genes, were expressed at an extremely high level starting at 6 hpi and decreased after 12 hpi. The AOS gene product (Fig. 7G) is involved in the upstream JA signaling pathway. Expression of this gene started to increase at 6 hpi, peaked at 12 hpi and dropped to basal levels at 24–48 hpi. The EIN3 and ERF1 genes (Fig. 7H,I), which are activated by ET, displayed a similar gene expression pattern to AOS, with induction observed at 6–12 hpi and the expression levels decreasing at 24–48 hpi. These genes were upregulated at 6–12 hpi and decreased to a lower level of expression at 24–48 hpi, except that PR1 gene expression dropped to the basal level at 48 hpi. The RBOHD gene (Fig. 7J) was highly expressed at 6–12 hpi and maintained a much lower expression at 24–48 hpi. Overall, At2OGO-KO plants displayed an amplified inducible defense gene response in the inflorescences, which may explain their enhanced disease resistant phenotype when inoculated with Fg.
Discussion
One of the major challenges for agribusiness and food security is to maintain sustainable agricultural practices while increasing both the quantity and quality of food produced. Pathogen attack is the leading cause of both yield loss and decreased crop quality globally. The discovery of disease susceptibility genes has made them an attractive target for gene manipulation since loss-of-function mutations may enhance plant internal resistance to infection. This potentially avoids problems of public acceptance associated with the creation of crops containing transgenes that enhance disease resistance. Moreover, loss-of-function mutations may also provide more resilient resistance under field conditions, since it is more difficult for the pathogen to revert to a virulent form, as is the case when specific R-genes are introduced or when single defense responses are enhanced. For these reasons, we are exploring the use of gene-edited, loss-of-function mutants in disease susceptibility genes as a path for creating stable and acceptable crops with a superior disease resistance profile.
The impairment of Arabidopsis DMR6 (2OGO) gene has been shown to confer broad-spectrum disease resistance37,38. This mutant was generated by a common mutagen, ethyl methane sulfonate (EMS), which may have caused mutations in other genes46. Therefore, in this study, we took advantage of the targeted mutagenesis afforded by CRISPR/Cas9 gene editing technology to generate At2OGO-KO mutants whose disease susceptibility phenotype could be determined upon Fg infection of inflorescences. Our At2OGO-KO plants were phenotypically similar to WT plants and produced viable seeds. It has been shown that the silencing of orthologous StDMR6 gene in potato resulted in normal looking plants that were resistant to late blight resistant plants47. Our results have shown that At2OGO-KO mutants are more resistant to Fg infection and proliferation than WT plants. We have also shown that the At2OGO orthologue of barley cv. Conlon (Hv2OGO) can restore the disease susceptible phenotype in At2OGO-KO mutants to almost WT levels, thus verifying its ability to function as a disease susceptibility gene and paving the way for editing Hv2OGO gene to study its effect on FHB resistance in barley.
DMR6 belongs to 2-oxoglutarate Fe(II)-dependent oxygenase (2OGO) superfamily whose members are largely involved in the biosynthesis of plant flavonoids and secondary metabolites48. The DMR6 orthologue in wheat was described as a flavanone-3-hydroxylase (F3H) gene49 whose expression has been shown to increase rapidly during incompatible interaction with Hessian fly, indicating its involvement in insect defense in wheat50. The discovery that the loss-of-function dmr6 mutant can confer broad spectrum disease resistance led us to identify potential homologues or orthologues of DMR6 in barley, with the goal of manipulating FHB susceptibility in that crop. In order to understand the role of DMR6 in barley, the cv. Conlon orthologue was identified, cloned and transformed into the At2OGO-KO mutant in order to verify whether the barley orthologue carries the same function in facilitating Fg invasion. Based on our results using quantitative assays for disease progression, At2OGO-KO plants were able to slow down fungal invasion on inflorescence tissues and delay the onset of spore proliferation, compared to WT and to Hv2OGO-complemented plants. Gene editing carried out in this study introduced frameshift mutations in exon 3 that truncated the C-terminal Fe(II) 2OG dioxygenase domain of the polypeptide and resulted in a protein that is predicted to lack dioxygenase enzyme activity. The disruption in this domain and the concomitant loss of 2OGO enzyme activity is further expected to affect the biosynthesis and catabolism of plant hormones, such as salicylic acid, ethylene, gibberellins and flavonoids48. Biochemical analysis of the 2OGO protein from previous reports indicated that it has a specific function in fine-tuning SA homeostasis40. Specifically, 2OGO was shown to convert SA to 2,5-dihydroxybenzoic acid (2,5-DHBA) while a related OGO encoded in Arabidopsis by the DLO1 gene converts SA to 2,3-dihydroxybenzoic acid (2,3-DHBA)40. Mutation of either OGO results in elevated SA levels and this is thought to form the basis for the enhanced, broad spectrum resistance found in either single or double mutants of these genes. These findings suggest that 2OGOs function as plant immunity suppressors for pathogen invasion. In this study, qPCR showed that At2OGO-KO floral tissues were able to restrict fungal invasion, compared to either WT or Hv2OGO-complemented floral tissues. Even though the more resistant At2OGO-KO plants might not have completely blocked the entry of Fg into host tissue, fungal progression and proliferation were greatly impeded and infection proceeded at a much slower rate. This observation provides solid evidence for an efficient defense mechanism within the resistant plants and points to the value in trying to enhance this response as an effective defense against FHB and, possibly, other disease as well. We further characterized this enhanced resistance by measuring spore production to measure the Fg disease progression and to determine the onset of Fg colonization and host wilting. Based on our results, At2OGO-KO floral tissues were able to delay the fungal progression for at least 48 hours and restrict spore production by approximately 30–60% compared to the WT and the Hv2OGO-complemented floral tissues. It seems that WT and the Hv2OGO-complemented floral tissues were compromised at the end of 2 dpi when Fg started to overwhelm the host defense system and exploit host nutrients for spore proliferation. On the other hand, At2OGO-KO floral tissues managed to strengthen their defense system and alleviate the disease symptoms. Our data showed that impairment of the At2OGO gene leads to enhanced FHB disease resistance in Arabidopsis.
In order to better understand the underlying molecular mechanism of the resistant plants, we performed RT-qPCR assay to examine the effect of At2OGO mutation on plant defense signaling pathways. In concert with the notion that enhanced resistance in At2OGO-KO plants results from elevated SA levels, we found significantly higher, constitutive levels of defense response- and defense-related genes in leaf tissues. However, the steady state levels of these genes in non-Fg inoculated floral tissues were not significantly induced, compared to WT plants. Instead, we found that the inducible response to infection was markedly enhanced in At2OGO-KO floral tissues, compares to WT or Hv2OGO-complemented floral tissues. Fg-inoculated floral tissues of At2OGO-KO mutant plants showed much higher levels of inducible gene expression in all the defense-related genes analyzed, with the most elevated levels at 3 hpi. In contrast, both WT and Hv2OGO-complemented floral tissues displayed much lower levels of gene expression upon fungal infection and showed no significant induction upon infection. These results suggest that enhanced resistance expressed in floral tissues is not the result of the prior activation of defense genes, as seen in leaf tissues. Instead, the ability of the floral tissue to rapidly and extensively react to the presence of the pathogen is markedly enhanced over the WT. The nature of such a priming effect remains to be determined, but may resemble those observed in other SA-related effects such as systemic acquired resistance (SAR)51,52. In barley and wheat, floral tissues represent the natural site of primary infection with Fg, fungal penetration occurring on the anthers and proceeding through the floral tissues and into neighboring tissues. It would be interesting to determine if this course of infection results from the opportunistic infection of floral tissues that, for whatever reason, either lack effective basal or inducible defenses.
DMR6 has been shown to encode a salicylate 5-hydrolase40 whose impairment results in the loss of hydrolase activity and the accumulation of high levels of SA which, in turn, stimulate systemic acquired resistance (SAR)53. SA is the central player in SAR and known to be a critical defense against biotroph infection. The accumulation of SA can enhance SAR responsiveness and therefore pathogen resistance. Based on this, we expected that during the early hemibiotrophic phase of Fg infection, defense responses of At2OGO-KO mutants would be expected to respond in a two-fold manner: first, by elevating the expression of SA-related genes, such as NPR1, and other SAR-associated markers (PR1, PR2, PR3, and PR5); second, by triggering an oxidative burst characterized by the production of reactive oxygen species (ROS). Our results support these predictions. Figure 7A–D showed that SA-related genes were expressed more strongly in At2OGO-KO plants, compared to WT, with accumulation starting from 3 hpi and peaking at 12 hpi, followed by a decrease thereafter. At2OGO-KO plants are predicted to have elevated levels of SA, as shown previously for dmr6 mutants. This provides a coherent explanation for the enhanced resistance seen in At2OGO-KO plants, based on higher levels of SA leading to either elevated levels of defense gene expression, as seen in leaf tissues, or the potentiation of the defense response, as seen in floral tissues which lack elevated basal defenses but which display a markedly enhanced inducible response. Either approach is likely to play a role in restricting fungal invasion and preventing further proliferation.
Following SA-related gene induction, it has been established that the JA/ET signaling pathways are activated to mediate the resistance against necrotrophic pathogen54. Our results have shown the sequential pattern in the activation of the defense pathways. The JA/ET signaling pathways-related genes (AOS, EIN3, ERF1, PR3 and PDF1.2) were induced rapidly starting from 6 hpi and their expression decreased after 12 hpi (Fig. 7E–J). The upregulation of JA/ET-related genes and the marked induction of PDF1.2 (approximately 56-fold compared to the WT and At2OGO-KO/Hv2OGO plants) indicate that these genes may also play a role in the enhanced resistance displayed by At2OGO-KO plants and that this, in turn, may be responsible for slowing down Fg infection and colonization. The RBOHD gene was highly expressed from 6–12 hpi, implicating it in regulating ROS production as a protective mechanism during attempted pathogen invasion55,56,57. Based on our qPCR analysis, JA signaling was activated in parallel with the SA signaling to attenuate damage caused by fungal infection, which was similar to responses observed in previous studies58. Overall, the much higher level of defense gene expression may be the underlying mechanism responsible for enhanced Fg resistance of At2OGO-KO plants. The overall pattern appears to involve the SA signaling pathway at the early stage of Fg infection followed by the activation of JA signaling pathway at later stages of infection. The impairment of DMR6 gene in At2OGO-KO mutant may be responsible for increasing SA levels which directly exerts its effects on defense responses via the SA/JA/ET signaling pathways. These defense pathways can then act, in turn, to augment natural levels of disease resistance during Fg infection. Plants complemented with the barley orthologue Hv2OGO were almost fully restored to WT levels of Fg susceptibility, strongly suggesting that Hv2OGO may be similarly involved in conditioning FHB susceptibility in barley plants. If so, CRISPR-editing the Hv2OGO gene in barley cultivars would represent a promising strategy for combating FHB in this, and other crop species.
Methods and materials
Arabidopsis thaliana plant growth
Arabidopsis thaliana Columbia ecotype (Col-0) seeds were sterilized with 30% bleach for 30 min and washed five times with sterile water. Sterilized seeds were stratified at 4 °C for two days and plated on growth medium [Murashige and Skoog (MS) medium containing 1% (w/v) sucrose and 0.3% (w/v) Gelzan]. The plated seeds grew at 22 °C under 16 h/8 h light-dark photoperiod. Germinated seedlings were transferred to Pro-Mix soil in an environmental-controlled chamber with a 16 h/8 h light-dark photoperiod at 22 °C and 40%-60% relative humidity.
Construction of At2OGO CRISPR-editing vector
The Arabidopsis thaliana genomic and mRNA sequence of 2-oxoglutarate and Fe(II)-dependent oxygenase (At2OGO) was obtained from TAIR (https://www.arabidopsis.org/) and NCBI (https://www.ncbi.nlm.nih.gov/) databases with accession number of AT5G24530 and NM_122361.4 respectively. The gRNA target sequence for At2OGO gene (5′- GGTCTCCAGATCTTGATCGACGG-3) was identified according to selection guidelines (www.addgene.org/crispr/guide) and included with 20 nucleotides upstream of a proto-spacer adjacent motif (PAM, CGG) sequence (5′-N20-NGG-3′) and a target sequence beginning with the G nucleotide base that includes a AGATCT BglII restriction site. The At2OGO-editing vector was constructed following previously published protocols29. The gRNA sense and anti-sense oligonucleotides (forward: 5′- GATTGGTCTCCAGATCTTGATCGA-3′; reverse: 5′- AAACTCGATCAAGATCTGGAGACC-3′) were phosphorylated and annealed under the following conditions: 37 °C for 30 min, 95 °C for 5 min and then ramping down to 25 °C at the rate of 5 °C/min, in a 10 μl reaction (1 μl of 10 × T4 ligation buffer, 0.5 μl of T4 PNK, 1 μl of each oligo at 100 μM). The annealed gRNA oligonucleotides were cloned into psgR-Cas9-At vector (kindly provided by Dr. J. Zhu28,). The resulting plasmid was sequenced and confirmed followed by subcloning into the plant expression vector pCAMBIA1300 (https://www.addgene.org/vector-database/5930/), resulting in pRD207, and introduced into Agrobacterium tumefaciens EHA105 strain for subsequent plant transformation.
Arabidopsis thaliana plant transformation and mutant selection
Arabidopsis thaliana Col-0 plants were grown for 3–4 weeks until flowering. Agrobacterium tumefaciens EHA105 strain harboring pRD207 (At2OGO gRNA/Cas9) was grown in 2 ml of LB liquid medium containing 50 μg/ml kanamycin and 50 μg/ml chloramphenicol. The 2 ml overnight culture was inoculated into 200 ml LB liquid medium containing the same antibiotics and was grown in a shaking incubator at 30 °C for approximately 5 h until the OD600 reached to 0.6. Agrobacterium cells were collected by centrifugation and rinsed twice with 5% sucrose before resuspending into 300 ml of 5% sucrose solution containing 0.05% Silwet L-77. Floral buds were dipped into Agrobacterium solution for 1–2 s59. The inoculated plants were covered with a clear plastic dome for 24 h before returning to growth chamber. The transformed plants were grown for another 3–4 weeks for seed harvesting. The collected T0 seeds were sterilized by 30% bleach and selected on MS growth medium containing 50 μg/ml hygromycin. The selected mutant plants were transferred to soil and acclimated growth in the growth chamber.
Genomic DNA (gDNA) isolation of Arabidopsis mutant plants
200 mg leaf tissues from each putative mutant plant were collected for gDNA isolation using CTAB extraction buffer (2% cetyl trimethylammonium bromide, 1% polyvinyl pyrrolidone, 100 mM Tris-HCl, 1.4 M NaCl, 20 mM EDTA). The CTAB-leaf tissue mix was vortexed vigorously for 30 s and then incubated at 65 °C for 1 h followed by phenol-chloroform isolation. The supernatant was then mixed with 100% ethanol to precipitate the DNA followed by a clean-up step with 70% ethanol. After centrifugation, DNA pellets were air-dried and resuspended in sterile deionized water. The concentration of DNA was measured with a Nanodrop spectrophotometer (Thermo Fisher, Waltham, MA, United States).
Mutant analysis: mutant confirmation, restriction fragment length polymorphism (RFLP) and T7 endonuclease 1 (T7E1) assays
The presence of the Cas9 gene in putative mutant plants was confirmed by PCR-amplification using Cas9 primers (forward: 5′- GAAGCGGAAGGTCGGTATCCACGG-3′; reverse: 5′- GGCCAGATAGATCAGCCGCAGGTC-3′) for its integration into the genome. The gDNA samples were then used to amplify a 420 bp fragment flanking the target sequence (forward: 5′- GTGCTTGGTGAACAAGGTCAACAC-3′; reverse: 5′- GTGCTTGGTGAACAAGGTCAACAC-3′). The amplified fragments were purified using phenol/chloroform clean-up method and resuspended in deionized water prior to analysis. RFLP assay was carried out by incubating BglII restriction enzyme with the purified 420 bp PCR fragment from each putative mutant line and the WT plant. Digested samples were electrophoresed on 1% agarose gel and the gel images were captured under UV illumination. Putative mutants were also analyzed using the T7E1 assay, in which formation of a heteroduplex between WT and mutant PCR products is followed by digestion of mismatched nucleotides by T7E160. The purified 420 bp PCR fragments were denatured and annealed in a thermocycler using the following program: 95 °C for 5 min, ramping down to 85 °C at the rate of 2 °C/s, 25 °C at the rate of 0.1 °C/s. The mixture was then digested with 0.5 μl of T7E1 enzyme (10 U/μl) at 37 °C for 60 min. Samples were electrophoresed on 1% agarose gel immediately after incubation and images captured under UV illumination.
Complementation of At2OGO-KO line with Hv2OGO
In order to clone the Hv2OGO cDNA from barley cv. Conlon, total RNA was isolated from four-week-old leaf tissues and subjected to RNAseq analysis by Novogen Co., Ltd.. The RNAseq data has been deposited in NCBI SRA database (accession number SRR10059574) under BioProject PRJNA563590. The RNAseq data from Conlon were aligned to cv. Morex 2OGO gene sequence (mRNA: MLOC_59596.1; gDNA: HORVU4Hr1G084810.2) using the NCBI BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Conlon Hv2OGO cDNA was amplified by PCR with gene specific primers (forward: 5′- GGTCTAGAATGGCGGAGCAGCTCATCTC-3′; reverse: 5′- GGGAGCTCCTAGGTTCTGAAGAGCTCCAGGC-3′) and cloned into plant expression vector pEL103 (kindly provided by Dr. E. Lam44,), resulting in pRD331, which was then transformed into Agrobacterium tumefacien EHA105 strain. The homozygous At2OGO-KO line (RD207-15-5) was complementarily transformed with pRD331 via the floral dip method. Seeds from T0 plants were selected on 70 μg/ml kanamycin. Segregation of kanamycin resistance in the confirmed T1 complemented plants was followed by plating seeds on 70 μg/ml kanamycin. Homozygousity of the T2 generation was similarly confirmed by antibiotic screening of segregants.
Fg inoculation on detached Arabidopsis inflorescences and quantitative assays for disease development
Fg tagged with sGFP (superfolder green fluorescent protein)45 was cultured on potato dextrose agar (PDA) for 7 days at 22 °C under UV light for 24/7. Fungal plug was extracted from PDA plate and cultured in 50 ml of mung bean soup for another 7 days to generate macroconidia61. The macroconidia were filtered out from mung bean soup using Miracloth (Millipore Sigma, Burlington, MA, United States) and washed twice with sterile deionize water before resuspension in sterile water. Macroconidia were counted using hemocytometer. Macroconidia were resuspended to an inoculum of 1 × 106 spores/ml. Bolting floral buds were detached from Arabidopsis plants and laid perpendicularly onto 0.7% water agar in petri dishes. Each inflorescence was inoculated with 2 μl of 1 × 106 spores/ml macroconidia solution along with a duplicate set of mock sample that was inoculated with sterile water. The inoculated detached inflorescences were maintained in a 100% humidity condition by spraying water on the inner side of the petri dish lids prior to plate sealing. The plates were left in environmentally controlled chamber with a 16 h/8 h light/dark photoperiod at 22 °C. Images were taken using the Dino-eye eyepiece (Dino-Lite, Hsinchu, Taiwan) attached to the Nikon SMZ 645 stereomicroscope (Nikon corp., Minato, Japan). Samples were collected every 24 h by pooling four inflorescences in a tube per time point until reaching 144 hours. After each collection, the pooled samples were rinsed with 1 ml of sterile water and mild shaking to remove Fg spores from the surface of inflorescence. At each time point, the first 1 ml of rinsed water was collected to quantitate the spore production at each time point using a hemocytometer. Pooled samples were rinsed for another two times with 1 ml of water to remove any surface-attached spores. Total DNA from these pooled samples were then isolated using the CTAB method. The amount of Fg DNA present in pooled samples was determined by quantifying the levels of the Fg-borne sGFP transgene. Quantitation of sGFP was performed by RT-PCR using an Applied Biosystems StepOne Plus thermocycler (Applied Biosystems, Thermo Fisher, Foster City, CA, United States) and primers specific for the Fg sGFP gene (forward: 5′- GTCCGCCCTGAGCAAAGA-3′; reverse: 5′- TCCAGCAGGACCATGTGATC-3′). Both quantitative assays were performed in triplicate and averages and standard deviations (SD) determined. Data collected from WT vs. At2OGO-KO and WT vs. At2OGO-KO/Hv2OGO groups were analyzed by Student’s t-test analysis using GraphPad software. Statistical significance was determined using the Holm-Sidak method, with alpha = 0.05. Each time point was analyzed individually, without assuming a consistent SD.
Relative gene expression profiling on plant defense mechanism
In order to understand the molecular mechanisms underlying the defense signaling in At2OGO-KO and At2OGO-KO/Hv2OGO plants infected with Fg, some of the key gene targets in the SA, JA, ET and ROS signaling pathways were selected for quantitation by RT-qPCR analysis. Arabidopsis actin-2 (ACT2) gene was used as the reference housekeeping gene in these studies. Primers for each selected gene were designed using the Primer Express software (Applied Biosystems, Thermo Fisher, Foster City, CA, United States). The primer sets for each gene were first validated to have similar amplification efficiency as the ACT2 gene. Detached inflorescences were each inoculated with 2 μl of Fg macroconidia solution as described above, and collected at 3, 6, 12, 24, 48 h time point. Four inflorescences were pooled into a tube and rinsed three times with 1 ml of sterile water. Total RNA was isolated from pooled samples were isolated using the TRIZOL method (Ambion Life Technologies, Thermo Fisher, Carlsbad, CA, United States) and the concentration was measured using the Nanodrop spectrophotometer (Thermo Fisher, Waltham, MA, United States). Reverse transcription (RT) reaction was carried out with the High Fidelity cDNA Synthesis Kit (Applied Biosystems, Thermo Fisher, Foster City, CA, United States) using approximately 2 μg RNA. cDNA generated in these RT reactions were used as templates in RT-qPCR reactions with the SYBR Green 2X Master Mix (Applied Biosystems, Thermo Fisher, Foster City, CA, United States). The RT-qPCR assay was run on the default setting at 95 °C for 3 min for initial denaturation and 40 cycles at 95 °C for 30 s followed by 60 °C for 30 s. The fold-change of gene expression was calculated by the 2−ΔΔCt method62. The RT-qPCR analysis was repeated three times with different batches of samples, and the gene expression levels were averaged. Primers used in the RT-qPCR gene expression assay are listed in Supplementary Table 1.
References
Munkvold, G. P. Cultural and genetic approaches to managing mycotoxins in maize. Annu. Rev. Phytopathol. 41, 99–116 (2003).
Manstretta, V. & Rossi, V. Effects of Temperature and Moisture on Development of Fusarium graminearum Perithecia in Maize Stalk Residues. Appl. Env. Microbiol. 82(1), 184–91 (2016).
McMullen, M. et al. A Unified Effort to Fight an Enemy of Wheat and Barley: Fusarium Head Blight. Plant. Dis. 96(12), 1712–1728 (2012).
Nganje, W. E. et al. Regional Economic Impacts of Fusarium Head Blight in Wheat and Barley. Appl. Economic Perspect. Policy 26(3), 332–347 (2004).
Wilson, W., Dahl, B. & Nganje, W. Economic costs of Fusarium Head Blight, scab and deoxynivalenol. World mycotoxin J. v.4 no.1(no. 2), 291–302 (2018).
Pestka, J. J. & Smolinski, A. T. Deoxynivalenol: toxicology and potential effects on humans. J. Toxicol. Env. Health B Crit. Rev. 8(1), 39–69 (2005).
Sobrova, P. et al. Deoxynivalenol and its toxicity. Interdiscip. Toxicol. 3(3), 94–9 (2010).
Dahleen, L. S., Okubara, P. A. & Blechl, A. E. Transgenic approaches to combat Fusarium head blight in wheat and barley. Crop. Sci. v.41(no. 3), 628–0 (2001).
Mackintosh, C. A. et al. Overexpression of defense response genes in transgenic wheat enhances resistance to Fusarium head blight. Plant. Cell Rep. 26(4), 479–488 (2007).
Makandar, R. et al. Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol. Plant. Microbe Interact. 19(2), 123–9 (2006).
Li, Z. et al. Expression of a radish defensin in transgenic wheat confers increased resistance to Fusarium graminearum and Rhizoctonia cerealis. Funct. Integr. Genomics 11(1), 63–70 (2011).
Han, J. et al. Transgenic expression of lactoferrin imparts enhanced resistance to head blight of wheat caused by Fusarium graminearum. BMC Plant. Biol. 12, 33 (2012).
Hallen-Adams, H. E. et al. Deoxynivalenol biosynthesis-related gene expression during wheat kernel colonization by Fusarium graminearum. Phytopathology 101(9), 1091–6 (2011).
Kimura, M. et al. Molecular and genetic studies of fusarium trichothecene biosynthesis: pathways, genes, and evolution. Biosci. Biotechnol. Biochem. 71(9), 2105–23 (2007).
Seong, K. Y. et al. Global gene regulation by Fusarium transcription factors Tri6 and Tri10 reveals adaptations for toxin biosynthesis. Mol. Microbiol. 72(2), 354–67 (2009).
Di, R. & Tumer, N. E. Expression of a truncated form of ribosomal protein L3 confers resistance to pokeweed antiviral protein and the Fusarium mycotoxin deoxynivalenol. Mol. Plant. Microbe Interact. 18(8), 762–70 (2005).
Shin, S. et al. Transgenic Arabidopsis thaliana expressing a barley UDP-glucosyltransferase exhibit resistance to the mycotoxin deoxynivalenol. J. Exp. Bot. 63(13), 4731–40 (2012).
Pasquet, J. C. et al. A Brachypodium UDP-Glycosyltransferase Confers Root Tolerance to Deoxynivalenol and Resistance to Fusarium Infection. Plant. Physiol. 172(1), 559–74 (2016).
Liu, L. & Fan, X. D. CRISPR-Cas system: a powerful tool for genome engineering. Plant. Mol. Biol. 85(3), 209–18 (2014).
Fogarty, N. M. E. et al. Erratum: Genome editing reveals a role for OCT4 in human embryogenesis. Nature 551(7679), 256 (2017).
Friedland, A. E. et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10(8), 741–3 (2013).
Hwang, W. Y. et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31(3), 227–9 (2013).
Sanchez, J. C. et al. Phenotypic and Genotypic Consequences of CRISPR/Cas9 Editing of the Replication Origins in the rDNA of Saccharomyces cerevisiae. Genetics, 2019.
Xue, W. et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514, 380 (2014).
Chen, K. et al. CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annu. Rev. Plant. Biol. 70, 667–697 (2019).
Endo, M., Mikami, M. & Toki, S. Multigene Knockout Utilizing Off-Target Mutations of the CRISPR/Cas9 System in Rice. Plant. Cell Physiol. 56(1), 41–47 (2014).
Feng, C. et al. Efficient Targeted Genome Modification in Maize Using CRISPR/Cas9 System. J. Genet. Genomics 43(1), 37–43 (2016).
Feng, Z. et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23(10), 1229–32 (2013).
Jiang, W. et al. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41(20), e188 (2013).
Li, Z. et al. Cas9-Guide RNA Directed Genome Editing in Soybean. Plant. Physiol. 169(2), 960–970 (2015).
Sanchez-Leon, S. et al. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant. Biotechnol. J. 16(4), 902–910 (2018).
Zhang, Z. et al. A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant. Cell Rep. 35(7), 1519–1533 (2016).
Gao, C. The future of CRISPR technologies in agriculture. Nat. Rev. Mol. Cell Biol. 19(5), 275–276 (2018).
Liang, Z. et al. Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleoproteins. Nat. Protoc. 13(3), 413–430 (2018).
Zhang, Y. et al. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617 (2016).
Van Damme, M. et al. Identification of arabidopsis loci required for susceptibility to the downy mildew pathogen Hyaloperonospora parasitica. Mol. Plant. Microbe Interact. 18(6), 583–92. (2005).
van Damme, M. et al. Arabidopsis DMR6 encodes a putative 2OG-Fe(II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant. J. 54(5), 785–93 (2008).
Zeilmaker, T. et al. DOWNY MILDEW RESISTANT 6 and DMR6-LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant. J. 81(2), 210–22 (2015).
Brewer, H. C., Hawkins, N. D. & Hammond-Kosack, K. E. Mutations in the Arabidopsis homoserine kinase gene DMR1 confer enhanced resistance to Fusarium culmorum and F. graminearum. BMC Plant. Biol. 14, 317 (2014).
Zhang, Y. et al. S5H/DMR6 Encodes a Salicylic Acid 5-Hydroxylase That Fine-Tunes Salicylic Acid Homeostasis. Plant. Physiol. 175(3), 1082–1093 (2017).
Chen, X. et al. Fusarium graminearum exploits ethylene signalling to colonize dicotyledonous and monocotyledonous plants. N. Phytol. 182(4), 975–83 (2009).
Nalam, V. J. et al. Facilitation of Fusarium graminearum infection by 9-lipoxygenases in Arabidopsis and wheat. Mol. Plant. Microbe Interact. 28(10), 1142–52 (2015).
Mascher, M. et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 544(7651), 427–433 (2017).
Mittler, R., Shulaev, V. & Lam, E. Coordinated Activation of Programmed Cell Death and Defense Mechanisms in Transgenic Tobacco Plants Expressing a Bacterial Proton Pump. Plant. Cell 7, 29–42 (1995).
Jansen, C. et al. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc. Natl Acad. Sci. USA 102(46), 16892–7 (2005).
Yang, J.F., et al., Identification of Light-Independent Anthocyanin Biosynthesis Mutants Induced by Ethyl Methane Sulfonate in Turnip “Tsuda” (Brassica rapa). Int J Mol Sci, 2017. 18(7).
Sun, K. et al. Silencing of six susceptibility genes results in potato late blight resistance. Transgenic Res. 25(5), 731–42 (2016).
Farrow, S. C. & Facchini, P. J. Functional diversity of 2-oxoglutarate/Fe(II)-dependent dioxygenases in plant metabolism. Front. Plant. Sci. 5, 524 (2014).
Himi, E., M. Maekawa & K. Noda, Differential expression of three flavanone 3-hydroxylase genes in grains and coleoptiles of wheat. Int J Plant Genomics, 369460 (2011).
Giovanini, M. P. et al. Gene-for-gene defense of wheat against the Hessian fly lacks a classical oxidative burst. Mol. Plant. Microbe Interact. 19(9), 1023–33 (2006).
Mauch-Mani, B. et al. Defense Priming: An Adaptive Part of Induced Resistance. Annu. Rev. Plant. Biol. 68, 485–512 (2017).
Yip, E. C. et al. Costs of plant defense priming: exposure to volatile cues from a specialist herbivore increases short-term growth but reduces rhizome production in tall goldenrod (Solidago altissima). BMC Plant. Biol. 19(1), 209 (2019).
Gao, Q. M. et al. Signal regulators of systemic acquired resistance. Front. Plant. Sci. 6, 228 (2015).
Yan, C. & Xie, D. Jasmonate in plant defence: sentinel or double agent? Plant. Biotechnol. J. 13(9), 1233–40 (2015).
Herrera-Vasquez, A., Salinas, P. & Holuigue, L. Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Front. Plant. Sci. 6, 171 (2015).
Kimura, M. & Kawano, T. Salicylic acid-induced superoxide generation catalyzed by plant peroxidase in hydrogen peroxide-independent manner. Plant. Signal. Behav. 10(11), e1000145 (2015).
Lehmann, S. et al. Reactive oxygen species and plant resistance to fungal pathogens. Phytochemistry 112, 54–62 (2015).
Makandar, R. et al. Involvement of salicylate and jasmonate signaling pathways in Arabidopsis interaction with Fusarium graminearum. Mol. Plant. Microbe Interact. 23(7), 861–70 (2010).
Zhang, X. et al. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1(2), 641–6 (2006).
Qiwei, S. et al. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 9(10), 2395 (2014).
Nalam, V., Sarowar, S. & Shah, J. Establishment of a Fusarium graminearum Infection Model in Arabidopsis thaliana Leaves and Floral Tissues. Bio-Protocol 6(14), e1877 (2016).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4), 402–8 (2001).
Acknowledgements
This work was supported in part by grants to R.D. from the U.S. Wheat and Barley Scab Initiative (FY18-DI-015 and FY18-DI-05) and to M.A.L. from USDA-NIFA Multistate Fund (NC1183).
Author information
Authors and Affiliations
Contributions
Y.C.L. designed and performed the experiments, analyzed the data and drafted the manuscript. R.D. and M.A.L. designed and advised on the experimental design and revised the manuscript. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Low, Y.C., Lawton, M.A. & Di, R. Validation of barley 2OGO gene as a functional orthologue of Arabidopsis DMR6 gene in Fusarium head blight susceptibility. Sci Rep 10, 9935 (2020). https://doi.org/10.1038/s41598-020-67006-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-67006-5
This article is cited by
-
Integrative systems biology of wheat susceptibility to Fusarium graminearum uncovers a conserved gene regulatory network and identifies master regulators targeted by fungal core effectors
BMC Biology (2024)
-
Genome-wide characterization of 2OGD superfamily for mining of susceptibility factors responding to various biotic stresses in Musa spp.
Physiology and Molecular Biology of Plants (2023)
-
Hormone biosynthesis and metabolism members of 2OGD superfamily are involved in berry development and respond to MeJA and ABA treatment of Vitis vinifera L
BMC Plant Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.